Abstract
Methylenetetrahydrofolate reductase (MTHFR) is a crucial enzyme that regulates nucleotide synthesis and DNA methylation. The MTHFR C677T gene polymorphism (rs1801133), a C → T transition at nucleotide 677 in exon 4, is a common gene variant of MTHFR and has been implicated in diabetic nephropathy, albeit with inconsistent results. Here, we performed a meta-analysis to assess the common effect size of this polymorphism on DN susceptibility. Case–control studies on the association of the MTHFR C677T gene polymorphism with DN risk were retrieved from databases up to August 1, 2013, and eligible studies were recruited into the meta-analysis and further analyzed. Of 132 studies, 33 were identified as suitable for this analysis. The results showed that T allele and TT genotype were distinctly associated with DN susceptibility in the overall population and Asians, and might be a risk factor in Caucasians and Africans (T allele: Overall population: p < 0.00001, Asians: p = 0.0002, Caucasians: p = 0.02, Africans: p < 0.00001; TT genotype: Overall population: p < 0.00001, Asians: p = 0.0003, Caucasians: p = 0.008, Africans: p = 0.0003). Furthermore, the analysis suggested that the CC genotype might play a protective role against DN onset in patients with type 2 diabetes for the overall population, Asians, Caucasian and Africans. However, due to the limited sample size in the African population, these results should be interpreted with care. In conclusion, the MTHFR C677T T allele or TT genotype might be a significant genetic molecular marker to determine the risk of DN in patients with type 2 diabetes and help to develop suitable disease prevention and management strategies.
Introduction
Diabetes is a metabolic disorder caused by loss of the insulin-producing beta cells of the Langerhans islets in the pancreas (type 1) or insulin resistance (type 2) and is characterized by hyperglycemia, which produces the classical symptoms of polyuria, polydipsia and polyphagia. Furthermore, diabetes negatively affects the hemodynamics in the macro and microvasculature of various organs, and the damage inflicted to the vasculature through various degenerate mechanisms results in a significantly increased risk of developing cardiovascular disease and ischemic stroke,Citation1 diabetic retinopathy,Citation2 diabetic neuropathyCitation3 and diabetic nephropathy (DN)Citation4–6.
DN is one of the most common microvascular complications of diabetes and the primary cause of end-stage renal disease and involves nearly 20–40% of diabetic patients.Citation4 DN affects numerous kidney cell types, including tubular and vascular epithelia, glomerular podocytes, interstitial fibroblasts, mesangial and endothelial cells,Citation4,Citation5 and is associated with extensive cellular dysfunction, hypoxia, oxidative stress and extracellular matrix accumulation.Citation6–8 Although both type 1 and type 2 diabetes are able to induce DN, the current DN epidemic has been shown to be predominantly associated with type 2 diabetes.Citation9 However, a fundamental understanding of the interplay between degenerate biological mechanisms in humans and exogenous influences that produce the clinical and pathologic alterations observed in DN remains incomplete. Next to environmental and life-style stressors, the susceptibility to DN has an intrinsic genetic basis, which is demonstrated by familial aggregation and ethnic-specific prevalence rates, and appears to be the most prominent in African-Americans and Pima Indians.Citation10 Recent investigations have implicated a myriad of gene polymorphisms as key players in the modulation of the pathogenesis of DN and have been distinctly associated with DN risk.Citation11–15 However, emerging evidence also suggests that additional genetic variants and derangements,Citation10 as well as alterations in epigenetic mechanisms, including DNA methylation variations, histone modifications and micro-RNA (miRNA) changes, are involved in diabetes and DN susceptibility.Citation16
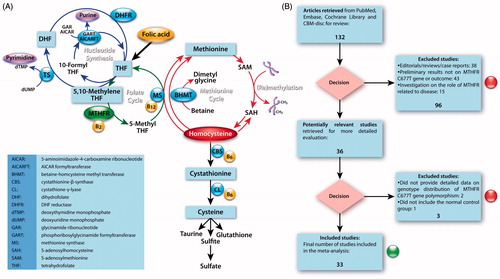
A candidate gene implicated to play a major role in DN susceptibility is the methylenetetrahydrofolate reductase (MTHFR) gene, located at the end of the short arm of chromosome 1 (1p36.3).Citation17 MTHFR codes for a crucial enzyme (flavoprotein) in folate and homocysteine metabolism () that catalyzes the flavin adenine dinucleotide (FAD)-dependent reduction of CH2-H4folate by NAD(p)H to CH3-H4folate and is involved in nucleotide synthesis, DNA repair and methylation.Citation18 Folate and vitamin B12 are required in the synthesis of methionine via remethylation of homocysteine, which results in the formation of S-adenosylmethionine; a methyl donor for the maintenance of methylation patterns, which determine gene expression and DNA conformation. Because of MTHFR’s pivotal role in many aspects of normal gene function (), including homocysteine levels (an established risk factor for vascular diseaseCitation19), it is not surprising that MTHFR gene polymorphisms may have dramatic effects on normal cellular function and that increasingly these are implicated in various diseases, including major depressive disorder, congenital heart diseases, breast cancer, leukemia and others.Citation20–22
Figure 1. (A) Schematic overview of the involvement of 5,10-methylenetetrahydrofolate reductase (MTHFR) in folate and homocystein metabolism, nucleotide synthesis and (re)methylation events. (B) Flow chart for study recruitment into the meta-analysis of the MTHFR C677T gene polymorphism and diabetic nephropathy relationship.
The MTHFR C677T single nucleotide polymorphism (rs1801133) involves a C → T transition at nucleotide 677 in exon 4 and is a common gene variant of MTHFR that includes CC, CT and TT genotypes and C and T alleles. MTHFR C → T transition induces enzyme thermolability and is distinctly associated with diminished enzyme activity.Citation23,Citation24 Even though the present epidemiologic evidence implicates the MTHFR C677T gene polymorphism in the etiology of DN in patients with type 2 diabetes mellitus, the available proof reported to date is inconclusive, predominantly due to sparseness of data and disagreements among studies. We performed this meta-analysis to investigate the relationship between the MTHFR C677T gene polymorphism and DN susceptibility in patients with type 2 diabetes mellitus, with the intention to provide a more reliable analysis of the significance of the purported association.
