Abstract
Objective: The aim of this study was to investigate the association between interleukin (IL)-10-1082 (G/A) promoter polymorphism and acute rejection (AR) in renal transplant recipients. Methods: We searched MEDLINE, EMBASE, Web of Science, and Cochrane Central Register from the inception to March 2015 for relevant studies. Data concerning publication information, population characteristics, and transplant information were extracted. Odds ratios (ORs) was calculated for the association between IL-10-1082 GG genotype (or IL-10-1082 G allele) and AR risk. Results: This meta-analysis included 22 case–control studies including 2779 cases of renal transplant recipients. The pooled estimate showed that the IL-10-1082 GG genotype was not significantly associated with AR risk (ORrandom=1.07, 95% CI 0.80–1.43, p = 0.64). Similarly, the pooled estimate showed that the IL-10-1082 G allele was not significantly associated with AR risk (ORfixed=1.02, 95% CI 0.90–1.16, p = 0.74). None of subgroup analyses yielded significant results in the association between IL-10-1082 GG genotype (or IL-10-1082 G allele) and AR risk. Meta-regression confirmed that there was no significant correlation between the pre-selected trial characteristics and our study results. Conclusions: This meta-analysis suggests that IL-10-1082 G/A polymorphism is not significantly associated with AR risk in renal transplant recipients.
Introduction
Kidney transplantation is often the best medical treatment for patients with end-stage renal disease. Despite recent improvement in immunosuppressive medication and HLA-matching, acute rejection (AR) continues to be problematic and remains the leading cause of graft loss after renal transplantation.Citation1 AR is mainly caused by T-cell-mediated immune responses. It has been suggested that graft microenvironment and cytokines play an important role in the precipitation of rejection.Citation2 In the last decade, several single-nucleotide polymorphisms (SNPs) in the genes encoding for cytokines have been identified.Citation3,Citation4 The production of cytokines is under genetic control and varies among individuals as a function of polymorphisms within the regulatory regions of the various genes that determine the transcriptional activation.Citation5 Although there is evidence that cytokine gene polymorphisms are associated with varying quantities of cytokine protein production, the data on the correlation between these gene polymorphisms and the risk of AR remains unclear.Citation6,Citation7
Cytokine interleukin-10 (IL-10) is thought to play a critical role in allograft tolerance, predominantly through its ability to suppress inflammation. IL-10 is encoded by IL-10 gene located on chromosome 1, while its promoter is a highly polymorphic region.Citation8 The promoter of the IL-10 gene contains three biallelic polymorphisms at positions -1082 (base G to A, dbSNP no. rs1800896), -819 (base C to T, dbSNP no. rs1800871), and -592 (base C to A, dbSNP no. rs1800872) from the transcription start site, and these influence the capacity of cells to produce IL-10.Citation5 In 2008, a review by NickersonCitation9 examines the accumulated literature implicating various cytokine (including IL-10) SNPs with renal or liver allograft outcomes, but it did not compute a pooled estimate results. Thakkinstian et al.Citation10 published findings from a meta-analysis suggesting possible association between a 3-SNP-haplotype of IL-10 and poor outcomes including chronic allograft nephropathy (CAN), graft rejection (GR), and graft failure (GF) in renal transplantation, but this needs to be confirmed in larger studies.
Over the last two decades, although a number of studiesCitation11–32 have shown varying results in the association between IL-10–1082 (G/A) promoter polymorphism and AR risk in renal transplant recipients, the results are inconsistent and inconclusive. Moreover, the conclusion of a single study is limited by many factors such as the small size, ethnicity, age, gender, donor type, immunosuppressive regimens, and Hardy–Weinberg equilibrium (HWE) deviations. In order to derive a more comprehensive estimation of the association between IL-10–1082 polymorphism and AR risk in renal transplant recipients, we conducted a meta-analysis to evaluate the association.
