Abstract
Purpose: Triplex-forming oligonucleotides (TFO) bind to the DNA double helix in a sequence-specific manner. Therefore, TFO seem to be a suitable carrier for Auger electron emitters to damage exclusively targeted DNA sequences, e.g., in tumor cells. We studied the influence of I-125 labeled TFO with regard to cell survival and induction of DNA double-strand breaks (DSB) using TFO with different genomic targets and target numbers. Furthermore, the ability of TFO to alter the gene expression of targeted genes was examined.
Materials and methods: TFO were labeled with I-125 using the primer extension method. DNA triplex formation and sequence-specific DSB were demonstrated in vitro. Cell survival was analyzed by colony-forming assay and DNA damage was assessed by microscopic quantification of protein 53 binding protein 1 (53BP1) foci in the human squamous carcinoma cell line II (SCL-II). Quantitative real-time polymerase-chain-reaction (qRT-PCR) was performed to analyze gene expression alterations.
Results: The sequence-specific induction of a single DSB in a 1695 bp long DNA double stranded fragment was demonstrated in vitro. I-125-labeled TFO binding to single and multiple targets were shown to induce a pronounced decrease in cell survival and an increase of DSB. TFO targeting multiple sites differing in the total target number showed a significant different cell killing per decay that is also in good accordance with the observed induction of DSB. Single gene targeting I-125-labeled TFO significantly decreased cell survival and altered gene expression in the targeted gene.
Conclusions: I-125-labeled TFO enable specific targeting of DNA in vitro as well as in a cellular environment and thus induce sequence-specific complex DNA lesions. Therefore I-125-labeled TFO might be a very useful tool for basic DNA repair research.
Introduction
Triplex-forming oligonucleotides (TFO) are short oligodeoxyribonucleotides able to bind complementary DNA sequences in a sequence-specific manner. They are major groove-binding ligands, which are associated with the DNA duplex via hydrogen bonds known as Hoogsteen or reverse Hoogsteen hydrogen bonds. Triplexes can only be formed with homopurine-homopyrimidine regions of the DNA (Frank-Kamenetskii and Mirkin Citation1995, Panyutin and Neumann Citation1996). The orientation to the target DNA duplex can be parallel and antiparallel depending on the base composition of the TFO. While the bases thymine, cytosine and guanine are able to bind in both orientations, triplexes with adenine as the third base can only form a triplex in antiparallel conformation. TFO containing cytosine form triplexes under acidic conditions as the cytosine has to be protonated (Praseuth et al. Citation1988). Regions containing homopurine-homopyrimidine sequences are quite common in the human genome, as has been demonstrated by Goni et al. (Citation2004). The presence of a third strand in the major groove can influence the conformation of the DNA, changing its ability to recognize specific proteins and consequently altering the mechanisms controlling DNA function (Kuznetsova et al. Citation1999). Due to their sequence-specific DNA binding capacity, TFO are regarded as promising molecular carriers in biotechnological and biomedical applications (Vekhoff et al. Citation2008).
During the decay of I-125, a shower of low-energy Auger electrons is emitted leading to a high energy deposition in a rather small volume (Pomplun et al. Citation1987). Panyutin and Neumann (Citation1994) showed that I-125-labeled TFO binding to DNA lead to a significant increase of DNA double-strand breaks (DSB). DNA DSB can be visualized by antibody staining of the protein 53 binding protein 1 (53BP1), which is an early participant in the cellular response to DNA DSB (Schultz et al. Citation2000).
As the energy deposition of the Auger electrons is limited to a 10–100 nm sphere at the decay site, the damage to the cellular environment will be only minor, even at a high activity of I-125, as long as the Auger emitter is not in the close vicinity of a radiation-sensitive target (Kassis et al. Citation1996, Lobachevsky and Martin Citation1996). In contrast, the decay of I-125 close to the DNA will induce significant damage, which is difficult to repair (Kassis et al. Citation1996, Lobachevsky and Martin Citation1996, Sedelnikova et al. Citation1998). Referring to these characteristics TFO can serve as a tool to silence genes very specifically without necessarily killing the cell itself. This application could be useful for antigene radiotherapy (Panyutin et al. Citation2003), which is the generic term for damaging selected genes by a high dose of radiation from radionuclides delivered to this gene by specific DNA-binding molecules.
The sequence specificity of TFO, the biological effectiveness of TFO labeled with Auger electron emitter binding to different targets and target numbers as well as the response of the targeted genes have still not yet been fully characterized. To further elucidate the biological effectiveness of I-125-labeled TFO in vitro, we used two different approaches. We examined the reduction of cell survival and DSB induction of a I-125-labeled multi-binding site (MBS) TFO with multiple targets (∼7000 targets) in the human genome (Sedelnikova et al. Citation2004) in comparison to a single-binding site (SBS) I-125-labeled TFO targeting the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene and a I-125-labeled TFO, which also binds to multiple targets, but as a part of a quadruplex structure in guanine-rich sequences (Onyshchenko et al. Citation2011). Based on a genome-wide survey, Todd et al. (Citation2005) identified 375,157 putative quadruplex sequences (PQS) in the human genome, although not all of them form quadruplexes in vivo. Huppert and Balasubramanian (Citation2007) identified 14,769 PQS in the promoter region of the 19,268 known human genes. Additionally, they observed an enrichment of such structures in the promoter region and evidence is emerging that guanine quadruplex motifs play a role in gene regulation (Verma et al. Citation2009, Kumar et al. Citation2011). In a further approach, we analyzed the general ability of TFO to alter the gene expression of targeted genes employing quantitative real-time polymerase-chain-reaction (qRT-PCR).