Materials and methods
Search strategy
Relevant studies were extracted from PubMed, Embase, Cochrane Library and CBM-disc (China Biological Medicine Database) on August 1, 2013, and association reports were identified. The retrieval strings entered into these databases were: “(methylenetetrahydrofolate reductase OR MTHFR) AND (DN OR DN OR diabetes mellitus nephropathy) AND (gene OR polymorphism OR allele OR genotype OR variant OR variation OR mutation)”. Additional reports were identified by scrutinizing the references cited in the recruited articles. If multiple publications for the same data from the same study group occurred, we only recruited the later dated paper into our final analysis.
Inclusion and exclusion criteria
Inclusion criteria
(1) A case–control study, (2) the outcome had to be DN; (3) there had to be at least two comparison groups (DN group vs. control group).
Exclusion criteria
(1) Review articles, editorials and case reports, (2) articles that did not provide detailed genotype data, (3) investigations on the association of other genes with DN, (4) investigations of the role of MTHFR related to diseases and (5) multiple publications of the same data from the same study group.
Data extraction and synthesis
The following information was extracted from each study independently by at least two investigators: first author’s surname, year of publication, ethnicity of study population, and the number of cases and controls for MTHFR genotype. Frequencies of T allele were calculated for case and control groups from the corresponding genotype distribution. The results were compared, and disagreements were resolved by discussion.
Statistical analysis
Available data were entered into Cochrane Review Manager Version 5 (Cochrane Library, UK) and analyzed. The pooled statistics were counted using the fixed effects model, but a random effects model was applied when the p value of the heterogeneity test was less than 0.1.Citation25,Citation26 Results were expressed with odds ratios (OR) for dichotomous data, and additionally 95% confidence intervals (CI) were calculated. p < 0.05 was required for the overall OR to be deemed statistically significant.Citation27–29 I2 was used to test the heterogeneity among the included studies. We classified the investigations into studies for Caucasians, Asians and African populations, because the genotype frequencies and prevalence of DN were quite different between the various ethnic groups. In order to avoid excessive comparisons, the OR was calculated using three methods,Citation30,Citation31 that is, method 1: allele comparison (T allele vs. C allele); method 2: comparison of TT homozygotes with the other two combinations (TT vs. TC + CC); and method 3: comparison of CC genotype with the other two combinations (CC vs. TC + TT). A chi-square test was used to determine if the reported genotype distribution of the control populations conformed to Hardy–Weinberg equilibrium (HWE; p < 0.05 was considered significant). Sensitivity analysis was performed when studies with controls not in HWE. Sensitivity analysis was also performed according to the source of the controls (healthy vs. hospital), genotyping methods (PCR-RFLP, etc.), by sequential omission of individual studies, and sample size of case or control (<100 vs. ≥100). Stata 11.0 (StataCorp LP, College Station, TX) was used to test for publication bias. The Begg adjusted rank correlationCitation32 and the Egger regression asymmetry testsCitation33 were used for exploring publication bias (p < 0.1 was considered significant) when the number of the included studies exceeded six.
Results
Study characteristics
The search yielded 132 studies, of which, after carefully applying the inclusion and exclusion criteria, 33 studies were identified as suitable for inclusion in the meta-analysis on the association between the MTHFR C677T gene polymorphism and DN susceptibility (). Eighteen reports were published in EnglishCitation34–51 and 15 reports were published in Chinese.Citation52–66 Twenty-two reports were conducted in Asian subjects,Citation42–44,Citation46–48,Citation50,Citation52–56 10 reports in CaucasiansCitation34–41,Citation45,Citation51 and 1 report in Africans.Citation49 The data of interest were extracted, as summarized in . In total, the 33 investigations contained 3959 DN patients and 3915 controls.
Table 1. General characteristics of the included studies in this meta-analysis.
The average distribution frequency of the MTHFR C677T T allele in the DN group was 42.14 and 38.41% in the control group. Overall, the results demonstrate that the average distribution frequency of the MTHFR C677T T allele in case group was markedly higher than frequency in the control group (case/control = 1.10). In the sub-group analysis, the average distribution frequency of the MTHFR C677T T allele was 45.46% in Asian patients with DN (41.09% in controls) and 33.65% in Caucasian patients with DN (31.65% in controls). However, in Africans, a significantly lower frequency was observed in controls, that is, 12.50%, with an average distribution frequency in cases that was in the same order of magnitude as the other ethnic groups, that is, 46.08%.
Overall, these results translate into case to control ratios for the average distribution frequency of the MTHFR C677T T allele that were larger than 1 for the overall population (case/control = 1.10), Asians (case/control = 1.11), Caucasians (case/control = 1.07) and Africans (case/control = 3.69). However, the results in Africans showed a large discrepancy in the control group, which resulted in a significantly higher ratio compared with other ethnic groups and might be a consequence of the limited number of studies and low-sampling frequency. Therefore, the results in the African group should be interpreted with care. Consequently, the MTHFR C677T T allele might only be considered a risk factor for DN disease in the overall population, Asians and Caucasians.
