Abstract
The presence of two basic amino acids strategically located within a single spanning transmembrane region has previously been shown to act as a signal for the endoplasmic reticulum associated degradation (ERAD) of several polypeptides. In contrast, the functionality of this degron motif within the context of a polytopic membrane protein has not been established. Using opsin as a model system, we have investigated the consequences of inserting the degron motif in the first of its seven transmembrane (TM) spans. Whilst these basic residue reduce the binding of the targeting factor, signal recognition particle, to the first TM span, this has no effect on membrane integration in vitro or in vivo. This most likely reflects the presence of multiple TM spans that can act as targeting signals within in the nascent opsin chain. We find that the degron motif leads to the efficient retention of mutant opsin chains at the endoplasmic reticulum. The mutant opsin polypeptides are degraded via a proteasomal pathway that involves the actions of the E3 ubiquitin ligase HRD1. In contrast, wild-type opsin remains stable for a prolonged period even when artificially accumulated at the endoplasmic reticulum. We conclude that a single dibasic degron motif is sufficient to initiate both the ER retention and subsequent degradation of ospin via an ERAD pathway.
| Abbreviations: | ||
| Endoglycosidase H | = | EndoH |
| Endoplasmic reticulum | = | ER |
| ER-associated degradation | = | ERAD |
| Peptide: N-glycosidase F | = | PNGaseF |
| transmembrane | = | TM |
Introduction
The endoplasmic reticulum (ER) enforces a strict quality control over newly synthesized proteins, ensuring that only correctly folded polypeptides that have attained their native conformation are allowed to reach their final destination [Citation1,Citation2]. Misfolded proteins that fail to pass the ER quality control system are ultimately delivered to cytosolic proteasomes for degradation by the ER-associated degradation (ERAD) pathway [Citation3]. Key events during ERAD include: Substrate selection, retro-translocation and proteolysis of the misfolded proteins. Selection of ERAD substrates is typically carried out by molecular chaperones, and retro-translocation is thought to occur via one or more specialized retro-translocation channels [Citation4–6].
The degron is a dibasic amino acid motif located within the hydrophobic transmembrane domains of certain membrane proteins, such as the alpha subunit of the T-cell antigen receptor complex (TCRα) [Citation7,Citation8]. This ER degron motif has been shown to target ‘orphan’ subunits and misassembled complexes to the ERAD pathway. The ability of the degron to promote degradation is well documented, but the precise molecular basis for its recognition and the route by which this substrate is delivered to the degradation pathway are unclear. Substitution of the arginine and lysine residues that form the degron motif prevented rapid degradation of TCRα [Citation7]. In contrast, insertion of the TCRα degron motif into the hydrophobic TM domain of the stable type I membrane glycoprotein haemagglutinin was shown to result in its rapid degradation via both proteasomal and lysosomal pathways, consistent with a second quality control ‘checkpoint’ for aberrant proteins that escape the ERAD pathway [Citation9].
To date, studies of the effect of the degron motif have focussed on its role in the context of single transmembrane spanning domains. We wished to establish whether the degon displays any activity when incorporated into a polytopic membrane protein. To this end, we have used bovine opsin as a model system to investigate the consequences of introducing the degron motif into the first transmembrane spanning region of this 7TM polypeptide [Citation10]. An in vitro analysis of opsin biogenesis revealed that the degron motif reduces the binding of the signal recognition particle (SRP) to opsin TM1 to almost half the level observed with the wild-type protein. Nevertheless, the membrane integration of the full-length degron mutant was comparable to the wild-type protein both in vitro and in vivo. When expressed in cultured mammalian cells, the degron mutant is efficiently retained at the ER, preventing the Golgi modification of N-linked glycans on the protein. ER retention of the degron mutant induces the rapid removal of the mutant opsin polypeptide via proteasomal degradation. This degradation is a specific feature of the degron containing protein, since when wild type opsin is artificially retained in the ER it remains stable and is not degraded. We therefore conclude that the insertion of a degron motif into a polytopic membrane protein leads to both its specific ER retention and subsequent degradation via the process of ER associated degradation.
