Abstract
Ectopic expression of recombinant human bone morphogenetic protein 2 (rhBMP2) induces osteogenesis, while ectopic expression of rhBMP12 and rhBMP13 induces the formation of tendon-like tissue. Despite their different in vivo activities, all three ligands bound to the type I bone morphogenic protein receptors (BMPRs), activin receptor-like kinase (ALK)-3 and ALK6, and to the type II BMPRs, activin receptor type-2A, activin receptor type-2B, and BMPR2, with similar affinities. Treatment of C3H10T1/2 cells with rhBMP2 activated SMAD signaling and induced expression of osteoblast markers including osteocalcin mRNA (Ocn). In contrast, treatment with rhBMP12 or rhBMP13 resulted in a dose-dependent induction of a tendon-specific gene (Thbs4) expression with no detectable activation of SMAD 1, 5, and 8. Differential regulation of Thbs4 and Ocn has potential utility as an in vitro biomarker for induction of tenogenic signaling. Such an assay also permits the ability to distinguish between the activities of different BMPs and may prove useful in studies on the molecular mechanisms of BMP tenogenic activity.
Introduction
Bone morphogenic proteins (BMPs) are members of the transforming growth factor beta superfamily, which is a group of growth factors known to induce growth and differentiation of various cell types (Reddi Citation1998). Originally identified as components of de-mineralized bone that can induce bone and cartilage formation in vitro and in vivo, the BMP family now includes more than 30 related members. On the basis of sequence similarity, BMPs have been grouped into several subtypes. BMP2, 4, 6, and 7 are members of a major BMP family subtype that are responsible for inducing bone and cartilage formation (Wozney et al. Citation1988). Ectopic expression of BMP2 in rat muscle, for example, initially induces mesenchymal stem cell migration, proliferation, and chondrocytic differentiation. Chondrocytes subsequently calcify and are then replaced by mature bone. Studies on the functions of rhBMP2 and rhBMP7 in various preclinical models demonstrated increases in bone growth and regeneration (Cook and Rueger Citation1996; Rosen and Wozney Citation2002).
A distinct subset of related and highly conserved BMPs (BMP12, BMP13, and growth/differentiation factor 5 (GDF5)) is characterized by a lack of propensity to induce bone formation. Instead, molecules of this class induce the formation of cartilage and tendon-like structures in vivo (Chang et al. Citation1994; Wolfman et al. Citation1997). Administration of these BMPs after tendon injury in animal models results in increased rates of tendon repair and increased strength and stiffness of injured tendons (Aspenberg and Forslund Citation1999; Lou et al. Citation2001; Bolt et al. Citation2007; Seeherman et al. Citation2008). GDF5 has been evaluated as an inducer of spine fusion in an animal model (Jahng et al. Citation2004; Walsh et al. Citation2004) and as a treatment for degenerative disc disease in the clinic. More recently, polymorphisms in Gdf5 were found to be associated with increased risk of tendinopathy (Posthumus et al. Citation2010).
BMP12, BMP13, and GDF5 share 82% amino acid sequence identity and similar functional properties (Wolfman et al. Citation1997). These molecules are also similar in sequence to osteogenic BMP family members (e.g. BMP12 and BMP2 share 55% amino acid sequence identity) and it has been suggested that BMP12, BMP13, and GDF5 can bind and signal through the same pathways as BMP2 and BMP4 (Mazerbourg et al. Citation2005). Both subclasses of BMPs initiate intracellular signaling through interaction with the type I receptors, activin receptor-like kinase (ALK)-3 and ALK6, and with the type II receptors, BMP receptor (BMPR)-2 and activin receptor 2B (ACVR2B; Mazerbourg et al. Citation2005). Specificity of response may in part be contributed to differential receptor ligand binding as ligand/receptor bindings has been found to be different among the BMP family members. BMPs bind their receptors as heterotetrameric complexes containing two type I and two type II receptors. Previous work has shown that BMP2 and BMP4 have higher binding affinity for the type I receptors, ALK3 and ALK6, and have lower binding affinity for the type II receptors, BMPR2, ACVR2A, and ACVR2B. BMP6 and BMP7 on the other hand can bind to the same receptors as BMP2 and BMP4, but bind the type II receptors with higher affinity than binding to the type I receptors. In contrast, GDF5 has been shown to preferentially bind ALK6 compared with ALK3 after forming a heteromeric complex with BMPR2 or ACVR2B (Nishitoh et al. Citation1996; Erlacher et al. Citation1998; Nickel et al. Citation2009). Activation of the ligand–receptor complex leads to signaling through the canonical SMAD pathway, but signaling can also proceed through the mitogen-activated protein (MAP) kinase pathway and possibly other pathways (Nohe et al. Citation2004).
Despite the high level of amino acid sequence identity and the strong in vivo phenotypes, the mechanism of action for BMPs which leads to the formation of tendon-like tissue vs. bone tissue is not well understood. In part, this is due to the lack of a specific and reproducible in vitro assay for measuring BMP tenogenic activity. The expression of two genes in particular, thrombospondin 4 (Thbs4) and tenomodulin (Tnmd), appears to be highly localized to tendon/ligament tissues, thereby making these genes potential tendon tissue-specific markers (Jelinsky et al. Citation2010). In contrast, biochemical markers of bone remodeling and growth have been well established and have been used for many years as useful tools to monitor bone health. Commonly, the detection of osteocalcin (Ocn), runt-related transcription factor 2 (Runx2), and osterix (Sp7) as markers has been used to determine early stages of bone formation. Additionally, alkaline phosphatase (ALP) activity and SMAD intracellular signaling have been monitored to quantify bone formation and BMP activity. In this study, we compare receptor binding affinities of tenogenic and osteogenic BMPs and show differential canonical BMPs signaling and activation of a novel tenogenic marker. These studies show that even though tenogenic (BMP12 and BMP13) and osteogenic (BMP2) BMPs bind the same receptors with high affinity they signal much differently and result in differential activation of osteogenic and tenogenic markers.
Methods
Ethics statement
Protocols for the use of animals in this work were approved by the Wyeth Animal Care and Use Committee, and were in accordance with the National Institute of Health's standards established by the Guidelines for the Care and Use of Experimental Animals.