Association of MTHFR C677T gene polymorphism with DN susceptibility
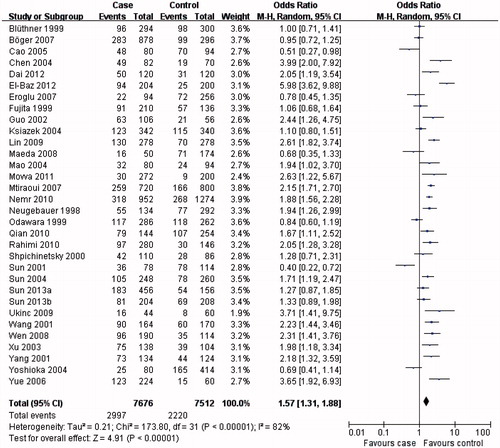
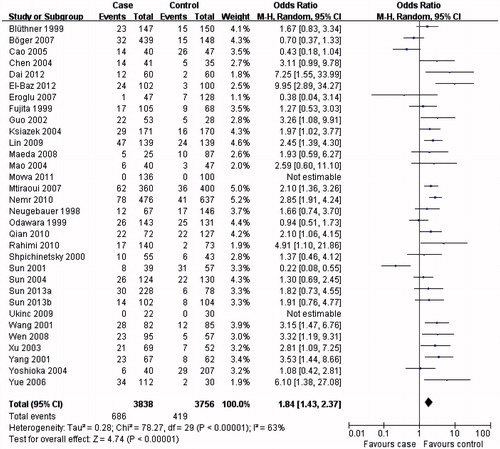
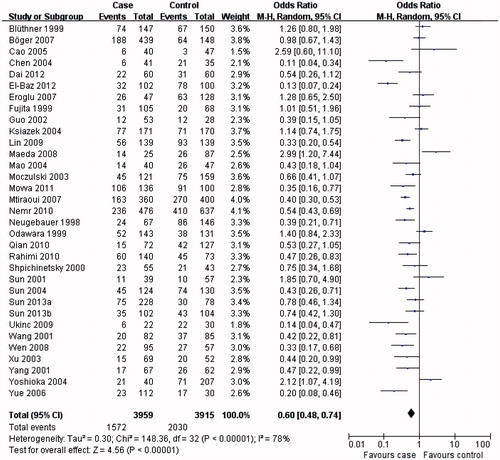
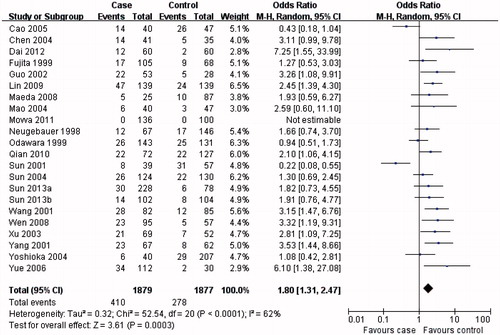
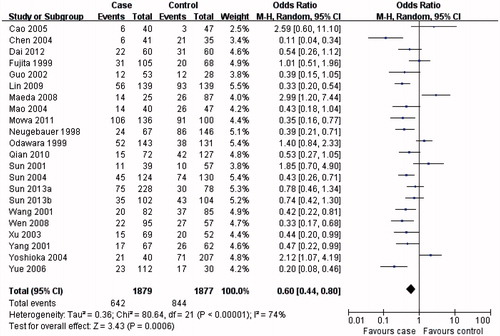
The results from this meta-analysis showed clear trends with respect to the association between MTHFR C677T genotypes and the risk of developing DN. As summarized in , the MTHFR C677T T allele () or TT genotype () was distinctly associated with DN susceptibility in the overall population (T allele: OR = 1.57, 95% CI 1.31–1.88, p < 0.00001; TT genotype: OR = 1.84, 95% CI 1.43–2.37, p < 0.00001). Conversely, evaluation of the MTHFR C677T CC genotype in the overall population () suggested a protective role for the MTHFR C677T CC genotype and a reduced risk of developing DN (OR = 0.60, 95% CI 0.48–0.74, p < 0.00001).
Table 2. Meta-analysis of the association of MTHFR C677T gene polymorphism with diabetic nephropathy risk.
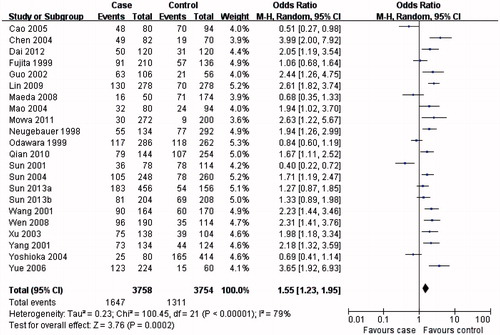
In the subgroup analysis, the MTHFR C677T T allele (), TT () and CC () genotypes were associated with DN susceptibility in the overall population and Asians, with similar results (T allele: OR = 1.55, 95% CI 1.23–1.95, p = 0.0002; TT genotype: OR = 1.80, 95% CI 1.31–2.47, p = 0.0003; CC genotype: OR = 0.60, 95% CI 0.44–0.80, p = 0.0006). In Caucasians, the MTHFR C677T T allele or TT genotype was associated with DN susceptibility (T allele: OR = 1.41, 95% CI 1.07–1.86, p = 0.02; TT genotype: OR = 1.73, 95% CI 1.15–2.60, p = 0.008; ), but not the MTHFR C677T CC genotype (OR = 0.70, 95% CI 0.04–0.96, p = 0.03; ). Therefore, this genotype might be a factor that reduces the risk of DN disease in Caucasians. Although the African sample size was relatively low, analysis of the available data showed that, equal to Caucasians, the MTHFR C677T T allele or TT genotype was associated with DN susceptibility (T allele: OR = 5.98, 95% CI 3.62–9.88, p < 0.00001; TT genotype: OR = 9.95, 95% CI 2.89–34.27, p = 0.0003; ), but the CC genotype was not (OR = 0.13, 95% CI: 0.07–0.24, p < 0.00001; ). Further studies are required to further clarify this trend and observations.
Sensitivity analysis
Sensitivity analysis examines data consistency, and if results are sensitive to restrictions on the included data to provide stronger evidence of an effect and of generalizability of the results. Therefore, the data included in this meta-analysis were subjected to further scrutiny via sensitivity analysis. Sensitivity analysis according to the genotype distribution of the control population reported for the MTHFR C677T gene polymorphism conformed to HWE, and the results were shown to be similar for the overall population, Asians and Africans compared with non-sensitivity analysis, but not for Caucasians ().