Materials and methods
Oligonucleotides and TFO labeling
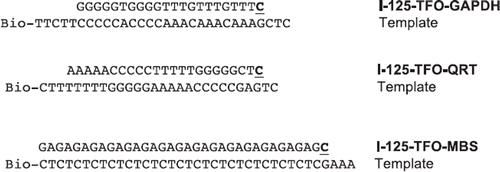
TFO and biotinylated oligonucleotides were designed employing TFO Target Sequence Search (http://spi.mdanderson.org/tfo/about.php) and synthesized by Metabion (Martinsried, Germany) in standard quality and are displayed in . The sequences of the I-125-labeled TFO (I-125-TFO-GAPDH, I-125-TFO-QRT and I-125-TFO-MBS) and the templates used for the primer extension are shown in . I-125-TFO-GAPDH binds specifically to a single target DNA sequence in the human GAPDH gene whereas I-125-TFO-QRT and I-125-TFO-MBS both bind to multiple targets. Labeling of TFO with I-125 was performed employing the primer extension method (Panyutin and Neumann Citation1996). Therefore the preTFO, which are the TFO used for the primer extension reaction and the biotinylated template (0.5 pmol each), were annealed in Klenow buffer (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM dithiothreitol (DTT)) for 10 min incubation at 95°C followed by slow cooling (1°C/min) to room temperature. Primer extension reaction was carried out in the presence of ∼2.5 MBq I-125-deoxycytidine triphosphate (I-125-dCTP) (Hartmann Analytic GmbH, Braunschweig, Germany) by 2.5 units of exonuclease-free Klenow fragment (Fermentas, St. Leon-Rot, Germany) in a total volume of 10 μl. After 15 min at 37°C, the reaction was stopped by heating to 75°C for 10 min. The duplex-DNA product was resuspended in TEN100 binding buffer (10 mM Tris-HCl, pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA), 100 mM NaCl). The labeled duplex DNA was bound to Streptavidin Magnetic Particles (Roche, Mannheim, Germany) by incubation for 15 min at room temperature followed by two washing steps with TEN1000 wash buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 M NaCl) and deionized water. The magnetic particles were resuspended in 20 μl 0.15 M NaOH and incubated for 15 min at room temperature to denature the DNA duplex. The magnetic particles were then removed with a magnet and the supernatant containing the I-125-TFO was collected. A further 2.2 μl of 10 × TFO binding buffer (100 mM MgCl2, 100 mM Tris-HCl, pH 5.8) and 2 μl 1.25 M acetic acid were added. In this step, the amount of incorporated I-125-deoxycytidine (I-125-dC) was determined by activity measurement using a 1480 automatic gamma counter (PerkinElmer, Rodgau, Germany). Labeled TFO were stored at −80°C for further use.
In vitro DNA triplex formation and binding assay
For in vitro verification of triplex formation, 2 pmole of specific target DNA fragments were mixed with 24 pmole of unlabeled TFO () in TFO binding buffer B (10 mM MgCl2, 10 mM Tris-HCl, pH 5.8, 16 μM coralyne chloride) and incubated at 37°C for 24 h. The target fragments, synthesized by Metabion, were exact duplicates of the target regions in the genome, containing six additional base pairs (bp) up- and downstream of the target region. Analysis was performed via electrophoretic mobility shift assay (EMSA). Aliquots of the samples were loaded on a 12% native polyacrylamide gel containing 10 mM MgCl2 and run at 150 V for 20 h in TBE buffer (89 mM Tris-Base, pH 8.0, 89 mM boric acid, 2 mM EDTA). The gels were subsequently silver-stained for visualization. Triplex formation was detected by band shift in the silver-stained gels.
Table I. Nucleotide sequence of investigated TFO and corresponding target sequence. Asterisks (*) indicate that no triplex formation could be detected for these TFO by EMSA.