Reagents and proteins
Mature forms of rhBMP12 and rhBMP13 were expressed in Escherichia coli in inclusion bodies. Inclusion bodies were solubilized in 8.0 M Urea, 100 mM DTT, 20 mM Tris, pH 8.4, and the pH was adjusted to 6.5. The unfolded protein was captured on an SP-Sepharose column equilibrated with 25 mM HEPES, 25 mM MES, 8.0 M Urea, and pH 6.5, and eluted with a linear 0–1.0 M NaCl gradient over 10 column volumes. Refolding was achieved by rapid dilution of the protein into refolding buffer (50 mM Tris, 5.0 mM EDTA, 1.0 M NaCl, 2% CHAPS, 0.03% Reduced Glutathione, 0.03% Oxidized Glutathione, pH 8.4). After 7 days, the refolded protein was purified using the SP Sepharose column, as described above, followed by size-exclusion chromatography on two 21.5 × 60-cm TOSOH G3000SWL columns in tandem in 6.0 M Urea, 300 mM NaCl, and 3% acetic acid. rhBMP2 was produced and purified from CHO cells as previously described (Wang et al. Citation1990). For initial quality control comparisons, some rhBMP12 was purified from CHO cells. The purified ligands were analyzed by reduced (with β-mercaptoethanol) and nonreduced SDS-PAGE analysis and Coomassie blue staining. Additionally ligand purity was determined by analytical HPLC gel filtration (data not shown). Proper refolding was determined by circular dichroism (CD) spectroscopy using mammalian expressed protein references. The spectra were taken at 0.1 mg/ml in 50 mM acetic acid, using a Jasco J-810 CD spectropolarimeter (Jasco, Easton, MD, USA) and a 1-mm quartz sample cell equilibrated at 20°C. Scans between 190 and 260 nm were performed at a scan rate of 50 nm per min.
Ectopic assays
Adult, male, Sprague-Dawley rats were obtained from Charles River Laboratory (Wilmington, MA, USA) and maintained on a 12-h:12-h light:dark cycle with food and water ad libitum. Rats were anesthetized using isoflorane (1–3% by inhalation). A small incision of approximately 0.5 cm in length was made on the medial aspect of the hamstring using a No. 15 scalpel blade. A small pocket was made in the intramuscular space with the blunt end of the scalpel to receive the implant. Implants consisted of 25 μg of rhBMP12, rhBMP13, or rhBMP2 lyophilized onto rat demineralized bone matrix particles. Implants were placed into the intramuscular space and sutured into position with 4-0 Vicryl cutting material. The surrounding soft tissue and skin were closed using absorbable suture material and skin staples.
Histology
Tissue sections (5–6 μm thick) were stained with Weigert's Safranin O stain (Polyscientific, Bay Shore, NY, USA) for histological analysis. Images were semi-quantitatively scored by a three-point scale based on density and fibroblast morphology. Multiple fields within an image were examined by three different blinded reviewers, and the final score was an average of all fields from all reviewers. A score of one was given to images containing primarily loosely packed fibroblast having a thin, spindle-shaped morphology, along with low proteoglycan and extracellular matrix content. A score of three was given for images showing densely packed fibroblasts with large cytoplasms, high fibrotic extracellular matrix levels, and high proteoglycan levels. Images with an “intermediate” number of fibroblasts were given a score of two. Prior to histological evaluation, ectopic implants were removed from animals, and a faxitron microradiograph apparatus (Faxitron X-ray Corp., Wheeling, IL, USA) was then used for the visualization of in vivo bone formation.
Cell culture conditions
The murine mesenchymal stem cell line C3H10T1/2 (clone 8) was purchased from ATCC (CCL-226; Manassas, VA, USA). Low passage cells were cultured in 1X Minimum Essential Media (MEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS) without the addition of antibiotics. Exponentially growing cells (2 × 105 cells at a density of 2 × 105/ml) from passages 3 to 6 were cultured in clear flat bottom 12-well plates overnight. The following day, the media was replaced with MEM supplemented with 1% FBS without antibiotics. A single dose of BMP was added to the culture, and the cells were incubated at 37°C in 5% CO2.
BMP activity assays
C3H10T1/2 cells were transiently transfected in 12-well plates using Fugene-6 (Roche, Basel, Switzerland) with a BMP-responsive firefly luciferase reporter (BRE-luc; Korchynskyi and ten Dijke Citation2002) vector, and a pRL-TK renilla luciferase transfection efficiency control vector. After 24 h, cells were treated with either 1, 10, or 100 nM of rhBMP12, rhBMP13, or rhBMP2 in Dulbecco's Modified Eagle Medium supplemented with 1% FBS or with BMP free control medium, and assayed for either luciferase activity (after 24 h) or ALP activity (after 4 days). Luciferase activity was determined after cell lysis using the Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA). All experiments were performed in triplicate. In all cases, firefly luciferase levels were normalized to the corresponding renilla luciferase levels in a given well, yielding final values in relative light units. ALP activity assays were performed as described previously (Thies et al. Citation1992).
Western blotting
Equal amounts of protein (80 μg/lane) or supernatants were concentrated (80% concentrated) using Microcon YM-10 (Millipore, Billerica, MA, USA) spin columns and underwent SDS-PAGE under reducing conditions using NuPage MOPS 4–12% gels (Invitrogen). Gels were transferred to nitrocellulose and probed with polyclonal antibodies against phospho-SMAD1/5/8 (Cell Signaling Technology, Beverly, MA, USA; 1:750), β-actin (Abcam, Cambridge, MA, USA), β-tubulin (Sigma-Aldrich, St. Louis, MO, USA), Thrombospondin-4 (R&D Systems, Minneapolis, MN, USA), or with the anti-HA.11 monoclonal antibody (Covance, Princeton, NJ, USA).
TaqMan real time quantitative PCR
RNA was isolated and purified from cells using RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Relative mRNA levels were measured by TaqMan real time quantitative PCR using Assay-on-Demand TaqMan reagents and primers (Applied Biosystems, Foster City, CA, USA). TaqMan was performed using an ABI PRISM 7900 Sequence Detection System (PE Applied Biosystems). Expression levels were determined by the ΔΔCT method, normalized to Gusb mRNA levels, and then presented as the ratio of expression of each target mRNA in BMP-treated samples compared with the expression of the same mRNA in the control sample.