When focusing on the source of the controls (healthy vs. hospital), sensitivity analysis according to the source of the controls from healthy showed that in the overall population (T allele: OR = 1.81, 95% CI 1.43–2.28, p < 0.00001; TT genotype: OR = 2.18, 95% CI 1.57–3.02, p < 0.00001; CC genotype: OR = 0.49, 95% CI 0.37–0.66, p < 0.00001) and Asians (Asians: T allele: OR = 1.88, 95% CI 1.48–2.38, p < 0.00001; TT genotype: OR = 2.20, 95% CI 1.44–3.38, p = 0.0003; CC genotype: OR = 0.45, 95% CI 0.35–0.59, p < 0.00001), both sensitivity and non-sensitivity analyses produced similar results. This was not the case for Caucasians, because the sensitivity analysis according to the source of the controls from healthy showed that the TT genotype was distinctly associated with DN risk, but T allele and CC genotype were not (). Furthermore, in the sensitivity analysis, according to the source of the controls from hospital, we found that the results in the overall population and in Africans were consistent with the non-sensitivity analysis, whereas this was not the case for the Asian population (). In this ethnic group, only the T allele and CC genotype were associated with DN risk (). Examination of the genotype determination method resulted in similar results for both sensitivity and non-sensitivity analyses.
Sensitivity analysis for the relationship between the MTHFR C677T gene polymorphism and DN risk was also performed according to sample size of case (<100 vs. ≥100). In the meta-analysis of sample size of case ≥100, we found that the results for the overall population, Asians and Africans were similar to those from non-sensitivity analysis, but in Caucasians, the results from T allele and CC genotype were inconsistent with the non-sensitivity analysis (). Similarly, when evaluating the sample size of case <100, the results of sensitivity and non-sensitivity analyses were consistent, and a clear association of the MTHFR C677T gene polymorphism with DN risk was established in the overall population and Asians, but inconsistent results were obtained for Caucasians (). Sequential omission of individual studies showed no significant deviation between both analyses (data not shown).
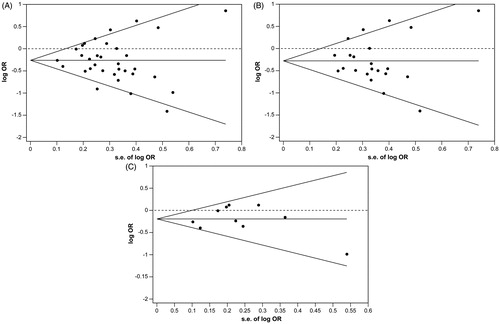
Evaluation of publication bias
Publication bias was tested (), and no publication bias was established for the MTHFR C677T gene polymorphism in the overall population (Begg p = 0.412; Egger p = 0.728), Asians (Begg p = 0.735; Egger p = 0.909) and Caucasians (Begg p = 1.000; Egger p = 0.645).
Discussion
Diabetes has been associated with renal disease in a large number of patients. However, diabetes-associated renal disease is neither simply determined by the duration nor severity of diabetes and, therefore, not an inevitable complication of diabetes. Consequently, other factors, such as environmental and life-style stressors and genetic factors, must play key roles in the pathogenesis of DN. Increasing scientific evidence suggests that genotype seems to be one of the major determinants of both the incidence and severity of DN. The probability of developing DN is significantly increased in both type 1 and type 2 diabetes patients with first degree relatives suffering from DN (familial aggregation), as clearly demonstrated in Pima Indian families with successive generations of diabetes sufferers.Citation67 Numerous studies have aimed to determine the genetic components that predispose patients with type 2 diabetes mellitus to developing DN and renal failure. However, very few genetic variants have conclusively been studied in relation to disease and population dynamics.
Alterations in the MTHFR gene have been implicated in the etiology of DN, especially because this gene code for an enzyme is intrinsically involved in nucleotide synthesis, DNA repair and DNA methylation and plays a pivotal role in folate and homocysteine metabolism (). Recently, Li et al.Citation68 reported that total plasma homocysteine concentration was independently associated with the occurrence of DN in Chinese type 2 diabetes patients at an early stage of the disease. Elevated homocysteine levels, predominantly caused by gene polymorphisms in key enzymes involved in homocysteine metabolism, are distinctly considered to be a risk factor for renal disease, including DN.Citation44,Citation47,Citation69 The C → T transition in the MTHFR C677T gene polymorphism results in an alanine to valine substitution, which causes thermolabilityCitation23,Citation24 and negatively affects enzyme activity and homocysteine levels. MTHFR TT homozygotes and CT heterozygotes have been shown to retain 30 and 65% enzyme activity compared with the wild-type CC genotype,Citation70 whereas T allele homozygosity equally results in reduced enzyme activity, but only mild to moderately increased homocysteine levels.Citation71 Overall, the MTHFR C677T gene polymorphism might represent an important risk factor for DN in diabetes.
Thirty-three suitable studies were recruited into our meta-analysis. Evaluation of the MTHFR C677T gene polymorphism in the overall population showed that the average distribution frequency of the MTHFR C677T T allele was 1.1-fold larger in the case group compared with the control group. Both T allele and TT genotype were associated with DN risk, whereas the analysis showed that CC genotype may be considered a protective factor against DN. Sensitivity analysis according to HWE, the source of the controls, genotyping methods (PCR-RFLP), by sequential omission of individual studies and sample size of case or control (<100 vs. ≥100) were similar to those in the non-sensitivity analysis for the overall population. No significant differences were observed between the sensitivity and non-sensitivity for the T allele and TT genotype according to the source of the controls from hospital. Furthermore, the pooled OR for the CC genotype from the sensitivity analysis according to the source of the controls from hospital was favorable with respect to the control group; although the difference was not statistically significant. Nonetheless, our results demonstrate a significant level of robustness in the analysis outcome and indicate that in the overall population, the T allele and TT genotype may be considered risk factors for DN.