DNA double-strand break analysis
Approximately 70 kBq of I-125-labeled TFO-GAPDH () (64 kBq/pmole) was added to 10 μg of a 1695 bp long DNA double-stranded target fragment in TFO binding buffer B (10 mM MgCl2, 10 mM Tris-HCl, pH 5.8, 16 μM coralyne chloride) to make up a total volume of 20 μl. The target fragment was a PCR amplificate from human squamous carcinoma cell line II (SCL-II) cell DNA isolates containing the TFO target sequence. The mixture was incubated at 37°C for 24 h and stored for at least 30 days (0.5 half-life of I-125) at −80°C for decay accumulation. Samples were thawed after accumulation of decays and electrophoresed in a 1% agarose gel using standard conditions. Gels were stained with ethidium bromide (EtBr) and transferred to a Magna Nylon Transfer Membrane (GE Water and Process Technology, Herentals, Belgium) using a downward capillary transfer blot in 20 × SSC transfer buffer (3 M NaCl, 0.3 M Na3C6H5O7, pH 7.0). Before transfer, the gels were washed twice in denaturation solution (1.5 M NaCl, 0.5 M NaOH) and neutralization solution (1.5 M NaCl, 0.5 M Tris-Cl, pH 7.0). The membrane was hybridized with P-32 labeled PCR-amplified fragments of the up- and downstream TFO target region (DecaLabel DNA labeling Kit, Fermentas) at 65°C for 20 h in CHURCH buffer (0.7 M Na2HPO4, 0.3 M NaH2PO4, pH 7.2, 0.5 M EDTA, 0.1% BSA, 0.7% sodium dodecyl sulfate [SDS]). Hybridization was followed by one washing step in washing solution 1 (2 × SSC Buffer) for 10 min, one washing step in washing solution 2 (2 × SSC Buffer + 0.1% SDS) for 10 min and two washing steps in washing solution 3 (0.1 SSC buffer + 0.1% SDS) for 20 min. All steps were carried out at 65°C. Visualization was done by exposure to MS imaging plates BAS-MS3543 and analyzed on a FLA-5000 imaging system (Fujifilm).
Cell line and culture conditions
SCL-II cells (Tilgen et al. Citation1983) were grown in minimum essential medium Eagle (MEM, PAA Laboratories GmbH, Cölbe, Germany) with L-glutamine, supplemented with 16% fetal bovine serum (FBS, PAA Laboratories GmbH) in a water-saturated atmosphere of 5% CO2 at 37°C as previously described (Kriehuber et al. Citation2004) and cultivated under standard conditions.
TFO transfection experiments
SCL-II cells were grown in cell culture flasks (PAA Laboratories GmbH) to 70–80% confluency, trypsinized and resuspended in MEM. The required number of cells (2 × 106) was centrifuged for 10 min at 90×g and resuspended in 100 μl Nucleofector Solution R (Lonza AG, Cologne, Germany). I-125-labeled TFO (∼70 kBq) was added and the electroporation reaction was performed on the Amaxa Nucleofector Device I (Lonza AG) employing transfection program L-013. Immediately after the reaction, 500 μl of MEM was added and the cell solution was transferred into a T25 cell culture flask (PAA Laboratories GmbH) and incubated at 37°C for 20–23 h prior to storage at −150°C.
Colony-forming assay
After the transfection procedure (previous section), the cells were washed twice in phosphate buffered saline (PBS, PAA Laboratories GmbH), trypsinized and aliquoted (2 × 105 cells each) in freezer vials (VWR International, Darmstadt, Germany) and stored in freezing medium (FBS + 10% dimethyl sulfoxide) for decay accumulation at −150°C. Prior to storage, the decays per sample were determined by gamma counting (1480 Gamma Counter, Perkin Elmer). After thawing, the cells were analyzed with respect to their colony-forming ability. For this purpose, the appropriate number of cells (500 cells/well) was plated in a six-well plate (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and incubated for 12 days. The cell culture medium was renewed after six days. For visualization, the cells were fixed (10 min, 100% MeOH) and Giemsa stained. Each colony consisting of 50 or more cells was counted as a survivor and the total number of colonies divided by the total number of colonies of controls was defined as the survival fraction. Finally, the survival fraction was plotted against the accumulated decays per cell.
The 53BP1 foci assay
SCL-II cells were treated for decay accumulation as already described (previous section). After thawing, an appropriate number of cells (5 × 104 cells/well) were plated in six-well plates and grown on sterilized glass coverslips (Menzel GmbH, Braunschweig, Germany) for 20–23 h under standard cell culture conditions. Afterwards cells were rinsed three times with PBS (PAA Laboratories GmbH), fixed with 4% paraformaldehyde for 20 min at room temperature and rinsed twice with TBP buffer (0.2% Triton X-100, 1% bovine serum albumin in PBS). Cover slips were then immersed with 1% Triton X-100 for 20 min at room temperature and rinsed twice with TBP buffer. Blocking was done in 5% goat serum for 1 h at room temperature followed by two washing steps with TBP buffer. The primary antibody (53BP1, H-300: sc-22760, Santa Cruz Biotechnology, Heidelberg, Germany) was added in a 500-fold dilution for 1 h at room temperature followed by three washing steps with TBP buffer and incubation for 45 min at room temperature with the secondary antibody (1:700, goat anti-rabbit immunoglobulin G-texas red (IgG-TR): sc-2780, Santa Cruz Biotechnology). Cover slips with cells were then washed twice in TBP buffer, once in PBS (PAA Laboratories GmbH) and finally rinsed in deionized water. For microscopy, the monolayers were mounted with ProLong Goldantifade reagent with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen, Darmstadt, Germany) on glass slides. Analysis was performed on a Zeiss LSM700 Observer Z1 microscope (Zeiss, Göttingen, Germany). For each sample, 100 randomly selected cells were analyzed by eye and the average number of foci was calculated.