Methods
Literature search strategy
Two reviewers (Q.W. Hu and P. Liao) independently searched the MEDLINE, EMBASE, Web of Science, and Cochrane Central Register up to 2 March 2015. To maximize the data search, the search strategy used relevant keywords and medical subject heading (MeSH) terms including “interleukin-10” or “IL-10” or “cytokine gene polymorphism”; “kidney transplant” or “renal transplant” or “allograft nephropathy”; “acute rejection” or “early graft rejection”. We also hand searched the reference lists of retrieved articles, case reports for relevant articles. The search was performed with restriction on English, and conducted on human subjects.
Inclusion and exclusion criteria
Two reviewers independently screened the titles and abstracts of studies to identify those that fulfilled the inclusion criteria: (1) evaluation of the association between IL-10–1082 G/A polymorphism and AR risk in renal transplant recipients and (2) IL-10–1082 GG and GA/AA genotype data (or IL-10–1082 G and A allele data) in AR and non-AR transplant recipients were respectively provided. Major reasons for exclusion of studies were: (1) an abstract, comment, review, or editorial and (2) no sufficient data were reported.
Data extraction and quality assessment
Three reviewers (Q.W. Hu, P. Liao, and H. Tian) independently performed the data extraction that met the inclusion criteria. The titles and abstracts were scanned to exclude any clearly irrelevant studies. The full texts of the remaining articles were read to determine whether they contained information on the useful topic. Any disagreement was resolved by discussion to reach a consensus among the investigators. The following data were collected from each study: (1) publication data: first author, year of publication, and country of origin; (2) characteristics of the study population: ethnicity, age, gender, IL-10–1082 GG and GA/AA genotype data in AR and non-AR transplant recipients, and IL-10–1082 G and A allele data in AR and non-AR transplant recipients; and (3) transplant information: donor type, AR definition, and HWE deviations.
The methodological quality of the included studies was assessed by three independent investigators using the Newcastle–Ottawa Quality Assessment Scale (NOS)Citation33 for observational trials, respectively. The NOS consists of eight questions with nine possible points. A points-scoring system was used to judge the data according to the selected populations, the comparability of the groups and exposure/outcome of interest (0–5: poor quality, 6–9: good quality).
Statistical analysis
Statistical analyses were performed using RevMan 5.0 (The Cochrane Collaboration, Copenhagen, Denmark) and STATA software 12.0 (StataCorp, College Station, TX). The outcome measure was biopsy-proven AR during the early post-transplantation period. There were two main outcome measurements in this meta-analysis: (1) odds ratios (OR) and 95% confidence interval (CI) of individual studies and pooled data for the association between IL-10–1082 GG genotype and AR risk; (2) ORs and 95% CI of individual studies and pooled data for the association between IL-10–1082 G allele and AR risk. We assessed for the heterogeneity between studies by calculating a Cochran Q statistic and an I2 statistic.Citation34 If significant heterogeneity (I2 > 50% or p < 0.05) was observed between studies, a random-effects model was used to pool the data; otherwise, a fixed-effects model was used. Sensitivity analysis and subgroup analysis for the primary outcome measurement were performed in a predefined manner to assess the impact of the following: (1) ethnicity, (2) region of nations, (3) donor type, (4) AR definition, and (5) HWE deviations. To explore the impact of pre-selected covariates on the association between IL-10–1082 GG genotype (or IL-10–1082 G allele) and AR risk, we performed a random-effects (or fixed-effects) meta-regression analysis. The logarithm of OR for IL-10–1082 GG genotype (or IL-10–1082 G allele), weighted by the inverse variance of each study, was regressed against age, male ratio, ethnicity, region of nations, donor type, AR definition, and HWE deviations. Begg’s rank correlation methodCitation34 and Egger’s weighted regression methodCitation35 were used to statistically assess publication bias (p < 0.05 was considered statistically significant). All p values were 2-tailed, and a p value < 0.05 was considered statistically significant.