Because geographic and ethnic difference might be important factors that affect the susceptibility to DN, we further evaluated the data according to ethnicity. The analysis showed that the average frequency of the T allele in controls was 41.09% for Asians and 33.65% for Caucasians and therefore a factor ∼1.22 higher in Asians. Conversely, the T allele frequency in Africans was only 12.50% for the control group (∼3.3- and ∼2.7-fold lower than in Asians or Caucasians). This low frequency might not necessarily reflect a lower occurrence of the polymorphism in this particular ethnic group, but might be a result of the small sample size. Furthermore, the data collected originated from patients geographically restricted to northern AfricaCitation49 and by no means reflected an ethnic cross-section of the whole continent. Nonetheless, a new principle finding can equally not be ruled out, but will require further prospective studies in Africans with various ethnic and geographic backgrounds.
Subgroup analysis showed that the average frequency of the T allele in the Asian case group was 1.11-fold higher compared with the control group and, therefore, the MTHFR C677T gene polymorphism might be a risk factor for DN in Asians. No publication bias was observed for the studies involving Asians, which largely validates the obtained results for the Asian population. In Caucasians, the average frequency of the T allele in the case group was 1.07-fold higher compared with the control group, which suggested and increased susceptibility to DN in patients with type 2 diabetes. In the sensitivity analysis according to the control source from healthy and the sensitivity analysis according to sample size of case (≥100), the TT genotype was associated with DN risk, and in the sensitivity analysis according the control source from hospital, the T allele was associated with DN risk. Sensitivity and non-sensitivity analyses were consistent, but when focusing on other factors, no such consistency could be established. Although we included 10 studies for the analysis in Caucasians and no publication bias was observed, the meta-analysis results showed that it was difficult to draw robust conclusions for this ethnic group. Consequently, more studies in Caucasians with significantly larger sample sizes are required to conclusively determine the exact MTHFR variants that represent a risk factor for DN in Caucasians. In Africans, the average frequency of T allele in the case group was 3.69-fold higher than the control group. Despite investing considerable efforts in searching for suitable published studies that focused on Africans, only one study fulfilled our selection criteria and was recruited into this meta-analysis, with the consequence that the sample size was low. As a result, our estimates of the role that the MTHFR C677T gene polymorphism plays in DN susceptibility in Africans are still likely to represent an overestimation, despite equal results from both sensitivity analysis and non-sensitivity analysis. Overall, information on the prevalence of nephropathy in African diabetic populations is scarce. With obesity on the incline (an established risk factor for diabetes), particularly in urbanized areas, diabetes and DN are likely to become an increasing health burden on the African population. Further investigations on the genetic predisposition of Africans are, therefore, urgently needed. For the overall population, sensitivity analysis according to HWE, the source of the controls (healthy), genotyping methods (PCR-RFLP), by sequential omission of individual studies and sample size of case or control (<100 vs. ≥100) were similar to those in non-sensitivity analysis. However, the results for T allele, TT genotype and CC genotype from sensitivity and non-sensitivity analyses according to the source of the controls from hospital were inconsistent. Interestingly, the patients from all the control groups from hospital were type 2 diabetes patients not suffering from DN. Further evaluation showed that no significant differences in genotype distribution between DN patients with type 2 diabetes and non-DN patients with type 2 diabetes could be established.
In recent years, several meta-analyses explored a possible association between the MTHFR C677T gene polymorphism DN susceptibility. Zintzaras et al.Citation72 included 11 studies involving subjects with DN and reported that the T allele was associated with DN risk in patients with type 2 diabetes. An evaluation by Niu et al.Citation73 included 22 studies and showed that the MTHFR TT genotype might confer a moderately augmented risk for DN. Chang et al.Citation74 recruited 13 studies focusing on the Chinese population in their meta-analysis and reported that the MTHFR C677T polymorphism might influence DN risk in this ethnic group. Yang et al.Citation75 performed a meta-analysis included nine reports in Caucasian individuals with type 2 diabetes, and this meta-analysis supported that there was an association between MTHFR C677T polymorphism and DN risk. Overall, these studies reported heterogonous results. Our meta-analysis (33 studies) showed that the MTHFR T allele or TT genotype was a distinct risk factor for the DN susceptibility in patient with type 2 diabetes in the overall population and Asians, and that the CC genotype might play a protective role against DN susceptibility, confirming the results by ZintzarasCitation72, ChangCitation74 and YangCitation75 and their co-workers. In Caucasians and Africans, there are some indications that an equal association exists. However, the results should be regarded cautiously, because of possible heterogeneity in the enrolled cases, limited statistical power, variable study designs and different interventions. Although a clear genetic component is observable in DN, it is likely that multiple