Relative gene expression analysis
After TFO transfection, cells were trypsinized, aliquoted in freezer vials (2 × 106 cells each) and stored for accumulation of decays at −150°C. After decay accumulation, cells were thawed, incubated for 20–23 h at 37°C and RNA isolation was performed using the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany). For qRT-PCR, the RNA concentration per sample was adjusted to 30 ng/μl and qRT-PCR was performed on a 7500 real time PCR System (Applied Biosystems, Darmstadt, Germany) using the Power SYBR® Green RNA-t-CT™ 1-Step Kit (Applied Biosystems) following the manufacturer's guidelines. Primers were designed for GAPDH as the gene containing the TFO target and beta-ACTIN as a housekeeping gene for quantification purpose.
Results
In vitro DNA triplex formation and binding assay
A variety of oligonucleotides were analyzed for their ability to form triplexes with their specific DNA target in vitro by the EMSA.
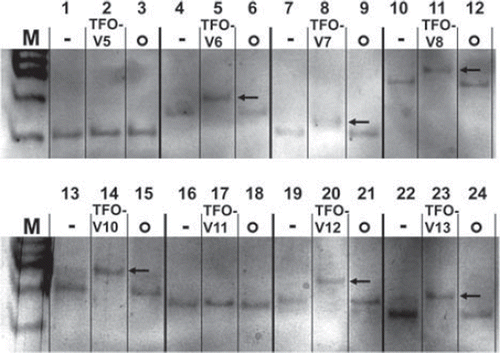
Of all the tested oligonucleotides having a suitable sequence to form a DNA triplex with their specific DNA targets, about 43% failed to form a DNA triplex in the EMSA in vitro (data not shown). Successful DNA triplex formation was indicated by a clear band shift of the target DNA/TFO construct (, lane 5, 8, 11, 14, 20 and 23) when compared to the target sequence only (, lane 1, 4, 7, 10, 13, 16, 19 and 22). DNA triplex formation in vitro can therefore not be predicted according to TFO and DNA target sequence only. However, all studied non-complementary TFO (non-sense TFO) failed, as expected, to form DNA triplexes (, lane 3, 6, 9, 12, 15, 18, 21 and 24).
Figure 2. Triplex formation in vitro visualized with electrophoretic mobility shift assay. Negative controls containing the specific TFO target fragment only, -; Triplex-forming oligonucleotides with their specific target sequences, TFO-V(x); Non-sense samples containing the target sequence with a non-complementary TFO, o; A band shift reflects DNA triplex formation, ![]()
DNA double-strand break analysis
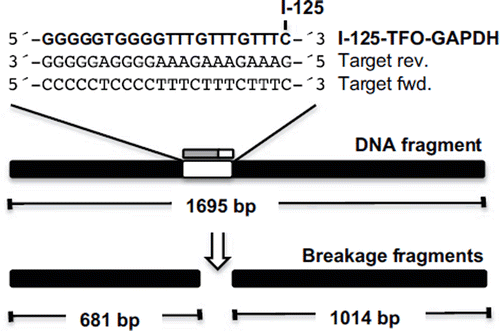
The target binding site and induced breakage fragments of I-125-TFO-GAPDH are shown in . The results of the DSB analysis revealed that the I-125-TFO-GAPDH induced a single DSB in the 1695 bp target fragment (, ![]() ) resulting in two expected breakage fragments of 1024 bp and 681 bp (,
) resulting in two expected breakage fragments of 1024 bp and 681 bp (, ![]() ). No DNA breakage whatsoever was observed for the non-binding, non-specific, non-sense I-125-labeled TFO (, lane 2). Southern blot analysis (), using probes which were P-32-labeled PCR amplificates of the region up- and downstream of the TFO target site, verified that the breakage fragments observed in the ethidium bromide stained agarose gels originated from the target fragment (, lane 1).
). No DNA breakage whatsoever was observed for the non-binding, non-specific, non-sense I-125-labeled TFO (, lane 2). Southern blot analysis (), using probes which were P-32-labeled PCR amplificates of the region up- and downstream of the TFO target site, verified that the breakage fragments observed in the ethidium bromide stained agarose gels originated from the target fragment (, lane 1).
Figure 3. Schematic diagram of the 1695 bp long DNA fragment containing the target sequence for TFO-GAPDH and the two expected breakage fragments of 681 bp and 1024 bp length. The TFO is I-125-labeled at the 3′-terminal cytosine and binds to the polypurine target sequence in reverse orientation.
The breakage efficiency in the target fragment was calculated as the ratio of the sum of the EtBr band intensities of the two breakage fragments to the sum of the intensities of all three fragments (Sedelnikova et al. Citation2001) leading to a percentage of breaks of ∼40% after 143 days of decay accumulation (∼ 1.5 × 109 decays/ng target DNA).