Materials and methods
Antibodies and reagents
Endoglycosidase H (EndoH) and peptide: N-glycosidase F (PNGaseF) were obtained from New England Biolabs (Hitchin, UK). Easytag L-[35S]methionine was purchased from NEN Dupont (Stevenage, UK). Reagents for cell culture, Lipofectamine 2000, Zeocin, and the expression vector pZeoSV2(+) were all obtained from Invitrogen (Paisley, UK), whilst all other chemicals were purchased from BDH/Merck (Poole, UK) and Sigma (Poole, UK). Proteasomal inhibitor I and kifunensine were obtained from Calbiochem (Nottingham, UK). The monoclonal antibody specific for the N-terminus of bovine opsin was from Paul Hargrave [Citation11], the αTGN-46 sheep polyclonal antibody was from Vas Ponnambalam (University of Leeds, UK), and the αCalnexin rabbit polyclonal serum was from Stressgen. The rabbit polyclonal sera for αERD2 and αGM130 were from Hans-Dieter Söling (University of Göttingen) and Martin Lowe (University of Manchester) respectively.
Plasmid constructs
Wild-type bovine opsin was excised from a previously described construct, pGEM3z [Citation12] using Eco RI and Pst I and cloned into the mammalian expression vector pZeoSV2(+) linearized with the same enzymes. The L46R and G51K mutations required to generate a degron motif were inserted using the QuikChange site-directed mutagenesis kit (Stratagene). Plasmids containing wild-type human HRD1, and a dominant negative mutant of the ring finger domain (HRD1-C1A) were as previously described [Citation13].
In vitro analysis of opsin derivatives
Truncated and full-length opsin transcripts were generated by PCR and in vitro transcription and translation in the presence or absence of ER microsomes, as described previously [Citation14]. Cross-linking of ribosome bound nascent chains to the 54 kDa subunit of SRP [Citation14] was induced using the heterobifuntional cross-linking reagent succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) at a 1 mM final concentration for 10 min at 30°C. For protease protection studies, the membrane fraction was recovered by ultracentrifugation through a high-salt sucrose cushion [Citation15], incubated with 125 μg/ml trypsin or buffer on ice for 30 min, and 1 mM phenylmethylsulfonyl fluoride was then added.
Electron microscopy
Cells were fixed with an equal volume of 4% electron microscopy grade PFA in 0.1 M HEPES at pH 7.2, blocked with PBS containing 1% BSA, 0.15 M glycine and 0.2% (w/v) saponin. Ultrasmall gold particles were enhanced with GoldEnhance kit (Nanoprobes, Yaphank, NY, USA) for approximately 5 min. Samples were postfixed with reduced osmium tetroxide, then processed for standard epon embedding and examined using a Technai 12 microscope (FEI, Eindhoven, The Netherlands) at 100 kV.
Fluorescence microscopy
COS-7 cells were fixed in methanol at −20°C followed by re-hydration in PBS. Stained samples were mounted in mowiol containing 25 mg/ml DABCO antifade (1,4-diazabicyclo[2,2,2]-octane) (Sigma). Samples were imaged using an Olympus BX-60 wide field microscope fitted with a 60× oil objective and a MicroMax-cooled slow scan CCD camera (Roper Scientific). Images were processed and assembled using Metamorph software (Univseral Imaging Corporation) and Adobe Photoshop.