genetic variants, which might isolated only have nominal effects, in conjunction with exogenous triggers, determine if diabetes patients ultimately will develop DN. Nonetheless, many questions regarding the role of MTHFR alleles in various diseases still remain. Alternate and more precise strategies would be required to obtain more accurate data. First, precise estimates of MTHFR genotype frequencies from large and representative samples among well-defined populations are required. Second, both relative and absolute risks as well as attributable fractions need to be determined for each disease and genotype of interest. Third, additional genes and alleles should be identified. Fourth, since the fundamental metabolic roles of folate and homocysteine suggest that genetic variation in the regulatory genes may also play a role in the etiology of DN, more outcomes should be evaluated. Fifth, the interplay between various genetic variants per se (gene-gene) and multiple genetic variants and exogenous factors (gene-environment) should be studied in more detail, in particular gene–nutrient interactions. Finally, in addition to study design considerations, special attention should be given to sample size requirements in order to maximize statistical significance. Such strategies would allow significantly more robust meta-analyses and minimize some of the limitations associated with meta-analyses. Alternatively, an integrated approach that incorporates datasets from multiple sources, including genetics and epigenetics, transcriptomic, proteomics, metabolomics and the effect of exogenous and life-style stressors might shed more light on which genetic elements contribute to this serious complication of diabetes. Prospective multilayer studies that address the interplay between various factors will enable researchers to identify promising candidate genes and pathways, develop better risk profiles, and allow a more targeted approach to both the management of diabetes and DN, and the development of novel therapeutic interventions. Various initiatives to collate data on renal disease from multiple sources of evidence have already been established, including the Kidney and Urine Pathway Knowledge Base,Citation76 the Human Kidney and Urine Proteome,Citation77 Nephromine (renal gene expression profiles),Citation78 and EuReGene (gene expression in mouse kidney).Citation79
Declaration of interest
This study was supported by the sub-item of 985 Project Foundation of Sun Yat-Sen (The Hundred Talents Program Foundation; No. 88000-3311300) and was supported by the Guangzhou Medical Key Subject Construction Project (2013--2015). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Sarwar N, Gao P, Seshasai SR, et al. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: A collaborative meta-analysis of 102 prospective studies. Lancet. 2010;375(9733):2215–2222
- Antonetti DA, Klein R, Gardner TW. Diabetic retinopathy. N Engl J Med. 2012;366(13):1227–1239
- Said G. Diabetic neuropathy — A review. Nat Clin Pract Neurol. 2007;3(6):331–340
- Dronavalli S, Duka I, Bakris GL. The pathogenesis of diabetic nephropathy. Nat Clin Pract Endocrinol Metab. 2008;4(8):444–452
- Kanwar YS, Wada J, Sun L, et al. Diabetic nephropathy: Mechanisms of renal disease progression. Exp Biol Med. (Maywood) 2008;233(1):4–11
- Singh DK, Winocour P, Farrington K. Mechanisms of disease: The hypoxic tubular hypothesis of diabetic nephropathy. Nat Clin Pract Nephrol. 2008;4(4):216–226
- Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol. 2010;21(3):438–447
- Krishan P, Chakkarwarb VA. Diabetic nephropathy: Aggressive involvement of oxidative stress. J Pharm Educ Res. 2011;2(1):35–41
- Hudkins KL, Pichaiwong W, Wietecha T, et al. BTBR Ob/Ob mutant mice model progressive diabetic nephropathy. J Am Soc Nephrol. 2010;21(9):1533–1542
- Palmer ND, Freedman BI. Insights into the genetic architecture of diabetic nephropathy. Curr Diab Rep. 2012;12(4):423–431
- Al-Kateb H, Boright AP, Mirea L, et al. Multiple superoxide dismutase 1/splicing factor serine alanine 15 variants are associated with the development and progression of diabetic nephropathy: The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Genetics study. Diabetes. 2008;57(1):218–228
- Maeda S, Araki S, Babazono T, et al. Replication study for the association between four Loci identified by a genome-wide association study on European American subjects with type 1 diabetes and susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes. Diabetes. 2010;59(8):2075–2079
- Ng DP, Tai BC, Lim XL. Is the presence of retinopathy of practical value in defining cases of diabetic nephropathy in genetic association studies? The experience with the ACE insertion/deletion polymorphism in 53 studies comprising 17,791 subjects. Diabetes. 2008;57(9):2541–2546
- Lim XL, Nurbaya S, Salim A, et al. KCNQ1 SNPS and susceptibility to diabetic nephropathy in East Asians with type 2 diabetes. Diabetologia. 2012;55(9):2402–2406
- Mooyaart AL, Valk EJ, van Es LA, et al. Genetic associations in diabetic nephropathy: A meta-analysis. Diabetologia. 2011;54(3):544–553
- Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299(1):F14–F25
- Goyette P, Pai A, Milos R, et al. Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR). Mamm Genome. 1998;9(8):652–656
- Trimmer EE. Methylenetetrahydrofolate reductase: Biochemical characterization and medical significance. Curr Pharm Des. 2013;19(14):2574–2593
- Clarke R, Collins R, Lewington S, et al. Homocysteine and risk of ischemic heart disease and stroke: A meta-analysis. JAMA. 2002;288(16):2015–2022
- Bousman CA, Potiriadis M, Everall IP, Gunn JM. Methylenetetrahydrofolate reductase (MTHFR) genetic variation and major depressive disorder prognosis: A five-year prospective cohort study of primary care attendees. Am J Med Genet B Neuropsychiatr Genet. 2014;165B(1):68–76