Colony-forming assay
The colony-forming assay (CFA) in SCL-II cells showed reduced cell survival with respect to accumulated decays/cell for all three I-125-labeled TFO investigated (). The survival data of all three I-125-labeled TFO were best fitted by using the straight exponential model of cell survival. The survival curve of I-125-TFO-GAPDH and the I-125-TFO-MBS showed a D37 (radiation dose at which approximately 63% of the starting entities have acquired the specified damage) value of 347 and 390 accumulated decays/cell, respectively, although I-125-TFO-GAPDH binds to a single site in the GAPDH gene only and the I-125-TFO-MBS binds to ∼7000 target sites in the genome. However, I-125-TFO-QRT, which binds to multiple target sequences in the human genome as well, induced a much more pronounced reduction of cell survival, showing a D37 value of 223 accumulated decays/cell, resulting in an almost 2-fold increase of cell killing when compared to I-125-TFO-GAPDH and I-125-TFO-MBS.
The 53BP1 foci assay
DSB caused by I-125-TFO-MBS, I-125-TFO-QRT and I-125-TFO-GAPDH were visualized and quantified using the 53BP1 foci assay (). Cells transfected with the corresponding unlabeled TFO served as negative controls. The negative controls showed on average 0.8 foci/cell. Transfection with the single-binding site I-125-TFO-GAPDH led to a linear increase of foci signals reaching ∼ 3 foci/cell at approximately 480 accumulated decays/cell. The I-125-TFO-MBS transfected cells showed a very similar increase in foci numbers of ∼ 3 foci/cell at approximately 450 accumulated decays/cell. In contrast, I-125-TFO-QRT transfected cells showed a more pronounced increase of foci per decay with ∼ 3 foci/cell at ∼ 300 accumulated decays/cell resulting in a 1.5-fold stronger foci induction per decay.
Relative gene expression analysis
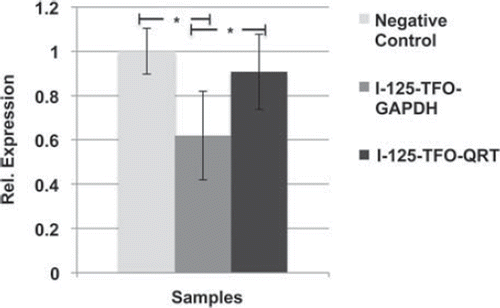
The relative GAPDH expression showed a significant down-regulation in I-125-TFO-GAPDH transfected SCL-II cells in comparison to the negative control cells which were transfected with non-labeled TFO-GAPDH or I-125-TFO-QRT transfected cells ().
Compared to the negative control, the I-125-TFO-GAPDH transfected cells showed a 0.6-fold reduction in GAPDH expression. Compared to I-125-TFO-QRT, the I-125-TFO-GAPDH transfected cells showed a 0.7-fold reduced GAPDH expression. In contrast, the I-125-TFO-QRT transfected cells displayed a non-significant 0.9-fold reduced GAPDH expression when compared to the negative control.
Discussion
The sequence of all investigated TFO, except the two multi-binding site TFO, was determined with the help of the TFO target sequence search tool provided by http://spi.mdanderson.org/tfo/about.php. Although all TFO were designed according to the following special criteria of a minimum length of 15 bp, a maximum guanine content of 50%, no pyrimidine interrupts allowed, and a default setting for TFO-target binding at near-physiological conditions (pH 7.2, 37°C, 10 mM MgCl2), only 43% of the TFO were found to form DNA triplexes in vitro. Therefore, we strongly recommend confirming DNA triplex formation of every TFO used with its specific target sequence by EMSA in vitro.
The site-directed and sequence-specific induction of DSB in a DNA target sequence, with a breakage percentage of 40% after 143 days (∼ 1.5 × 109 decays/ng target DNA) as shown in is in good agreement with the results of Sedelnikova et al. (Citation2001), who described a percentage of 50% TFO-induced DSB in plasmid and genomic DNA after 136 days of decay accumulation. Additionally, we showed that no DSB occur at similar accumulated decays when the target DNA is exposed to non-specific I-125 labeled TFO (; lane 2), demonstrating that the DNA damage caused by non-DNA bound I-125 can be neglected. Based on the in vitro results, we conclude that Auger electron emitters (AEE) exhibit their DNA-damaging potential only when bound or attached closely to the DNA target. Moreover, the in vitro results prove that the AEE labeled TFO specifically induce DSB exclusively at the targeted site in the DNA target molecule.