Metabolic labelling and pulse-chase analysis
COS-7 cells were transfected with plasmids encoding wild-type or degron opsin alone ( to ), or co-transfected with degron opsin and wild-type human HRD1 or the mutant HRD1-C1A () [Citation13], using Lipofectamine 2000 according to the manufacturers instructions. One day after transfection, cells were starved for 20–60 min in methionine/cysteine free MEM (Sigma) with 2 mM glutamine, then metabolically labelled for 1 h with the same medium containing 22 μCi/ml of [35S]methionine/cysteine protein labelling mix. Cells were rinsed twice with PBS then chased in normal growth medium supplemented with 5 mM unlabelled methionine/cysteine mix. At the end of the chase period (0, 3, 5 or 8 h) cells were rinsed twice with PBS and solubilized in immunoprecipitation buffer (10 mM Tris/HCl, pH 7.6, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with 1mM phenylmethylsulfonyl fluoride. Where appropriate, transfected cells were pre-treated with inhibitors for 12 h before the pulse chase analysis (lactacystin at 50 µM and kifunensine at 5µg/ml) and these inhibitors were also included during subsequent starvation, pulse and chase incubations. Immunoprecipitation was carried out in the absence of SDS [Citation16], using an excess of monoclonal anti-opsin antibody (1 µg) to ensure efficient recovery (data not shown). EndoH or PNGase F cleavage of N-linked glycans from the labelled opsin polypeptides was carried out after immunoprecipitation, using 500 U of EndoH or PNGase F and the appropriate buffers added to the immuno-isolated material according to the manufacturers instructions, and incubating for 30 min at 37°C prior to the addition of an equal volume of 2× SDS-PAGE sample buffer.
Sample analysis
Samples were analysed on 14%polyacrylamide gels containing SDS unless otherwise stated, dried gels exposed to a phosphorimaging plate for one to three days, and samples visualized with a Fuji BAS 3000 system. Quantitative analysis was carried out using AIDA version 4.00 (Raytest, Isotoopenmeßgeräte GmbH). Statistical analysis (Paired t-tests, standard error of the mean) was carried out using GraphPad Prism 4 for the Macintosh.
Results and discussion
Construction and characterization of a degron containing opsin mutant
The TM segment of the TCRα subunit has been shown to act as a portable signal for ER degradation, and two basic residues present within this segment of the protein have been shown to act as the primary mediators of this effect [Citation8,Citation9,Citation17]. To date, the effectiveness of this dibasic degron motif within the context of a polytopic membrane protein had not been investigated and we therefore introduced it into the first TM segment of the seven TM spanning integral membrane protein opsin (). It is worth noting that the alterations to opsin TM1 did not remove a highly conserved asparagine located at residue 55 that is involved in interhelical hydrogen bonds that stabilise the tertiary structure of the protein [Citation10,Citation18].
Figure 1. In vitro characterization of a novel degron mutant. (A) The amino acid sequence of the first transmembrane region (residues 39–61 inclusive) of wild-type opsin was modified to include the degron motif from TCRα, equating to R46 and K51 mutations. The predicted ΔGpredapp for the membrane integration of the three transmembrane segment shown was calculated according to Hessa et al. [Citation22], the more negative the value the more favourable membrane integration. (B) Ribosome bound, 91 residue long, fragments of wild-type and degron opsin with a single cysteine at position 48, wtOp91[C48] and degOp91[C48], respectively, were synthesized in a rabbit reticulocyte translation system for 30 minutes. Ribosome/nascent chain complexes were stabilized by the addition of cycloheximide and interactions with the SRP examined by cross-linking with SMCC or a solvent control (DMSO). The resulting products were analysed directly (lanes 1, 2, 5 and 6) or following immunoprecipitation with antisera specific for the SRP54 subunit (lanes 3,4, 7 and 8). Quantitative phosphorimaging indicated that ∼ 4.0% of the OP91wt chains were cross-linked to SRP54 but only ∼ 2.3% of the OP91deg chains formed adducts. (C) Full-length wild-type or degron opsin were translated in vitro in the presence of ER microsomes and subjected to limited proteolysis with trypsin, either in the presence or absence of 1% triton X100 as indicated. Reactions were either analysed directly (upper panel) or after immunoprecipitation with antibodies specific for the N-terminus of the opsin protein (lower panel). Opψ indicates the fully glycosylated form of both the wild-type and degron variant of opsin, ΔN-Opψ shows the trypsin-trimmed opsin fragments and Op any non-glycosylated chains. (D) Diagram showing the topology of opsin in the ER membrane, the relative location of the trypsin-sensitive lysine residues (66 and 67) in the loop between TM1 and TM2, and the two N-glycosylation sites on the N-terminus.