Receptor binding
Fusion proteins consisting of the BMPR extracellular domain linked to the human IgG Fc region (ALK1-Fc, ALK2-Fc, ALK3-Fc, and ACVR2A-Fc) were either purchased (R&D Systems), or synthesized in-house (ALK6-Fc, ACVR2B-Fc, and BMPRII). Receptor Fc chimeras were immobilized on anti-human IgG Fc biosensor tips (ForteBio, Menlo Park, CA, USA). Binding assays using sixfold serial dilutions of human BMPs in solution were performed using Octet RED (ForteBio). Association time was set at 10 min and dissociation time was set at 30 min. Binding affinities were calculated using ForteBio Data Acquisition 6.3 software (ForteBio). Affinities were derived by fitting the kinetic data to a 1:1 Langmuir binding model as described previously (Heinecke et al. Citation2009) utilizing global fitting algorithms.
Generation and use of ALK3 kinase dead adenovirus
cDNAs encoding HA-tagged, dominant negative ALK3 (K261R; Gupta et al. Citation2000), or GFP were used to create adenoviral vectors in 293 cells. Large scale purification by CsCl gradient ultracentrifugation was done at Viraquest (North Liberty, IA, USA). C3H10T1/2 cells were infected with a multiplicity of infection of 250 and then incubated with adenoviruses for 6 h at 37°C in MEM supplemented with 1% FBS without antibiotics. Media were replenished, rhBMP12 or rhBMP13 (10 or 100 nM) was added, and cells incubated for an additional 3 days.
Results
Purification Of Rhbmp2, Rhbmp12, And Rhbmp13
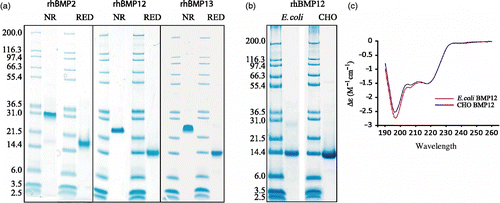
The mature domain of rhBMP2 was purified from CHO cells overexpressing rhBMP2.The mature domains of rhBMP12 and rhBMP13 were purified from E. coli inclusion bodies and refolded. Small quantities of CHO expressed rhBMP12 was purified and used as reference material to validate refolded E. coli expressed proteins. Purified proteins (5 μg) were characterized by SDS-PAGE under reducing conditions (with β-mercaptoethanol) or nonreducing conditions (). No glycosylations are predicted by the primary sequence of BMP12 and both CHO-expressed and E. coli expressed rhBMP12 had the same apparent molecular mass (approximately 14 KDa), on reduced SDS-PAGE gel consistent with the predicted molecular mass from the amino acid sequence of native BMP12 without any post translational modifications (). The apparent size of the proteins determined by unreduced SDS-PAGE analysis is consistent with homodimer formation. The purity of the protein was determined by SDS-PAGE and HPLC (data not shown) and suggested that all three proteins were greater than 95% pure. CD spectroscopy suggested that E. coli-derived and CHO-derived rhBMP12 have similar secondary structure confirming that E. coli-derived protein is properly folded ().
Figure 1. Characterization of purified rhBMP2, rhBMP12, and rhBMP13. (a) The purified proteins were collected and analyzed under reducing (RED) and nonreducing (NR) conditions using 12% polyacrylamide slab gels. (b) SDS-PAGE analysis of rhBMP12 purified from E. coli or CHO cells reduced with β-mercaptoethanol. (c) CD spectroscopy of BMP12 purified from E. coli or CHO cells.
Distinct tissues mediated by ectopic expression of rhBMP2, rhBMP12, and rhBMP13
After 14 days of implantation, microscopic evaluation of ectopic implants containing rhBMP2, rhBMP12, or rhBMP13 showed increased fibroblast content and activity in a dose-dependent manner (, ). rhBMP2-containing implants induced the proliferation or migration of many more cells than either rhBMP12 or rhBMP13-containing ectopic implants. Fibroblasts in rhBMP12- and rhBMP13-containing implants aligned in parallel to each other and displayed a wave-like pattern, which is characteristic of embryonic or neonatal tendon. In contrast, rhBMP2-containing implants contained endochondral bone that was encompassing small islands of hypertrophic cartilage. rhBMP2 formed ectopic bone in all implants, whereas only high doses of rhBMP12 or rhBMP13 in implants induced ectopic bone formation. However, the ectopic bone induced by rhBMP12- and rhBMP13-containing implants was visibly less than that induced by rhBMP2-containing implants. The ectopic bone formed by rhBMP2 encompassed most of the implant area, whereas only small spicules of ectopic bone were formed by rhBMP12 and rhBMP13 (, and ). Low doses of rhBMP2 induced low levels of ectopic bone and the accumulation of loosely packed quiescent fibroblasts. In rhBMP2 implants, the fibroblasts did not exhibit any observable parallel alignment as was observed in the rhBMP12- and rhBMP13-containing implants.
Figure 2. Histology and X-ray analysis of BMP-treated implants after 14 days. Saffranin O stain of representative sections of implants containing (a) 25 μg of rhBMP2, (b) 25 μg of rhBMP12, or (c) 25 μg of rhBMP13. Faxitron X-ray images of representative implants containing (d) 25 μg of rhBMP2, (e) 25 μg of rhBMP12, or (f) 25 μg of rhBMP13. The implants containing rhBMP2 consists of mostly bone, whereas the implants containing rhBMP12 or rhBMP13 consists of only small little bone spicules (arrows).
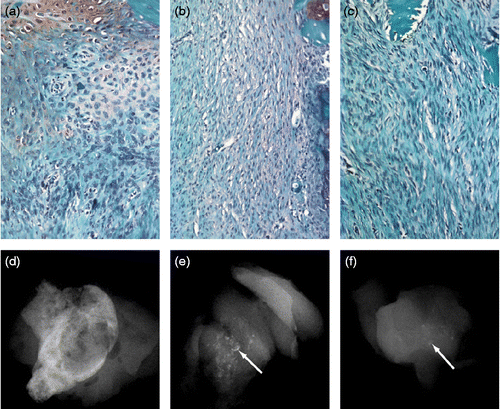
Table I. Ectopic cell assay.