The single-target site-specific I-125-TFO-GAPDH displayed a pronounced cytotoxic effect in the CFA. Interestingly, the D37 value of ∼ 350 decays/cell was very similar to the data observed for I-125-TFO-MBS transfected cells, a TFO which binds to multiple targets of approximately ∼ 7000 sites in the human genome. This probably reflects the biological importance of the selected target gene and suggests that targeting one single essential target in the genome can be at least as cytotoxic as targeting multiple, but non-essential targets. This hypothesis is underlined by the work of Sedelnikova et al. (Citation2004), who compared the cytotoxicity of a multi-binding TFO to a single binding site TFO. Sedelnikova et al. (Citation2004) used a single-binding site TFO in the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene, which is part of the purine recycling pathway in human cells (Stout and Caskey Citation1985) and which was previously shown to be a nonessential enzyme for cells growing in culture (Stout and Caskey Citation1985). By comparing the non-essential single-binding site TFO-HPRT to a multiple- binding site TFO, Sedelnikova et al. (Citation2004) observed a 1.7 times higher cytotoxicity of the multi-binding TFO. In contrast, the observed strong cytotoxicity of the TFO-GAPDH in our study might be explained by the crucial importance of GAPDH in glycolysis (Alberts Citation2010), which is essential for growing cells in cell culture. This leads to the conclusion that target quality plays an important role for the cytotoxicity of a TFO. However, I-125-TFO-QRT with a D37 value of ∼ 220 decays/cell showed the highest cytotoxicity displaying an almost twice as high cytotoxicity per decay when compared to TFO-GAPDH and TFO-MBS. This particular TFO possesses the highest target number in the genome of all three investigated TFO, with more than ∼ 370,000 putative target sequences overall, and ∼ 14,700 located in promoter and regulatory regions (Todd et al. Citation2005, Huppert and Balasubramanian Citation2007). Therefore, the question arises whether it is the target quantity or the target quality that determines the observed cytotoxicity. The obtained results allow conclusions in both directions. It could be shown that a single binding TFO (TFO-GAPDH) can be as cytotoxic as a multiple binding TFO (TFO-MBS), which is probably due to the target quality. However, the TFO-QRT, which possesses the highest target number of all three investigated TFO, induced the most pronounced cytotoxic effect per decay, which in turn indicates that the target quantity might be of major importance. Taken into consideration that I-125-TFO-QRT induced almost twice as much 53BP1 foci per decay when compared to TFO-GAPDH and TFO-MBS it is highly probable that the amount of targets and hence induced DSB per decay is most relevant for cell killing rather than the quality of the target. It should also be taken into account that I-125-TFO-QRT was not originally designed to form DNA triplexes but as an oligonucleotide with a high probability of forming secondary structures caused by the guanine motif contained in it (Onyshchenko et al. Citation2011). It is well known that numerous genes possess guanine-rich sequences in promoter regions as well as in other regulatory and non-regulatory structures of the genomic DNA, which enables them to form various secondary structures, including guanine quadruplexes (Onyshchenko et al. Citation2011). We conclude therefore that I-125-TFO-QRT binds within regions as a part of a quadruplex structure. The observed strong cytotoxicity can by explained by multiple binding and disruption of these sequences, leading to the enhanced cytotoxic effect of TFO-QRT. Longe et al. (Citation2009) and Forsha et al. (Citation2010) found that non-labeled oligonucleotides capable of forming intramolecular G-quadruplex structures were also able to cause significant cell death and cell cycle arrest in transfected cells. The question thus arises of whether the effect found in the present work is due to the oligonucleotide (TFO) itself or the I-125 brought in close proximity to the DNA. Since a pronounced decay-dependent effect could be shown, we have reason to believe that the observed cytotoxicity is caused by the decay of the I-125 rather than by the carrier TFO alone.
Additionally, it has to be stated that the overall cytotoxicity of the TFO in the present work (D37 of ∼ 220 and ∼ 390 decays/cell) is about 40-times higher compared to the results in the work of Sedelnikova et al. (Citation2004) reporting a D37 of ∼ 8400 and ∼ 14,200 decays/cell. We assume that this effect is caused by the different transfection methods that were used. In the present work, transfection was performed with the Nucleofector system (Lonza AG), which allows immediate nucleic acid delivery directly into the nucleus, whereas the oligofectamine (Invitrogen)-based transfection used by Sedelnikova et al. (Citation2004) is limited to cytoplasmic delivery and depends on cellular mechanisms for subsequent TFO transfer into the nucleus. To achieve comparable TFO concentration in the nucleus, the overall amount of labeled TFO transferred into the cells has to be considerably higher when using oligofectamine (Invitrogen)-based transfection compared to the Nucleofector-based transfection system.
DNA DSB were quantified using the 53BP1 foci assay. The single binding I-125-TFO-GAPDH transfected cells induced ∼3 foci/cell at 480 decays/cell and the I-125-TFO-MBS transfected cells displayed the same amount of foci/cell at about 450 accumulated decays/cell. Cells after transfection with I-125-TFO-QRT showed a much steeper increase of foci induction and induced 3 foci/cell after approximately 300 decays/cell. The observed 1.5-fold increase of DSB in comparison to I-125-TFO-GAPDH or I-125-TFO-MBS transfected cells is in very good agreement with the increased cytotoxicity of I-125-TFO-QRT observed in the CFA ().
Targeting the GAPDH gene with the specific binding I-125-TFO-GAPDH resulted in a significant down-regulation of GAPDH (). We suggest that this reflects the DNA damage caused by GAPDH-bound I-125-TFO-GAPDH and the subsequent decay of I-125. This is supported by the observed induction of site-specific DSB in our in vitro studies () and the general cyto- and genotoxicity of I-125-TFO-GAPDH in the CFA and 53BP1 foci assay ( and ). Also, this correlates very well with the findings of Sedelnikova et al. (Citation2000), who were able to show in their study the induction of specific DSB in the human MDR1 gene in KB-V1 cells by a specific binding I-125-labeled TFO. Furthermore, these results underline the high target specificity of I-125-TFO-GAPDH in comparison to I-125-TFO-QRT, which possesses a much higher cytotoxicity but merely affects the gene expression of GAPDH.