![Figure 1. In vitro characterization of a novel degron mutant. (A) The amino acid sequence of the first transmembrane region (residues 39–61 inclusive) of wild-type opsin was modified to include the degron motif from TCRα, equating to R46 and K51 mutations. The predicted ΔGpredapp for the membrane integration of the three transmembrane segment shown was calculated according to Hessa et al. [Citation22], the more negative the value the more favourable membrane integration. (B) Ribosome bound, 91 residue long, fragments of wild-type and degron opsin with a single cysteine at position 48, wtOp91[C48] and degOp91[C48], respectively, were synthesized in a rabbit reticulocyte translation system for 30 minutes. Ribosome/nascent chain complexes were stabilized by the addition of cycloheximide and interactions with the SRP examined by cross-linking with SMCC or a solvent control (DMSO). The resulting products were analysed directly (lanes 1, 2, 5 and 6) or following immunoprecipitation with antisera specific for the SRP54 subunit (lanes 3,4, 7 and 8). Quantitative phosphorimaging indicated that ∼ 4.0% of the OP91wt chains were cross-linked to SRP54 but only ∼ 2.3% of the OP91deg chains formed adducts. (C) Full-length wild-type or degron opsin were translated in vitro in the presence of ER microsomes and subjected to limited proteolysis with trypsin, either in the presence or absence of 1% triton X100 as indicated. Reactions were either analysed directly (upper panel) or after immunoprecipitation with antibodies specific for the N-terminus of the opsin protein (lower panel). Opψ indicates the fully glycosylated form of both the wild-type and degron variant of opsin, ΔN-Opψ shows the trypsin-trimmed opsin fragments and Op any non-glycosylated chains. (D) Diagram showing the topology of opsin in the ER membrane, the relative location of the trypsin-sensitive lysine residues (66 and 67) in the loop between TM1 and TM2, and the two N-glycosylation sites on the N-terminus.](/cms/asset/4a56ed9a-f7c8-415a-940e-1215e37241dd/imbc_a_433561_f0001_b.jpg)
The first TM domain of opsin appears to act as the default ER targeting signal, and binds the signal recognition particle (SRP) prior to delivery of the nascent polypeptide to the ER membrane [Citation12,Citation19,Citation20]. A key feature of signal sequence recognition and binding by the 54 kDa subunit of SRP (SRP54) is hydrophobicity [Citation21]. Hence, the introduction of the degron motif into opsin TM1, and the resulting alteration in its biological hydrophobicity [Citation22,Citation23], may affect its interaction with SRP (, see ΔGpredapp). We therefore used an in vitro approach to address the effect of the degron motif on the association of SRP with opsin TM1 [Citation24]. Short, ribosome bound, opsin chains were synthesized in a cell free system that lacked ER derived membrane, and the interactions of the nascent polypeptides with cytosolic SRP was analysed by cross-linking. In both cases, specific adducts between the ribosome bound opsin polypeptides and the SRP54 subunit were formed upon the addition of the hetero-bifunctional reagent SMCC, and these products could be visualized after immunoprecipitation (, lanes 3, 4, 7 and 8). Quantification indicates that the degron motif decreases the association of opsin TM1 with SRP54 to almost half the wild-type level (from 4% to 2.3% of total nascent chains present).
The reduction in SRP binding to opsin TM1 raised the possibility that the targeting of the nascent opsin degron polypeptide to the ER may be inefficient, resulting in little or no membrane integrated protein. To address this issue, we analysed the membrane integration of full-length wild-type and degron opsin using an in vitro translation system supplemented with canine pancreatic microsomes as a source of ER membrane. Similar levels of membrane-associated polypeptides are detected for both the wild-type and degron versions of opsin, and the majority of each species is correctly N-glycosylated (, c.f. Op and Opψ in lanes 1 and 4). We therefore conclude that although the binding of SRP54 is perturbed by the introduction of the degron motif, this does not impact on the SRP dependent delivery of the protein to the ER membrane [Citation25]. This is most likely due to the ability other opsin TM segments, for example TM2, to mediate SRP-dependent targeting of the nascent polypeptide, consistent with previous studies showing five opsin TM segments can function as independent ER signal sequences [Citation15,Citation26]. Alternatively, the binding of SRP to opsin TM1, as reported by cross-linking to the SRP54 subunit, may not be rate limiting in relation to the ER targeting of nascent opsin chains (cf. [Citation27]).