All implants formed a hard callus and were easily isolated from surrounding tissues. RNA expression analysis identified many tendon marker genes including Col1a1, Col1a2, Eya1, Eya2, Scx, Epha4, Tnmd, and Thbs4 that exhibited changes in relative level in rhBMP12- and rhBMP13-induced ectopic tissues (). In contrast, many bone marker genes including Bglap (osteocalcin) Sp7 (osterix) and Alpl (ALP) exhibited changes in relative level in rhBMP2-induced ectopic tissues (). Ectopic tissue induced by both rhBMP12 and rhBMP2 increased expression of cartilage marker genes including Sox9, Acan, and Col2a (). The increased expression of tendon-related genes was not specific to rhBMP12 treatment as most of these genes were also increased in the rhBMP2-treated ectopics. However, increases in bone-related genes appeared to be selective to rhBMP2-treated ectopics.
Table II. Fold change in RNA expression of musculoskeletal markers in rat ectopic tissue.
rhBMP2, rhBMP12, and rhBMP13 bind the same receptors
To determine whether the observed differences in BMP-mediated gene expression were due to a difference in canonical BMPR–ligand binding, receptor-ligand binding affinities were calculated. Using Bio-Layer Interferometry (Octet) analysis, binding curves were generated from experiments in which immobilized receptor extracellular domain—IgG Fc fusion proteins were exposed to purified BMPs. rhBMP2, rhBMP12, and rhBMP13 bound the same type I and type II BMPRs. The type I receptors ALK3 and ALK6 bound to rhBMP2, rhBMP12, and rhBMP13 with high affinity (KD) and bound all three BMPs with similar association (kon) and dissociation (koff) rates (). While ALK2 did bind to rhBMP2, but not to rhBMP12 or rhBMP13, the affinity of this interaction was orders of magnitude weaker than the affinity of rhBMP2 for ALK3 or ALK6. rhBMP2, rhBMP12, and rhBMP13 also bound the type II receptors ACVR2A, ACVR2B, and BMPRII with comparable levels of affinity. Each of the three BMPs interacted with ACVR2A and BMPRII with a binding affinity (KD) that is more than 10-fold less than the affinity observed with the type I receptors. Contrastingly, ACVR2B exhibits affinity for rhBMP12 and rhBMP13 that is similar to that observed for type I receptors. Calculated binding affinities for receptor–ligand interactions in which all three BMP ligands bound tightly were very similar, thus distinguishing this state of interaction from weaker interactions, such as those observed with ACVR2A and BMPR2.
Table III. Binding affinities (kD, nM) of BMP ligands as determined by surface plasmon resonance.
Distinct activation of bone, cartilage, and tendon gene markers with rhBMP2, rhBMP12, and rhBMP13 in C3H10T1/2
Analysis of ectopic tissues is complicated by the multiple distinct cell types present including skeletal muscle, inflammatory cells, and fibroblasts. To begin to distinguish the reproducible differential activity of rhBMP2, rhBMP12, and rhBMP13, activation of musculoskeletal marker genes was investigated in the easily cultured BMP-responsive mesenchymal cell line, C3H10T1/2. C3H10T1/2 cells cultured with rhBMP2 but not rhBMP12 or rhBMP13 produced high levels of the bone marker mRNAs Ocn, Runx2, Alpl, and Sp7. Cells treated with relatively high concentrations of rhBMP12 or rhBMP13 (100 nM) made relatively low levels of Ocn mRNA and did not express detectable levels of the other bone markers examined (). Each of the three BMPs tested had the effect of increasing levels of the collagen marker mRNAs Sox9 and Col2a1, but rhBMP2 treatment increased Col2a1 by approximately 2.5-fold over that observed with rhBMP12 or rhBMP13 treatment. Furthermore, rhBMP2 treatment increased Sox9 mRNA levels at much lower doses than did rhBMP12 and rhBMP13 ().
Figure 3. Expression of bone, tendon, and cartilage markers by BMPs in C3H10T1/2 cells. mRNA levels were determined by Taqman for the bone markers (a) Ocn (osteocalcin), (b) Runx2 (runt-related transcription factor 2, (c) Alpl (ALP), and (d) Sp7 (osterix), the tendon markers (e) Thbs4 (thrombospondin 4) and (f) Tnmd, (tenomodulin) and the cartilage markers (g) Sox9 (SRY-box containing gene 9), (h) Col2a1 (collagen, type II, alpha 1), (i) Acan (Aggrecan 1) and (j) Col11a1 (collagen, type 11, alpha 1).
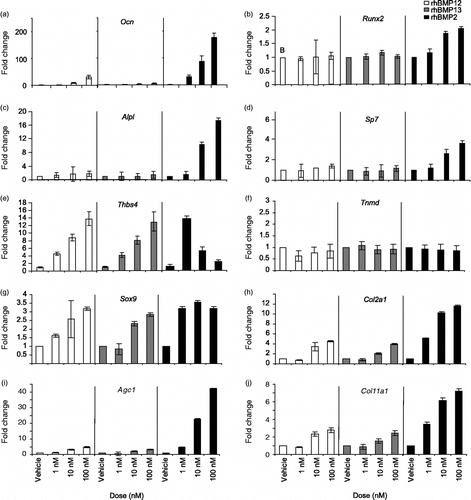
Thbs4 and Tnmd were previously identified as tendon-specific markers in rat and human tissues (Jelinsky et al. Citation2010). mRNA and protein analyses support the tissue specificity of Thbs4 in mouse tendon (Supplemental Figure 1). A 3-day treatment of C3H10T1/2 with rhBMP12 or rhBMP13 increased Thbs4 mRNA levels in a dose-dependent manner up to 15-fold (). Treatment of C3H10T1/2 cells with low concentrations of rhBMP2 (1 and 5 nM) resulted in increased Thbs4 mRNA levels, but unlike rhBMP12 and rhBMP13, higher concentrations of rhBMP2 failed to increase Thbs4 mRNA levels further (). Neither of the BMPs appeared to affect Tnmd mRNA levels, and Scx was not expressed under the conditions tested.