Results
Studies selection
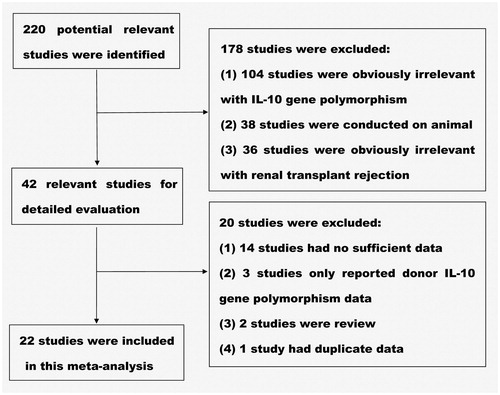
is a flow diagram of the selection of eligible studies. Two hundred twenty records were identified by the primary literature search. However, after screening the titles and abstracts, 178 irrelevant studies were excluded. We therefore retrieved 42 potentially relevant articles for a detailed review. Of these studies, 14 studies having no sufficient data, three studies reporting donor IL-10 gene polymorphism, two reviewed studies, and one study having duplicate data were further excluded. Consequently, the remaining 22 studies were included for this meta-analysis ().
Study characteristics
All of the 22 included studies, a total of 2779 renal transplant patients were grouped for case–control design. One studyCitation27 is divided into Caucasian group and African-American group, so there were 23 sets of data altogether. The baseline characteristics of the included studies are summarized as follows: first author; year of publication (range, 2000–2014); country of origin (10 countries in Europe, 6 countries in Asia, 5 countries in America, 1 country in Africa); ethnicity (Caucasian or African-American); average age (range, 25.6–50.8 years); male ratio (range, 47–84%); number of IL-10–1082 GG and GA/AA genotype in AR and non-AR transplant recipients; number of IL-10–1082 G and A allele in AR and non-AR transplant recipients (data in two studiesCitation14,Citation20 were not informed); donor type (living, deceased, living or deceased); AR definition (biopsy except two studiesCitation14,Citation22); and HWE deviations (HWE deviations were studied using appropriate Chi-square tests and p-values < 0.05 were considered significant) (). Average NOS scores of the included 22 studies were 6.36 (range, 5–8 points), and only one studyCitation23 scored below 6 points was rated as poor quality study (Supplementary Table available online at http://informahealthcare.com/doi/suppl/10.3109/0886022X.2015.1106770).
Table 1. Characteristics of the studies included in the meta-analysis.
Meta-analysis
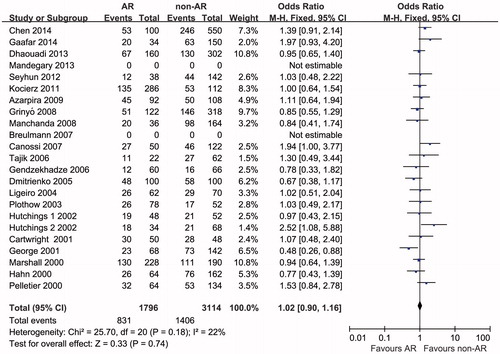
The outcome measurements of this meta-analysis are summarized in and . Twenty-two studies reported IL-10–1082 GG genotype were analyzed for OR. There was significant heterogeneity among the studies (pheterogeneity=0.03, I2 = 40%); thus, a random-effects model was used to pool the data. The pooled estimate showed that the IL-10–1082 GG genotype was not significantly associated with AR risk (ORrandom=1.07, 95% CI 0.80–1.43, p = 0.64) (). Twenty studies reported IL-10-1082 G allele were analyzed for OR. There was no significant heterogeneity among the studies (pheterogeneity=0.18, I2 = 22%); thus, a fixed-effects model was used to pool the data. The pooled estimate showed that the IL-10-1082 G allele was not significantly associated with the AR risk (ORfixed=1.02, 95% CI 0.90–1.16, p = 0.74) ().
Figure 2. Odds ratios and 95% CI of individual studies and pooled data for the association between interleukin-10–1082 GG genotype and acute rejection risk in renal transplant recipients.