Whilst the substantial level of N-glycosylation of the opsin N-terminus observed for the opsin degron mutant is consistent with its efficient integration at the ER membrane, it was also possible that the mutant form of opsin TM1 failed to correctly engage the Sec61 translocon and was erroneously fully translocated into the ER lumen [Citation22,Citation23,Citation28,Citation29]. To distinguish between the possibility of stable integration and complete translocation of TM1, we exploited the protease sensitivity of the cytosolically exposed loop linking opsin TM1 and TM2 that would normally be available when opsin TM1 assumes an authentic transmembrane topology [Citation12]. To this end, wild-type and degron opsin were synthesized in parallel and subjected to a limited digestion with trypsin. Proteolysis resulted in the formation of a faster migrating product in both the wild-type and degron variants ( upper panel, c.f. Opψ and ΔN-Opψ in lanes 1and 4 with lanes 2 and 5). Solublization of the ER membrane with detergent before the trypsin treatment abolished the formation of this trypsin clipped product in each case and resulted in an identical pattern of limit digest products ( upper panel, lanes 3 and 6). Specific immunoprecipitation of the reactions with a monoclonal antibody that recognizes the N-terminus of the opsin protein failed to recover the membrane dependent lower molecular weight product, consistent with the N-terminal trimming of opsin in the presence of trypsin ( lower panel, cf. lanes 1, 2, 4 and 5). Taken together these data indicate that the wild-type and degron opsin both attain the same transmembrane topology when synthesized at the ER membrane (see ). This conclusion is consistent with studies of TCRα, which suggest that despite potentially significant compromises arising from the degron motif (cf. ), the protein is nevertheless integrated into the ER membrane [Citation7]. During the early phases of the biosynthesis of opsin, the first TM domain is stalled at the ER translocon site [Citation29], an effect suggested to promote the folding and assembly of the protein. Thus, in the case of degron opsin, it is possible that chaperoning by the ER translocon may facilitate the normal association of TM1 with other opsin TM segments and thereby enhance its stability within the membrane.
The opsin degron mutant is primarily retained at the endoplasmic reticulum
Having confirmed the opsin degron mutant could be efficiently and authentically membrane integrated in vitro, the protein was next expressed in cultured mammalian cells in order to study its subcellular localization and stability [Citation30,Citation31]. COS-7 cells have proved a robust platform for previous studies of opsin [Citation32], and immuno-electron microsocopy showed that the wild-type protein is efficiently trafficked to the plasma membrane, in addition to being present at intracellular structures such as the ER, that are en route to the cell surface (, see WT Opsin). The opsin degron mutant was clearly expressed in COS-7 cells, and immuno-labelling was readily apparent. However, this protein appeared to be almost entirely intracellular and primarily located in a compartment that resembled the ER (, see Deg Opsin). Fixation of the COS-7 cells prior to antibody labelling in the absence of detergent, followed by immunofluorescence based detection, confirms that the N-terminus of the opsin degron mutant is not detectable outside the cell (Supplementary Figure 1, online version only). We conclude that the artificially generated opsin degron mutant does not reach the plasma membrane.
Figure 2. Degron opsin remains intracellular and is primarily located at the ER. (A) COS-7 cells plated on plastic coverslips were transfected with wild-type opsin or the degron mutant and incubated overnight, followed by fixation, blocking and incubation with a monclonal anti-opsin antibody and then a gold-conjugated secondary antibody. En face sections were obtained using a Reichert-Yung ultramicrotome. For wild-type opsin (upper panels), labelling was evenly distributed along the plasma membrane (PM). For the degron mutant (lower panels), the majority of cells examined showed strong labelling of a compartment presumed to be the endoplasmic reticulum (ER) with little immunoreactive material visible at the cell surface (PM). Other discrete intracellular structures identifiable by their morphology were mitochondria (M), the nucleus (N), and the nuclear envelope (NE). Scale bars represent 2.3 μM. (B) Transiently transfected COS-7 cells were fixed in ice-cold methanol and labelled using the anti-opsin antibody and an antibody recognising the ER localized component calnexin. Species specific secondary antibodies were then used to reveal opsin (red in merge) and calnexin (green in merge) and determine the extent of their co-localization (merge). Scale bars represent 10 μM. This Figure is reproduced in colour in Molecular Membrane Biology online.