Distinct intracellular pathway activation with rhBMP2, rhBMP12, and rhBMP13
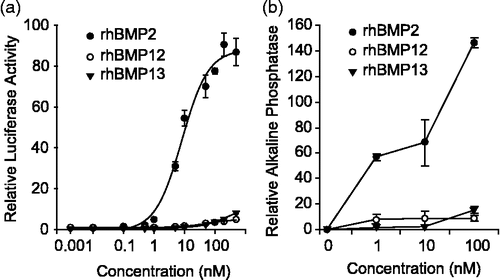
As C3H10T1/2 cells were responsive to all three BMPs and replicate many of the molecular changes observed in vivo after ectopic expression, we explored differences in intracellular signaling mechanisms in this cell line. BMP ligands differentially activate a SMAD 1, 5, 8 dependent BRE-luciferase reporter (BMP response element) in C3H10T1/2 cells. rhBMP2 treatment led to a more than 100-fold induction of BRE-luciferase reporter activity (EC50, ∼8 nM) compared with control. rhBMP12 induced the lowest levels of BRE-luciferase reporter activity at 6-fold (EC50, ∼52 nM) that of control, whereas rhBMP13 induced only slightly more BRE-luciferase reporter activity at 10-fold (EC50, ∼272 nM; ). In a similar trend, neither rhBMP12 (10 nM) nor rhBMP13 (10 nM) caused considerable increases in ALP activity, whereas rhBMP2 treatment (10 nM) did result in a clear increase in ALP activity (60-fold increase). At 100 nM rhBMP2, there was a 14-fold higher increase in ALP activity compared with changes in ALP activity observed with rhBMP12 or rhBMP13 ().
Figure 4. BMP activity in C3H10T1/2 cells. (a) C3H10T1/2 cells infected with BRE-Luc construct were treated with increasing concentration of rhBMP2, rhBMP12, or rhBMP13. (b) ALP activity in C3H10T1/2 cells following 2-day incubation with 0, 1, 10, or 100 nM of rhBMP2, rhBMP12, or rhBMP13.
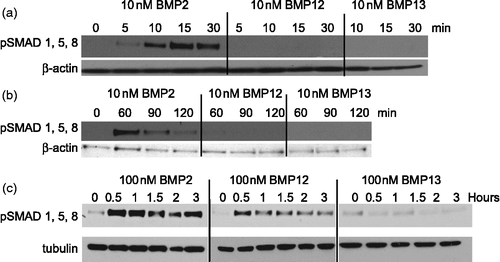
rhBMP2 induced phosphorylation of SMADs 1, 5, and 8 much more potently than rhBMP12 or rhBMP13. C3H10T1/2 cells treated with 10 nM rhBMP2 exhibited induced SMAD 1, 5, and 8 phosphorylation 30 min after treatment, but this decreased after 60 min (,b). In contrast, C3H10T1/2 cells treated with rhBMP12 (10 nM) or rhBMP13 (10 nM) did not exhibit any detectable changes in phosphorylated SMADs after up to 2 h of treatment (). At higher doses of rhBMP2 (100 nM), high levels of phosphorylated SMADs were maintained for at least 3 h. Higher doses of rhBMP12 (100 nM) induced SMAD 1, 5, and 8 phosphorylation, but this effect was much less than that observed with rhBMP2 ().
Figure 5. Induction of SMAD signaling by BMPs in C3H10T1/2 cells. Cell lysates from rhBMP2, rhBMP12, or rhBMP13 treated C3H10T1/2 cells were analyzed by western blot with antibodies to phosphorylated SMAD 1/5/8 and to β-actin or tubulin. Cells were treated with (a)10 nM BMPs for 0 to 30 min, (b) 10 nM BMPs for 0 to 120 min or (c) treated with 100 nM BMPs for 0 to 3 h.
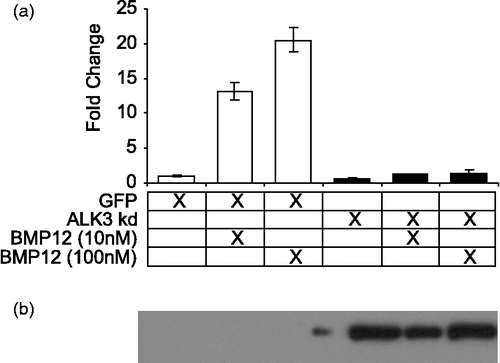
Kinase dead ALK3 expression blocks BMP12 induction of Thbs4
Thbs4 is the only tendon enriched gene that showed differential activity between rhBMP12, 13, and 2 in C3H10T1/2 cells. Expression of a kinase-dead dominant-negative type I BMPR (ALK3) completely abrogated the increased Thbs4 mRNA levels observed with rhBMP12 (). Our data also indicate that rhBMP12 and 13 can cause increased Thbs4 expression at concentrations that do not result in considerable increases in phosphorylated SMADs, but in a way that involves signaling through type I receptors. We have not yet examined other type I receptor possibilities because these cells are not known to express any other relevant receptor. On the basis of quantitative PCR analysis, C3H10T1/2 cells express high levels of Alk1, Alk2, and Alk3, but do not express detectable levels of Alk6 (Supplemental Figure 2). As the BMP ligands tested in this study do not bind ALK1, and only BMP2 binds ALK2 with very low affinity (), our efforts were focused on ALK3.
Discussion
Despite high amino acid sequence identity among BMPs 2, 12, and 13, divergent in vivo activities are observed for these ligands. Ectopic expression of rhBMP2 results in endochondral bone growth, while ectopic expression of rhBMP12 or rhBMP13 induces the formation of aligned, collagen-secreting fibroblasts. Here, we show that, despite very similar binding affinities for the same BMPRs, tenogenic and osteogenic BMPs signal through the canonical BMP SMAD pathway in different ways that result in greater levels of characteristic bone marker mRNAs, or increases in tendon marker Thbs4 mRNA. These results demonstrate the utility of Thbs4 as a specific in vitro tenogenic BMP marker that can be regulated by BMP12 and BMP13.