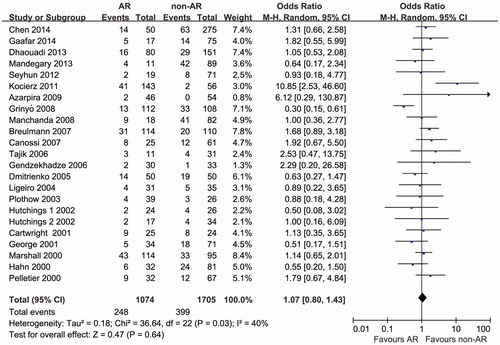
Figure 3. Odds ratios and 95% CI of individual studies and pooled data for the association between interleukin-10–1082 G allele and acute rejection risk in renal transplant recipients.
When stratified for ethnicity, no significant association was observed among either Caucasian group or non-Caucasian group (for IL-10-1082 GG genotype: Caucasian group, ORrandom=0.88, 95% CI 0.41–1.89, p = 0.74; non-Caucasian group, ORrandom=1.23, 95% CI 0.96–1.59, p = 0.11 and for IL-10-1082 G allele: Caucasian group, ORfixed=0.95, 95% CI 0.78–1.16, p = 0.65; non-Caucasian group, ORfixed=1.07, 95% CI 0.91–1.26, p = 0.42). None of the other subgroup (region of nations, donor type, AR definition, and HWE deviations) analyses yielded significant results in the association between IL-10-1082 GG genotype (or IL-10–1082 G allele) and AR risk (). Meta-regression confirmed that there was no significant correlation between the pre-selected trial characteristics (age, male ratio, ethnicity, region of nations, donor type, AR definition, and HWE deviations) and our study results.
Table 2. Subgroup analyses for the primary outcome measurement.
Publication bias
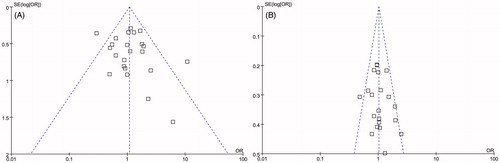
Publication bias was examined by evaluating funnel plots for studies of AR in renal transplant recipients. Egger and Begg tests showed that no potential publication bias existed among the included studies for the two end-points analyzed: Egger test p = 0.483, Begg test p = 0.712 for IL-10-1082 GG genotype (); Egger test p = 0.397, Begg test p = 0.194 for IL-10-1082 G allele ().
Discussion
The balance between pro- and anti-inflammatory cytokines determines the inflammatory response which mediates renal allograft outcome.Citation36 In the context of allograft rejection, IL-10 as an anti-inflammatory cytokine can inhibit the production of tumor necrosis factor-α, IL-1, IL-6, IL-8, and IL-12 in monocytes/macrophages and interferon-γ in T cells.Citation37 Some studies have suggested that IL-10 mRNA levels are increased just before a rejection episode.Citation38 Suarez et al. showed that allelic variants at position -1082 were the most important genetic factor in the regulation of the constitutive mRNA levels of IL-10.Citation39 Our study is the first meta-analysis ever published on this topic assessing the association between IL-10–1082 (G/A) promoter polymorphism and AR risk in renal transplant recipients. There were a total of 2779 renal transplant patients in 22 case–control studies enrolled in this meta-analysis, but it was unable to demonstrate any statistically significant association.