- Liang H, Yan Y, Li T, et al. Methylenetetrahydrofolate reductase polymorphisms and breast cancer risk in Chinese population: A meta-analysis of 22 case–control studies. Tumour Biol. 2014;35(2):1695–1701
- Sahiner UM, Alanay Y, Alehan D, Tuncbilek E, Alikasifoglu M. Methylene tetrahydrofolate reductase polymorphisms and homocysteine level in heart defects. Pediatr Int 2014;56(2):167--172
- Engbersen AM, Franken DG, Boers GH, Stevens EM, Trijbels FJ, Blom HJ. Thermolabile 5,10-methylenetetrahydrofolate reductase as a cause of mild hyperhomocysteinemia. Am J Hum Genet. 1995;56(1):142–150
- Kang SS, Wong PW, Susmano A, Sora J, Norusis M, Ruggie N. Thermolabile methylenetetrahydrofolate reductase: An inherited risk factor for coronary artery disease. Am J Hum Genet. 1991;48(3):536–545
- Zhou TB, Xu HL, Yin SS. Association between endothelial nitric oxide synthase Glu298Asp gene polymorphism and diabetic nephropathy susceptibility. Ren Fail. 2013;35(1):173–178
- Zhou TB, Yin SS. Association of endothelial nitric oxide synthase Glu298Asp gene polymorphism with the risk of end-stage renal disease. Ren Fail. 2013;35(4):573–578
- Qin YH, Zhou TB, Su LN, Lei FY, Huang WF, Zhao YJ. Association between ACE polymorphism and risk of IgA nephropathy: A meta-analysis. J Renin Angiotensin Aldosterone Syst. 2011;12(3):215–223
- Zhou TB, Ou C, Qin YH, et al. Association of angiotensin converting enzyme insertion/deletion gene polymorphism with idiopathic nephrotic syndrome susceptibility in children: A meta-analysis. J Renin Angiotensin Aldosterone Syst. 2011;12(4):601–610
- Zhou TB, Qin YH, Su LN, et al. The association between angiotensin-converting enzyme insertion/deletion gene variant and risk of focal segmental glomerulosclerosis: A systematic review and meta-analysis. J Renin Angiotensin Aldosterone Syst. 2011;12(4):624–633
- Zhou TB, Liu YG, Lin N, et al. Relationship between angiotensin-converting enzyme insertion/deletion gene polymorphism and systemic lupus erythematosus/Lupus nephritis: A systematic review and metaanalysis. J Rheumatol. 2012;39(4):686–693
- Zhou TB, Qin YH, Su LN, Lei FY, Huang WF, Zhao YJ. ACE I/D gene polymorphism can't predict the steroid responsiveness in Asian children with idiopathic nephrotic syndrome: A meta-analysis. PLoS One. 2011;6(5):e19599
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634
- Bluthner M, Bruntgens A, Schmidt S, Strojek K, Grzeszczak W, Ritz E. Association of methylenetetrahydrofolate reductase gene polymorphism and diabetic nephropathy in type 2 diabetes? Nephrol Dial Transplant. 1999;14(1):56–57
- Shpichinetsky V, Raz I, Friedlander Y, et al. The association between two common mutations C677T and A1298C in human methylenetetrahydrofolate reductase gene and the risk for diabetic nephropathy in type II diabetic patients. J Nutr. 2000;130(10):2493–2497
- Ksiazek P, Bednarek-Skublewska A, Buraczynska M. The C677T methylenetetrahydrofolate reductase gene mutation and nephropathy in type 2 diabetes mellitus. Med Sci Monit. 2004;10(2):BR47–BR51
- Mtiraoui N, Ezzidi I, Chaieb M, et al. MTHFR C677T and A1298C gene polymorphisms and hyperhomocysteinemia as risk factors of diabetic nephropathy in type 2 diabetes patients. Diabetes Res Clin Pract. 2007;75(1):99–106
- Boger CA, Stubanus M, Haak T, et al. Effect of MTHFR C677T genotype on survival in type 2 diabetes patients with end-stage diabetic nephropathy. Nephrol Dial Transplant. 2007;22(1):154–162
- Eroglu Z, Erdogan M, Tetik A, et al. The relationship of the methylenetetrahydrofolate reductase C677T gene polymorphism in Turkish type 2 diabetic patients with and without nephropathy. Diabetes Metab Res Rev. 2007;23(8):621–624
- Ukinc K, Ersoz HO, Karahan C, et al. Methyltetrahydrofolate reductase C677T gene mutation and hyperhomocysteinemia as a novel risk factor for diabetic nephropathy. Endocrine. 2009;36(2):255–261
- Nemr R, Salman RA, Jawad LH, Juma EA, Keleshian SH, Almawi WY. Differential contribution of MTHFR C677T variant to the risk of diabetic nephropathy in Lebanese and Bahraini Arabs. Clin Chem Lab Med. 2010;48(8):1091–1094
- Maeda M, Yamamoto I, Fukuda M, et al. MTHFR gene polymorphism is susceptible to diabetic retinopathy but not to diabetic nephropathy in Japanese type 2 diabetic patients. J Diabetes Complications. 2008;22(2):119–125
- Movva S, Alluri RV, Venkatasubramanian S, et al. Association of methylene tetrahydrofolate reductase C677T genotype with type 2 diabetes mellitus patients with and without renal complications. Genet Test Mol Biomarkers. 2011;15(4):257–261
- Yoshioka K, Yoshida T, Umekawa T, et al. Methylenetetrahydrofolate reductase gene polymorphism is not related to diabetic nephropathy in Japanese Type 2 diabetic patients. Diabet Med. 2004;21(9):1051–1052
- Moczulski D, Fojcik H, Zukowska-Szczechowska E, Szydlowska I, Grzeszczak W. Effects of the C677T and A1298C polymorphisms of the MTHFR gene on the genetic predisposition for diabetic nephropathy. Nephrol Dial Transplant. 2003;18(8):1535–1540
- Odawara M, Yamashita K. A common mutation of the methylenetetrahydrofolate reductase gene as a risk factor for diabetic nephropathy. Diabetologia. 1999;42(5):631–632
- Neugebauer S, Baba T, Watanabe T. Methylenetetrahydrofolate reductase gene polymorphism as a risk factor for diabetic nephropathy in NIDDM patients. Lancet. 1998;352(9126):454
- Fujita H, Narita T, Meguro H, et al. No association between MTHFR gene polymorphism and diabetic nephropathy in Japanese type II diabetic patients with proliferative diabetic retinopathy. J Diabetes Complications. 1999;13(5–6):284–287
- El-Baz R, Settin A, Ismaeel A, et al. MTHFR C677T, A1298C and ACE I/D polymorphisms as risk factors for diabetic nephropathy among type 2 diabetic patients. J Renin Angiotensin Aldosterone Syst. 2012;13(4):472–477