We have attempted to describe how BMP ligands bind the same canonical receptors but illicit distinct in vivo responses. Although our binding affinity data were only investigated biochemically, our results clearly identified differences in binding affinities between the ligands and the different receptors tested. These differences lead to hypothesis on how these ligands illicit their biological responses. It is however important to note that interaction with additional cellular receptors, cofactors, or inhibitors may alter the in vivo binding affinities.
It is possible that low levels of BMP activity are necessary to signal tendon differentiation and that high levels are required for bone formation. In support of this, treatment of cells with a relatively high dose (100 nM) of rhBMP12 or rhBMP13 can result in low Ocn mRNA levels and in the formation of endochondral bone, indicating that high levels of BMP12 or BMP13 might have an effect that is similar to comparably low levels of BMP2 (1 nM). Although bone and mRNA markers for bone are clearly detected following rhBMP12 or rhBMP13 treatment, the amount of actual bone and levels of markers for bone are relatively low compared with that observed upon rhBMP2 treatment. Moreover, ectopic implants treated with low doses of rhBMP2 do not resemble ectopic implants treated with rhBMP12 and rhBMP13 based on histological examination. Fibroblasts found in ectopic tissue treated with low doses of rhBMP2 do not display the characteristic wave-like pattern seen after ectopic expression of rhBMP12 or rhBMP13. It may be possible that the development of bone prevents the development of tendon tissue.
Expression of the tendon marker mRNA Thbs4 is relatively high shortly after treatment with low concentrations of rhBMP2, whereas treatment with higher concentrations of this ligand results in no detectible Thbs4 mRNA. As an interesting parallel, during muscle development, signaling through the BMP2 pathway can lead to decreased smooth muscle cell marker expression (Hayashi et al. Citation2006) and can block development of muscle from forelimb bud mesenchyme (Duprez et al. Citation1996; Amthor et al. Citation1998). BMP2 can also inhibit terminal differentiation of myoblasts and is capable of converting their differentiation pathway into the osteoblast lineage (Katagiri et al. Citation1994). Similarly, BMP12 and BMP13 can block myogenic differentiation, but these ligands fail to initiate osteoblast differentiation (Inada et al. Citation1996).
Our receptor binding data correlate well with others' showing that BMP2/4 and BMP12/13/GDF5 have high affinity for the type I receptors and lower affinity for the type II receptors (Nickel et al. Citation2009). Specificity may in part be due to the nature of the interaction. BMP2, BMP4, and GDF5 initially bind type I receptors followed by binding to the lower affinity type II receptors while BMP6 and BMP7 have higher affinity for the type II receptors and bind type II receptors followed by binding to type I receptors (Nishitoh et al. Citation1996; Heinecke et al. Citation2009; Nickel et al. Citation2009).
Although rhBMP2 and rhBMP12/13 bind the same BMPRs, their biological effects are very different. BMP2 signals through the canonical SMAD pathway leading to bone formation, while BMP12 and 13 induce expression of the tendon marker, Thbs4, at doses where there is little SMAD 1, 5, or 8 signaling. There are a number of possible explanations for why these ligands have different in vitro and in vivo signaling properties. Our data suggests that the tenogenic BMP12 and BMP13 have higher affinity for ALK6 compared with ALK3, while BMP2 appears to have similar affinities for ALK3 and ALK6. It is possible this slight affinity change could alter downstream SMAD signaling. It has also been suggested that slight changes in the orientation of the extracellular ligand/receptor complex formation are enough to change the conformation of the intracellular kinase domains to drastically affect downstream signals (Nickel et al. Citation2009). Additionally, BMP12 and BMP13 may interact with a different set of receptors/co-receptors that may be less than optimal for efficient downstream canonical SMAD signaling compared with that observed for BMP2, but signaling still may occur through a noncanonical pathway. BMPs are known to signal through MAP kinases and p38 in bone (Guicheux et al. Citation2003; Noth et al. Citation2003). We have not yet evaluated the potential role of this pathway, but it represents interesting potential future work. Furthermore, there may be additional unknown cofactors that could interact with ligand–receptor complexes in ways that impact canonical signaling and there may be crosstalk between canonical and noncanonical BMP signaling pathways that affects relative levels of osteogenic vs. tenogenic gene expression. Given that rhBMP2 is capable of binding ALK2 weakly, it is possible that BMP2 signaling through ALK2 results in sufficient differentiation to promote bone formation. Activating mutations in ALK2 increase SMAD phosphorylation and can lead to fibrodysplasia ossificans progressiva, a rare disabling disease characterized by heterotopic ossification (Fukuda et al. Citation2009; Kaplan et al. Citation2009). Finally, specificity maybe based on receptor expression patterns. While the exact cells that responded to BMP ligands are not known, six of the receptors (Alk1, Alk2, Alk3, Acvr2, Acvr2b and Bmpr2) are expressed in the three major musculoskeletal tissues likely to be the sight of action, bone, muscle, and tendon (Jelinsky et al. Citation2010). The expression of Alk6 is below the sensitivity of the assay (Jelinsky et al. Citation2010). Assuming that the protein expression correlates to mRNA expression, this suggests that BMP2, BMP12, and BMP13 have the capacity to signaling through ALK3 in the target tissue of interest.
We also report on the characterization of a molecular marker for monitoring the activity of rhBMPs involved in neotendon/ligament formation. The assay relies on changes in tendon-selective Thbs4 mRNA levels (Jelinsky et al. Citation2010). Thbs4 mRNA levels are high in normal tendon cells, and this marker appears to be selective for tendon tissue. Therefore, Thbs4 mRNA levels might be a good surrogate for the formation of tendon tissues. We optimized assay conditions to obtain reproducible effects on changes in Thbs4 mRNA levels, and show that the dose-dependent increase in Thbs4 mRNA is specific for BMPs that exhibit in vivo neotendon/ligament formation ability.