Most of the included studies revealed no significant correlation between IL-10–1082 (G/A) promoter polymorphism and AR risk in the recipients except for 3 articles.Citation21,Citation27,Citation29 Canossi et al.Citation21 reported that more incidence of AR episodes (ARE)-positive patients (35.8% vs. 18.2%) in the high producers group (GCC/GCC,GCC/ACC,GCC/ATA) was observed compared with the low producers. Hutchings et al.Citation27 found African-American patients who were IL-10 A homozygous had a significantly lower probability of experiencing one or more AREs (p = 0.006), although there was no significant relationship in the Caucasian patients. Nevertheless, George et al.Citation29 draw a different conclusion that the frequency of IL-10–1082 AA, a genotype associated with low production of IL-10, was high in the rejection group when compared to the rejection-free group (p = 0.01). Moreover, there were only two studiesCitation13,Citation32 showed some correlation tendency between IL-10–1082 (G/A) polymorphism and AR risk. Dhaouadi et al.Citation13 suggested that there was a greater prevalence of ARE among kidney transplant recipients carrying the IL-10–1082 GG genotype than those with other levels, albeit that the differences were no longer significant. Pelletier et al.Citation32 found an increase in AR incidence as the IL-10 production phenotype increased (low, intermediate, high).
Lack of statistical power to detect small or moderate gene effects is a major limitation for many gene-association studies. Meta-analyses based on published data might be helpful to overcome this problem. However, some key factors may influence the results of meta-analyses, such as the AR definition. AR was defined as “biopsy proven AR” in most of the 22 included studies except for two studiesCitation14,Citation22 in which AR was defined as “increased serum creatinine” or “clinical diagnostic criteria”. However, subgroup analyses for AR definition did not yield any significant results. In addition, the occurrence time of AR was also defined inconsistently. Among the included studies, eight studiesCitation14,15,18,20,21,27,28,32 did not refer to the occurrence time of AR, other studies defined it varies from one month to one year after transplantation. These different definitions and standards might make our result bias.
Another key factor “ethnicity” was also a potential source of bias. Although the article of Pallet et al.Citation40 demonstrated that ethnic origin did not affect outcome after renal transplantation, previous research had shown that ethnicity can strongly influence the distribution of cytokine gene polymorphisms.Citation41 For example, Hutchings et al.Citation27 performed a study to determine the distribution of IL-10–1082 (G/A) promoter polymorphism in African-American and Caucasian renal transplant recipients, then he found that there was a significant link between IL-10 genotype and AREs, but only in African-American patients (p < 0.01). Among our included studies, six studiesCitation11,16,18,24,26,31 were in research of Caucasian recipients, two studiesCitation27,Citation32 were in research of both Caucasian and African-American recipients, and the others did not refer to ethnicity. Nevertheless, when stratifying for ethnicity, no significant association was observed.
Different donor types were other factors affecting the results of this analysis. Among the 22 studies, living kidney donor was researched in five studies,Citation12,14,15,19,22 deceased kidney donor was researched in five studies,Citation16,21,25,28,30 and living or deceased kidney donor was researched in the remaining case studies. But subgroup analyses for donor type did not yield significant results. Moreover, in the 22 case–control studies, although all of the researches took an immunosuppressive regimen in patients after renal transplantation, differences in immunosuppressive regimens, such as type of induction therapy, the scheme of maintenance therapy and dosage of immunosuppressants, were also a potential source of bias. Other important factors for the association between IL-10–1082 polymorphism and AR risk in renal transplant recipients included age, gender, region of nations, and HWE deviations. Nevertheless, none of the subgroup analyses and meta-regression analyses for these factors yielded any significant results.
It is necessary to mention the relatively limited power of our meta-analysis to find a different result. For example, the country zones of the 22 studies were unevenly distributed, in which, 10 researchesCitation11,16,18,20,21,25,28–30,32 were from Europe, six researchesCitation12,14,15,17,19,22 were from Asia, 5 researchesCitation23,24,26,27,31 were from America, and only one researchCitation13 was from Africa. Another drawback that might influence our study results is the insufficient information among the included studies, encompassing ethnicity, age, gender, HWE deviations, etc. In summary, this meta-analysis suggests that the IL-10-1082 (G/A) promoter polymorphism is not significantly associated with AR risk in renal transplant recipients. In the future, larger and well-designed multicenter case–control studies should be performed to reevaluate the association.