Figure 4. DSB analysis of a 1695 bp DNA fragment ![]()
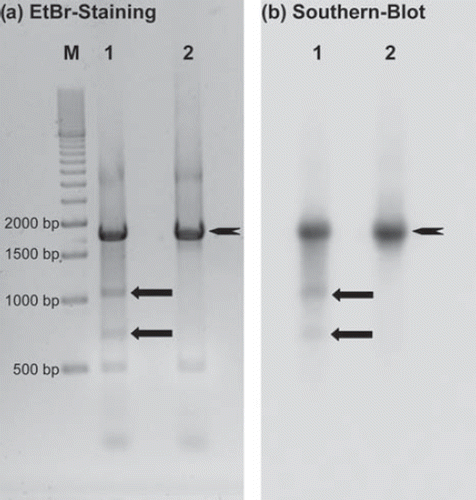
Figure 5. Cell survival curves of SCL-II cells after transfection with I-125-TFO-MBS (♦, —), I-125-TFO-QRT (■, ----) or I-125-TFO-GAPDH (Δ, -----). For decay accumulation, the transfected samples were stored at − 150°C. The data were fitted according to the exponential model of cell survival.
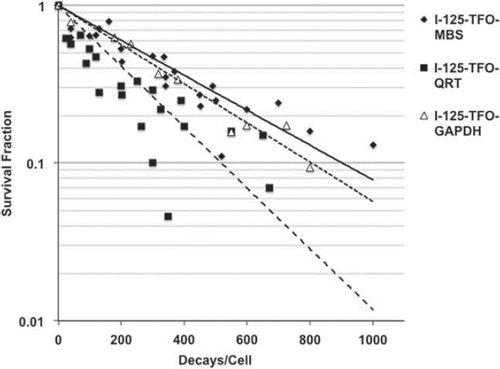
Figure 6. Double-strand break detection by the 53BP1 foci assay in SCL-II cells after transfection with I-125-TFO-MBS (♦, — ), I-125-TFO-QRS (■, -----) and I-125-TFO-GAPDH (Δ, -----). Negative controls (not displayed) were transfected with the corresponding unlabeled TFO and showed on average ∼ 0.8 foci/cell. For decay accumulation the samples were stored at − 150°C for decay accumulation. Data points were fitted according to a linear model
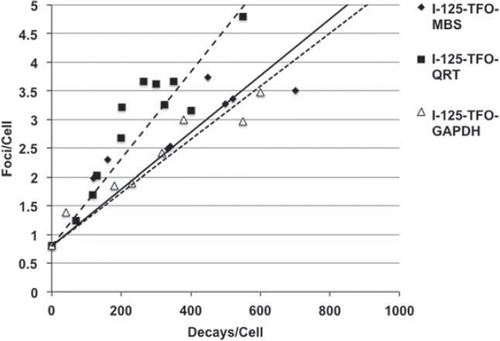
Figure 7. Relative gene expression of GAPDH after transfection with I-125-TFO-GAPDH (n = 7), I-125-TFO-QRT (n = 3) and unlabeled TFO-GAPDH (n = 3) as negative control. The (I-125-)TFO-GAPDH binds to a single target sequence in the GAPDH gene. I-125-TFO-QRT binds to multiple targets in the whole human genome. Error bars indicate the standard error of the mean (SEM) of n independent experiments. (* = p-value 0.05).
Summary
We demonstrated that TFO are able to form stable DNA triplexes with their specific targets and that site-specific DNA DSB can be induced by I-125-labeled TFO in vitro. Single-binding site as well as multi-binding site I-125 labeled TFO possess a pronounced cytotoxicity and induce an increased DSB frequency as a function of delivered decays per cell. The observed cyto- and genotoxic effects seem to depend more on target quantity rather than quality. I-125-labeled TFO possessing the highest target number also induced the most pronounced cytotoxic effect and the highest number of 53BP1 foci. A significant gene expression alteration can be induced with specific single-binding site I-125-labeled TFO.
Conclusions
We conclude from our results that specific I-125-labeled TFO cause site-specific DNA damage, leading to decreased gene expression in the targeted gene and that they have cytotoxic and DSB-inducing capacity. The observed differences in cytotoxicity per decay suggest that the effects depend on the target quantity rather than quality. I-125-labeled TFO targeting single genes can alter gene expression probably due to site-specific induction of DSB and might therefore be a very useful tool to induce site-specific complex DNA lesions in a cellular environment, making this approach very interesting and desirable for DNA repair research.
Acknowledgements
We would like to thank Dr Igor Panyutin for helpful discussions.
Declaration of interest
The authors report no declaration of interest. The authors alone are responsible for the content and writing of the paper.
V.D. and R.K. were funded by the German Federal Ministry of Education and Research (BMBF), grant number 02NUK005.
References
- Alberts B. 2010.Cell biology: The endless frontier. Molecular Biology of the Cell 21:3785–3785.
- Forsha SJ, Panyutin IV, Neumann RD, Panyutin IG. 2010. Intracellular traffic of oligodeoxynucleotides in and out of the nucleus: Effect of exportins and DNA structure. Oligonucleotides 20:277–284.
- Frank-Kamenetskii MD, Mirkin SM. 1995. Triplex DNA structures. Annual Review of Biochemistry 64:65–95.