Additionally, we show that BMP-induced Thbs4 is blocked by overexpression of a dominant negative BMP type I receptor ALK3. Although our results suggest that ALK3 is important, inhibiting BMP12-induced Thbs4 mRNA may be the result of a number of different mechanisms including blocked receptor activity or loss of receptor specificity/selectivity due to titration of ALK3 interacting proteins (for example type II receptors). While the exact mechanism is not known, it is clear that altering BMPR-mediated signaling affects the ability of BMP12 to induce Thbs4 mRNA expression.
Related members of the BMP family are capable of affecting Thbs4 mRNA levels, but in different ways. rhBMP2 causes an increase in Thbs4 levels only at low ligand concentrations, while a dose-dependent increase in Thbs4 mRNA levels is observed with rhBMP12. As treatment with rhBMP2 showed a bell-shaped response for the induction of Thbs4 but not for other endpoints, one might expect that a larger dose range of rhBMP12 might show a similar response for induction of Thbs4. However, BMP12 is relatively insoluble and higher concentrations than that reported here result in an insoluble protein fraction which is predicted to decrease activity on all end points.
We also note that under our culture conditions, these cells are not differentiating into tendon cells and little change in cell morphology is observed. Other tendon-associated markers including Scx (scleraxis), Col1a1, Six1, and Six2 are not induced under these conditions (data not shown). Thbs4, as a marker that is highly and selectively expressed in tendon cells, is essential for dissecting the different functions of various BMP ligands. In particular, for the tenogenic BMPs, Thbs4 mRNA levels provide a more sensitive way to assess BMP activity compared with the BRE-luciferase reporter assay, which may be limited to detecting canonical BMP signaling through SMAD activation. Our work compared the characterization of in vivo ectopic gene expression and cultured cell lines. We note that many parallels exist in genes expressed in both systems but many differences exist. We have used a cell culture system to begin to elucidate distinct mechanism of activity between the different BMP molecules and to use an easily cultured cell line to accurately measure activity. More work is needed to determine if these differences exist in vivo as well as to determine the exact cell type that responds to BMPs in vivo.
Most established in vitro BMP assays monitor bone related formation activity, such as increases in ALP activity and Ocn mRNA levels. While the tenogenic BMPs studied in this work impact levels of ALP activity and Ocn mRNA levels, their activity is considerably reduced compared with true osteogenic BMP family members such as BMP2. Rat osteosarcoma cell ALP activity increases by 50% following treatment with BMP12 (Furuya et al. Citation1999). However, in C3H10T1/2 cells, the increases in ALP activity resulting from rhBMP12 and rhBMP13 treatments are 100-fold less than the activity induced by rhBMP2, and this is consistent with previous work in C3H10T1/2 and C2C12 cells using recombinant adenovirus (Luu et al. Citation2007). Measuring changes in Thbs4 mRNA levels represents a much more sensitive and specific way to measure tenogenic activity from BMPs compared with other established BMP-related assays. This assay has the potential to be used to supplement traditional methods in screening for tenogenic activity and in evaluating specific functions of different BMPs. The utility of Thbs4 mRNA levels in the study of tendon development may be equal to, or greater than, the utility of Ocn in studies of bone development due to the tissue specificity and sensitivity that we describe.
Supplementary Material
Download PDF (643.6 KB)Supplementary Material
Download PDF (528.5 KB)Acknowledgements
The authors thank Donna Gavin for histological processing. The authors thank Clifford Entrican and Stephen Raso for the circular dichroism spectroscopy analysis.
Declaration of interest: Financial support of this work was provided by Wyeth/Pfizer Research. All authors were employees of Wyeth/Pfizer at the time this research was conducted. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Amthor H, Christ B, Weil M, Patel K. 1998. The importance of timing differentiation during limb muscle development. Curr Biol. 8:642–652.
- Aspenberg P, Forslund C. 1999. Enhanced tendon healing with GDF 5 and 6. Acta Orthop Scand. 70:51–54.
- Bolt P, Clerk AN, Luu HH, Kang Q, Kummer JL, Deng ZL, Olson K, Primus F, Montag AG, He TC, Haydon RC, Toolan BC. 2007. BMP-14 gene therapy increases tendon tensile strength in a rat model of Achilles tendon injury. J Bone Joint Surg Am. 89:1315–1320.
- Chang SC, Hoang B, Thomas JT, Vukicevic S, Luyten FP, Ryba NJ, Kozak CA, Reddi AH, Moos MJr. 1994. Cartilage-derived morphogenetic proteins. New members of the transforming growth factor-beta superfamily predominantly expressed in long bones during human embryonic development. J Biol Chem. 269:28227–28234.
- Cook SD, Rueger DC. Osteogenic protein-1: Biology and applications. Clin Orthop Relat Res. 199629–38.
- Duprez DM, Coltey M, Amthor H, Brickell PM, Tickle C. 1996. Bone morphogenetic protein-2 (BMP-2) inhibits muscle development and promotes cartilage formation in chick limb bud cultures. Dev Biol. 174:448–452.
- Erlacher L, McCartney J, Piek E, ten Dijke P, Yanagishita M, Oppermann H, Luyten FP. 1998. Cartilage-derived morphogenetic proteins and osteogenic protein-1 differentially regulate osteogenesis. J Bone Miner Res. 13:383–392.
- Fukuda T, Kohda M, Kanomata K, Nojima J, Nakamura A, Kamizono J, Noguchi Y, Iwakiri K, Kondo T, Kurose J, Endo K, Awakura T, Fukushi J, Nakashima Y, Chiyonobu T, Kawara A, Nishida Y, Wada I, Akita M, Komori T, Nakayama K, Nanba A, Maruki Y, Yoda T, Tomoda H, Yu PB, Shore EM, Kaplan FS, Miyazono K, Matsuoka M, Ikebuchi K, Ohtake A, Oda H, Jimi E, Owan I, Okazaki Y, Katagiri T. 2009. Constitutively activated ALK2 and increased SMAD1/5 cooperatively induce bone morphogenetic protein signaling in fibrodysplasia ossificans progressiva. J Biol Chem. 284:7149–7156.
- Furuya K, Nifuji A, Rosen V, Noda M. 1999. Effects of GDF7/BMP12 on proliferation and alkaline phosphatase expression in rat osteoblastic osteosarcoma ROS 17/2.8 cells. J Cell Biochem. 72:177–180.