Supplementary material available online
Acknowledgments
The authors thank all the participants in the study.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Djamali A, Premasathian N, Pirsch JD. Outcomes in kidney transplantation. Semin Nephrol. 2003;23:306–316
- Jazrawi SF, Zaman A, Muhammad Z, et al. Tumor necrosis factor-alpha promoter polymorphisms and the risk of rejection after liver transplantation: a case control analysis of 210 donor-recipient pairs. Liver Transpl. 2003;9:377–382
- Burckart GJ, Amur S. Update on the clinical pharmacogenomics of organ transplantation. Pharmacogenomics. 2010;11:227–236
- Vu D, Tellez-Corrales E, Shah T, et al. Influence of Cyclooxygenase-2 (COX-2) gene promoter-1195 and allograft inflammatory factor-1 (AIF-1) polymorphisms on allograft outcome in Hispanic kidney transplant recipients. Hum Immunol. 2013;74:1386–1391
- Liu F, Li B, Wang WT, et al. Interleukin-10-1082G/A polymorphism and acute liver graft rejection: a meta-analysis. World J Gastroenterol. 2012;18:847–854
- Kruger B, Schroppel B, Murphy BT. Genetic polymorphisms and the fate of the transplanted organ. Transplant Rev (Orlando). 2008;22:131–140
- Azarpira N, Aghdai MH, Raisjalali GA, et al. Influence of recipient and donor IL-10, TNFA and INFG genotypes on the incidence of acute renal allograft rejection. Mol Biol Rep. 2009;36:1621–1626
- Karimi MH, Ebadi P, Pourfathollah AA. Association of cytokine/costimulatory molecule polymorphism and allograft rejection: a comparative review. Expert Rev Clin Immunol. 2013;9:1099–1112
- Nickerson P. The impact of immune gene polymorphisms in kidney and liver transplantation. Clin Lab Med. 2008;28:455–468
- Thakkinstian A, Dmitrienko S, Gerbase-Delima M, et al. Association between cytokine gene polymorphisms and outcomes in renal transplantation: a meta-analysis of individual patient data. Nephrol Dial Transplant. 2008;23:3017–3023
- Chen Z, Bouamar R, Van Schaik RH, et al. Genetic polymorphisms in IL-2, IL-10, TGF-β1, and IL-2RB and acute rejection in renal transplant patients. Clin Transplant. 2014;28:649–655
- Gaafar A, Iqniebi A, Sheereen A, et al. Study of the cytokine polymorphisms in correlation to rejection and graft survival in renal allograft donors and recipients from a homogenous Saudi population. Transpl Immunol. 2014;30:34–39
- Dhaouadi T, Sfar I, Bardi R, et al. Cytokine gene polymorphisms in kidney transplantation. Transplant Proc. 2013;45:2152–2157
- Mandegary A, Azmandian J, Soleymani S, et al. Effect of donor tumor necrosis factor-alpha and interleukin-10 genotypes on delayed graft function and acute rejection in kidney transplantation. Iran J Kidney Dis. 2013;7:135–141
- Seyhun Y, Mytilineos J, Turkmen A, et al. Influence of cytokine gene polymorphisms on graft rejection in Turkish patients with renal transplants from living related donors. Transplant Proc. 2012;44:1670–1678
- Kocierz M, Siekiera U, Kolonko A, et al. -174G/C interleukin-6 gene polymorphism and the risk of transplanted kidney failure or graft loss during a 5-year follow-up period. Tissue Antigens. 2011;77:283–290
- Azarpira N, Aghdai MH, Raisjalali GA, et al. Influence of recipient and donor IL-10, TNFA and INFG genotypes on the incidence of acute renal allograft rejection. Mol Biol Rep. 2009;36:1621–1626
- Grinyó J, Vanrenterghem Y, Nashan B, et al. Association of four DNA polymorphisms with acute rejection after kidney transplantation. Transpl Int. 2008;21:879–891
- Manchanda PK, Mittal RD. Analysis of cytokine gene polymorphisms in recipient's matched with living donors on acute rejection after renal transplantation. Mol Cell Biochem. 2008;311:57–65