- Goni JR, de la Cruz X, Orozco M. 2004. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Research 32:354–60.
- Huppert JL, Balasubramanian S. 2007. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Research 35:406–413.
- Kassis AI, Harapanhalli RS, Adelstein SJ. 1996. Comparison of strand breaks in plasmid DNA following positional changes of Auger- electron-emitting iodine-125. Journal of Nuclear Medicine 37:397–397.
- Kriehuber R, Riedling M, Simkó M, Weiss DG. 2004. Cytotoxicity, genotoxicity and intracellular distribution of the Auger electron emitter (65)Zn in two human cell lines. Radiation and Environmental Biophysics 43:15–22.
- Kumar P, Yadav VK, Baral A, Kumar P, Saha D, Chowdhury S. 2011. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Research 39:8005–8016.
- Kuznetsova S, Ait-Si-Ali S, Nagibneva I, Troalen F, Le Villain JP, Harel-Bellan A, Svinarchuk F. 1999. Gene activation by triplex-forming oligonucleotide coupled to the activating domain of protein VP16. Nucleic Acids Research 27:3995–4000.
- Lobachevsky PN, Martin RF. 1996. DNA strand breakage by 125I-decay in a synthetic oligodeoxynucleotide-2. Quantitative analysis of fragment distribution. Acta Oncol 35:809–815.
- Longe HO, Romesser PB, Rankin AM, Faller DV, Eller MS, Gilchrest BA, DenisGV. 2009. Telomere homolog oligonucleotides induce apoptosis in malignant but not in normal lymphoid cells: Mechanism and therapeutic potential. International Journal of Cancer 124:473–482.
- Onyshchenko MI, Gaynutdinov TI, Englund EA, Appella DH, Neumann RD, Panyutin IG. 2011. Quadruplex formation is necessary for stable PNA invasion into duplex DNA of BCL2 promoter region. Nucleic Acids Research 39:7114–7123.
- Panyutin IG, Neumann RD. 1994. Sequence-specific DNA double-strand breaks induced by triplex forming 125I labeled oligonucleotides. Nucleic Acids Research 22:4979–4982.
- Panyutin IG, Neumann RD. 1996. Sequence-specific DNA breaks produced by triplex-directed decay of iodine-125. Acta Oncologica 35:817–823.
- Panyutin IG, Sedelnikova OA, Karamychev VN, Neumann RD. 2003. Antigene radiotherapy: Targeted radiodamage with 125I-labeled triplex-forming oligonucleotides. Annals of the New York Academy of Sciences 1002:134–140.
- Pomplun E, Booz J, Dydejczy kA, Feinendegen LE. 1987. A microdosimetric interpretation of the radiobiological effectiveness of 125I and the problem of quality factor. Radiation and Environmental Biophysics 26:181–188.
- Praseuth D, Perrouault L, Doan TL, Chassignol M, Thuong N, Helene C. 1988. Sequence-specific binding and photocrosslinking of alpha-oligodeoxynucleotides and beta-oligodeoxynucleotides to the major groove of DNA via triple-helix formation. Proceedings of the National Academy of Sciences of the USA 85:1349–1353.
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. 2000. p53 Binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. Journal of Cell Biology 151:1381–1390.
- Sedelnikova OA, Panyutin IG, Thierry AR, Neumann RD. 1998. Radiotoxicity of Iodine-125-labeled oligodeoxyribonucleotides in mammalian cells. Journal of Nuclear Medicine 39:1412–1418.
- Sedelnikova OA, Luu AN, Karamychev VN, Panyutin IG, Neumann RD. 2001. Development of DNA-based radiopharmaceuticals carrying Auger-electron emitters for antigene radiotherapy. International Journal of Radiation Oncology*Biology*Physics 49:391–396.
- Sedelnikova OA, Panyutin IV, Neumann RD, Bonner WM, Panyutin IG. 2004. Assessment of DNA damage produced by 125I-triplex- forming oligonucleotides in cells. International Journal of Radiation Biology 80:927–931.
- Sedelnikova OA, Panyutin IG, Luu AN, Reed MW, Licht T, Gottesman MM, Neumann RD. 2000. Targeting the human mdr1 gene by 125I-labeled triplex-forming oligonucleotides. Antisense and Nucleic Acid Drug Development 10:443–452.
- Stout JT, Caskey CT. 1985. HPRT: Gene structure, expression, and mutation. Annual Review of Genetic 19:127–148.
- Tilgen W, Boukamp P, Breitkreutz D, Dzarlieva RT, Engstner M, Haag D, Fusenig NE. 1983. Preservation of morphological, functional, and karyotypic traits during long-term culture and in vivo passage of two human skin squamous cell carcinomas. Cancer Research 43:5995–6011.
- Todd AK, Johnston M, Neidle S. 2005. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Research 33:2901–2907.
- Vekhoff P, Ceccaldi A, Polverari D, Pylouster J, Pisano C, Arimondo P. 2008. Triplex formation on DNA targets: How to choose the oligonucleotide. Biochemistry 47:12277–12289.
- Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. 2009. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Research 37:4194–4204.