- Sun J, Xu Y, Zhu Y, Lu H. Genetic polymorphism of methylenetetrahydrofolate reductase as a risk factor for diabetic nephropathy in Chinese type 2 diabetic patients. Diabetes Res Clin Pract. 2004;64(3):185–190
- Rahimi M, Hasanvand A, Rahimi Z, et al. Synergistic effects of the MTHFR C677T and A1298C polymorphisms on the increased risk of micro- and macro-albuminuria and progression of diabetic nephropathy among Iranians with type 2 diabetes mellitus. Clin Biochem. 2010;43(16–17):1333–1339
- Qian D. Matched case–control study on the association between AGT and MTHFR gene polymorphism and diabetic nephropathy. Tianjing: Tianjing Medical University Press; 2010: 17 p
- Lin R, Wang C, Liu X. Matched case–control study on the association between polymorphism of methylenetetrahydrofolate reductase (MTHFR) gene and diabetic nephropathy in type 2 diabetic patients. Modern Prevent Med. 2009;36(20):3801–3804
- Wen J, Chen Z, Li X, et al. Correlation of MTHFR gene polymorphism and plasma homocysteine with microalbuminuria in type 2 diabetes. Shanghai Med J. 2008;31(1):47–51
- Yue H, Liu J, Kang W, et al. Relationship between plasma level of homocysteine and urine microalbumin in incipient type 2 diabetic nephropathy. Chin J Gen Pract. 2006;5(12):725–729
- Mao L. A correlation between 5,10-methylenetetrahydrofolate reductase gene polymorphism, serum homocysteine levels and vascular complications in patients type 2 diabetic mellitus. Jiangsu: Nantong Medical College Press; 2004: 27 p
- Chen A, Ning Y, Zhu X, Li L, Shi H. Study on the relationship between gene polymorphisms of N5,10-methylenetetrahydrofolate reductase and nephropathy in type 2 diabetes mellitus in Gansu Han Chinese of China. Clin J Prev Contr Chron Non-Commun Dis. 2004;12(5):195–197
- Xu J, Zhang J, Shan B, Ma H. Relationship between methylenetetrahydrofolate reductase gene polymorphism and diabetic nephropathy in type 2 diabetes mellitus in the Hans of Hebei Province. Clin Focus. 2003;18(14):787–789
- Guo Q, Lu J, Qin H, Sheng C, Yin S, Pan C. Changes of the plasma homocysteine and its mechanism in type 2 diabetes with microangiopathy. Chin J Diabetes. 2002;10(1):32–36
- Yang G, Lu J, Pan C. Study on the relationship between N5,10-methylenetetrahydrofolate reductase gene polymorphism and the susceptibility to microangiopathy in type 2 diabetes mellitus. Chin J Endocrinol Metab. 2001;17(4):224–227
- Wang L, Wang J, Xue Y, Cheng Y, Zou H. Relationship between methylenetetrahydrofolate reductase gene polymorphism and diabetic nephropathy. Chin J Med Genet. 2001;18(4):226–228
- Cao H, Huang D, Mao L, Gao Y. Association of homocysteine, methyleneterahydrofolate reductase gene polymorphism with nephropathy in type 2 diabetes mellitus. ACTA Universitatis Medicinalis Nanjing (Natural Science). 2005;25(4):249–251
- Sun L, Wang S, Shi X, Yang Z. Interactions between APOE and MTHFR mutations is associated with the risk for type 2 diabetic nephropathy. J Med Mol Biol. 2013;10(2):95–99
- Dai H, Zhang Y. An association study of MTHFR and eNOS genes polymorphism with diabetic nephropathy. Chin J Trauma Disabil Med 2012;20(6):4–6
- Sun L, Wang S, Wang X, Shi X, Yang Z. Association study between MTHFR C677T polymorphism and levels of Scr and BUN in diabetic nephropathy patients. Chinese J Health Lab Technol. 2013;23(5):1167–1169
- Sun J, Xu Y, Zhu Y, et al. An association study of methylenetetrahydrofolate reductase gene polymorphism with diabetic nephropathy. J Nephrol Dial Transplant. 2001;10(1):33–35
- Pettitt DJ, Saad MF, Bennett PH, Nelson RG, Knowler WC. Familial predisposition to renal disease in two generations of Pima Indians with type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia. 1990;33(7):438–443
- Li J, Shi M, Zhang H, et al. Relation of homocysteine to early nephropathy in patients with Type 2 diabetes. Clin Nephrol. 2012;77(4):305–310
- Sun J, Xu Y, Zhu Y. Genetic polymorphism of methylenetetrahydrofolate reductase as a risk factor for diabetic nephropathy in Chinese type 2 diabetic patients. Diabet Res Clin Pract. 2004;64(5):185–190
- James SJ, Pogribna M, Pogribny IP, et al. Abnormal folate metabolism and mutation in the methylenetetrahydrofolate reductase gene may be maternal risk factors for Down syndrome. Am J Clin Nutr. 1999;70(4):495–501
- Frosst P, Blom HJ, Milos R, et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10(1):111–113
- Zintzaras E, Uhlig K, Koukoulis GN, Papathanasiou AA, Stefanidis I. Methylenetetrahydrofolate reductase gene polymorphism as a risk factor for diabetic nephropathy: A meta-analysis. J Hum Genet. 2007;52(11):881–890
- Niu W, Qi Y. An updated meta-analysis of methylenetetrahydrofolate reductase gene 677C/T polymorphism with diabetic nephropathy and diabetic retinopathy. Diabetes Res Clin Pract. 2012;95(1):110–118
- Chang WW, Zhang L, Yao YS, Su H, Jin YL, Chen Y. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and susceptibility to diabetic nephropathy in Chinese type 2 diabetic patients: A meta-analysis. Ren Fail. 2013;35(7):1038–1043
- Yang S, Zhang J, Feng C, Huang G. MTHFR 677T variant contributes to diabetic nephropathy risk in Caucasian individuals with type 2 diabetes: A meta-analysis. Metabolism. 2013;62(4):586–594
- Kidney & Urinary Pathway Knowledge Base. Renal Fibrosis Laboratory (INSERM, France) and the Bio-health Informatics Group (University of Manchester, UK). Available at: www.kupkb.org/
- Human Kidney and Urine Proteome Project. Human Proteome Organization (Santa Fe, US). Available at: http://www.hkupp.org/
- Nephromine. Applied Systems Biology Core O'Brien Renal Center (University of Michigan, US) and Compendia Bioscience (Life Technologies, US). Available at: www.nephromine.org/
- Willnow T, Antignac C, Brandli A, et al. The European renal genome project: An integrated approach towards understanding the genetics of kidney development and disease. Organogenesis. 2005;2(2):42–47