- Guicheux J, Lemonnier J, Ghayor C, Suzuki A, Palmer G, Caverzasio J. 2003. Activation of p38 mitogen-activated protein kinase and c-Jun-NH2-terminal kinase by BMP-2 and their implication in the stimulation of osteoblastic cell differentiation. J Bone Miner Res. 18:2060–2068.
- Gupta IR, Macias-Silva M, Kim S, Zhou X, Piscione TD, Whiteside C, Wrana JL, Rosenblum ND. 2000. BMP-2/ALK3 and HGF signal in parallel to regulate renal collecting duct morphogenesis. J Cell Sci. 113 Pt 2:269–278.
- Hayashi K, Nakamura S, Nishida W, Sobue K. 2006. Bone morphogenetic protein-induced MSX1 and MSX2 inhibit myocardin-dependent smooth muscle gene transcription. Mol Cell Biol. 26:9456–9470.
- Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, Nickel J. 2009. Receptor oligomerization and beyond: A case study in bone morphogenetic proteins. BMC Biol. 7:59.
- Inada M, Katagiri T, Akiyama S, Namiki M, Komaki M, Yamaguchi A, Kamoi K, Rosen V, Suda T. 1996. Bone morphogenetic protein-12 and -13 inhibit terminal differentiation of myoblasts, but do not induce their differentiation into osteoblasts. Biochem Biophys Res Commun. 222:317–322.
- Jahng TA, Fu TS, Cunningham BW, Dmitriev AE, Kim DH. 2004. Endoscopic instrumented posterolateral lumbar fusion with Healos and recombinant human growth/differentiation factor-5. Neurosurgery. 54:171–180 discussion 180–171.
- Jelinsky SA, Archambault J, Li L, Seeherman H. 2010. Tendon-selective genes identified from rat and human musculoskeletal tissues. J Orthop Res. 28:289–297.
- Kaplan FS, Xu M, Seemann P, Connor JM, Glaser DL, Carroll L, Delai P, Fastnacht-Urban E, Forman SJ, Gillessen-Kaesbach G, Hoover-Fong J, Koster B, Pauli RM, Reardon W, Zaidi SA, Zasloff M, Morhart R, Mundlos S, Groppe J, Shore EM. 2009. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum Mutat. 30:379–390.
- Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. 1994. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 127:1755–1766.
- Korchynskyi O, ten Dijke P. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 277:4883–4891.
- Lou J, Tu Y, Burns M, Silva MJ, Manske P. 2001. BMP-12 gene transfer augmentation of lacerated tendon repair. J Orthop Res. 19:1199–1202.
- Luu HH, Song WX, Luo X, Manning D, Luo J, Deng ZL, Sharff KA, Montag AG, Haydon RC, He TC. 2007. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res. 25:665–677.
- Mazerbourg S, Sangkuhl K, Luo CW, Sudo S, Klein C, Hsueh AJ. 2005. Identification of receptors and signaling pathways for orphan bone morphogenetic protein/growth differentiation factor ligands based on genomic analyses. J Biol Chem. 280:32122–32132.
- Nickel J, Sebald W, Groppe JC, Mueller TD. 2009. Intricacies of BMP receptor assembly. Cytokine Growth Factor Rev. 20:367–377.
- Nishitoh H, Ichijo H, Kimura M, Matsumoto T, Makishima F, Yamaguchi A, Yamashita H, Enomoto S, Miyazono K. 1996. Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor-5. J Biol Chem. 271:21345–21352.
- Nohe A, Keating E, Knaus P, Petersen NO. 2004. Signal transduction of bone morphogenetic protein receptors. Cell Signal. 16:291–299.
- Noth U, Tuli R, Seghatoleslami R, Howard M, Shah A, Hall DJ, Hickok NJ, Tuan RS. 2003. Activation of p38 and Smads mediates BMP-2 effects on human trabecular bone-derived osteoblasts. Exp Cell Res. 291:201–211.
- Posthumus M, Collins M, Cook J, Handley CJ, Ribbans WJ, Smith RK, Schwellnus MP, Raleigh SM. 2010. Components of the transforming growth factor-beta family and the pathogenesis of human Achilles tendon pathology—a genetic association study. Rheumatology (Oxford). 49:2090–2097.
- Reddi AH. 1998. Role of morphogenetic proteins in skeletal tissue engineering and regeneration. Nat Biotechnol. 16:247–252.
- Rosen V, Wozney JM. 2002. Bone morphogenetic proteins. In: Bilezikian JP, Raisz LG, Rodan GA. editors. Principles of bone biology. San Diego, CA: Academic Press. p 919–928.
- Seeherman HJ, Archambault JM, Rodeo SA, Turner AS, Zekas L, D'Augusta D, Li XJ, Smith E, Wozney JM. 2008. rhBMP-12 accelerates healing of rotator cuff repairs in a sheep model. J Bone Joint Surg Am. 90:2206–2219.
- Thies RS, Bauduy M, Ashton BA, Kurtzberg L, Wozney JM, Rosen V. 1992. Recombinant human bone morphogenetic protein-2 induces osteoblastic differentiation in W-20-17 stromal cells. Endocrinology. 130:1318–1324.
- Walsh AJ, Bradford DS, Lotz JC. 2004. In vivo growth factor treatment of degenerated intervertebral discs. Spine (Phila Pa 1976). 29:156–163.
- Wang EA, Rosen V, D'Alessandro JS, Bauduy M, Cordes P, Harada T, Israel DI, Hewick RM, Kerns KM, LaPan P, . 1990. Recombinant human bone morphogenetic protein induces bone formation. Proc Natl Acad Sci USA. 87:2220–2224.
- Wolfman NM, Hattersley G, Cox K, Celeste AJ, Nelson R, Yamaji N, Dube JL, DiBlasio-Smith E, Nove J, Song JJ, Wozney JM, Rosen V. 1997. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5. 6, and 7, members of the TGF-beta gene family. J Clin Invest. 100:321–330.
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. 1988. Novel regulators of bone formation: Molecular clones and activities. Science. 242:1528–1534.