- Breulmann B, Bantis C, Siekierka M, et al. Influence of cytokine genes polymorphisms on long-term outcome in renal transplantation. Clin Transplant. 2007;21:615–621
- Canossi A, Piazza A, Poggi E, et al. Renal allograft immune response is influenced by patient and donor cytokine genotypes. Transplant Proc. 2007;39:1805–1812
- Tajik N, Kazemi T, Delbandi A, et al. The predictive value of HLA-DR matching and cytokine gene polymorphisms in renal allograft acute rejection: A living-unrelated donor (LURD) study. Iran J Immunol. 2006;3:150–156
- Gendzekhadze K, Rivas-Vetencourt P, Montano RF. Risk of adverse post-transplant events after kidney allograft transplantation as predicted by CTLA-4 + 49 and TNF-alpha -308 single nucleotide polymorphisms: a preliminary study. Transpl Immunol. 2006;16:194–199
- Dmitrienko S, Hoar DI, Balshaw R, Keown PA. Immune response gene polymorphisms in renal transplant recipients. Transplantation. 2005;80:1773–1782
- Ligeiro D, Sancho MR, Papoila A, et al. Impact of donor and recipient cytokine genotypes on renal allograft outcome. Transplant Proc. 2004;36:827–829
- Plothow A, Bicalho MG, Benvenutti R, Contieri FL. Interleukin-10 and acute rejection in renal transplantation. Transplant Proc. 2003;35:1338–1340
- Hutchings A, Guay-Woodford L, Thomas JM, et al. Association of cytokine single nucleotide polymorphisms with B7 costimulatory molecules in kidney allograft recipients. Pediatr Transplan. 2002;6:69–77
- Cartwright NH, Keen LJ, Demaine AG, et al. A study of cytokine gene polymorphisms and protein secretion in renal transplantation. Transpl Immunol. 2001;8:237–244
- George S, Turner D, Reynard M, et al. Significance of cytokine gene polymorphism in renal transplantation. Transplant Proc. 2001;33:483–484
- Marshall SE, McLaren AJ, Haldar NA, et al. The impact of recipient cytokine genotype on acute rejection after renal transplantation. Transplantation. 2000;70:1485–1491
- Hahn AB, Kasten-Jolly JC, Constantino DM, et al. TNF-alpha, IL-6, IFN-gamma, and IL-10 gene expression polymorphisms and the IL-4 receptor alpha-chain variant Q576R: effects on renal allograft outcome. Transplantation. 2001;72:660–665
- Pelletier R, Pravica V, Perrey C, et al. Evidence for a genetic predisposition towards acute rejection after kidney and simultaneous kidney-pancreas transplantation. Transplantation. 2000;70:674–680
- Wells GA, Shea B, O’Connell D, et al The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. Ottawa Health Research Institute. Available from: URL: http//www.ohri.ca/programs/clinical_epidemiology/oxford. Asp
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634
- Misra MK, Pandey SK, Kapoor R, et al. HLA-G gene expression influenced at allelic level in association with end stage renal disease and acute allograft rejection. Hum Immunol. 2014;75:833–839
- Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765
- Platz KP, Mueller AR, Rossaint R, et al. Cytokine pattern during rejection and infection after liver transplantation-improvements in postoperative monitoring? Transplantation. 1996;62:1441–1450
- Suarez A, Castro P, Alonso R, et al. Interindividual variations in constitutive interleukin-10 messenger RNA and protein levels and their association with genetic polymorphisms. Transplantation. 2003;75:711–717
- Pallet N, Thervet E, Alberti C, et al. Kidney transplant in black recipients: are African Europeans different from African Americans? Am J Transplant. 2005;5:2682–2687
- Hoffmann SC, Stanley EM, Cox ED, et al. Ethnicity greatly influences cytokine gene polymorphism distribution. Am J Transplant. 2002;2:560–567