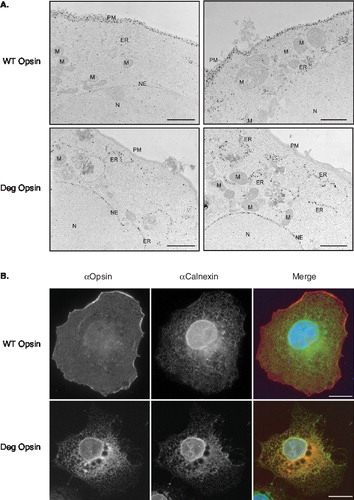
The ER retention of single spanning membrane proteins containing the degron motif is a well-characterized phenomenon [Citation7], and we therefore addressed the location of the opsin-degron mutant by indirect immuno-fluorescence microscopy. When COS-7 cells expressing the wild-type protein are analysed, further evidence that a substantial amount of bovine opsin reaches the cell surface is obtained (, WT Opsin, see αOpsin and merge panels), whilst only modest co-localization of the protein and the ER component calnexin is apparent (, WT Opsin, αCalnexin and Merge panels). For the degron version of opsin, the staining pattern for this mutant protein and that of calnexin show substantial overlap, especially in the perinuclear region, with little evidence of cell surface labelling (, see Deg Opsin panels). Hence, both electron and immuno-fluoresence microscopy confirm that wild-type and degron opsin are efficiently expressed and membrane integrated in COS-7 cells, but show quite distinct fates in terms of membrane trafficking. Interestingly, expression of the degron mutant consistently resulted in the appearance of distinctive ‘vacuole-like’ structures within the ER staining pattern (Supplementary Figure 2, online version only). These structures are typically located near to the nucleus and may reflect changes in ER morphology resulting from the expression of aberrant opsin chains (c.f. [Citation33]).
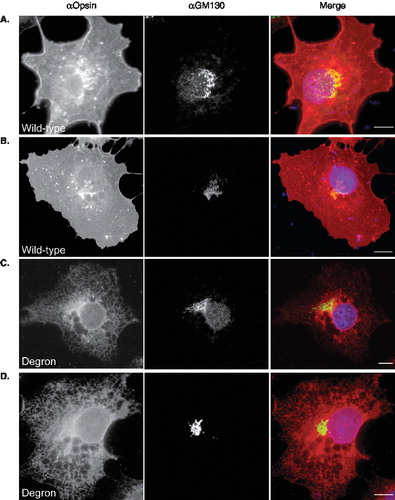
We also used immunofluorescence microscopy to determine whether any of the degron mutant could reach post ER locations within the secretory pathway. Using the KDEL receptor, ERD2 as a marker for the ER-Golgi intermediate compartment, we find that the wild-type protein can be found here consistent with its delivery to the cell surface via the classical secretory pathway (, panel A), although detectable levels are not always apparent (, panel B). Likewise, the degron mutant is absent from the ERGIC of some COS-7 cells (, panel C) but clearly present in other cases (, panel D). When COS-7 cells expressing wild-type and degron opsin are co-stained for the cis-Golgi marker, GM130, co-localization of the wild-type protein is consistently seen (, panels A and B). In contrast, we find no evidence that degron opsin can reach a GM130 positive compartment (, panels C and D). Likewise, co-staining for the more distal location of the trans-Golgi network also showed the presence of wild- type opsin but not the degron mutant (Supplementary Figure 3, online version only). Hence, we conclude that wild-type opsin is efficiently delivered from the ER to the plasma membrane, whilst the degron mutant is efficiently retained at very early locations within the secretory pathway, primarily the ER. This most likely reflects the recognition of degron opsin as aberrant by one or more ER components. The escape of some opsin degron polypeptides to the ERGIC appears to correlate with higher levels of protein expression, and is consistent with a proposed role for this compartment in the normal cellular quality control process [Citation34,Citation35].
Figure 3. Degron opsin can reach the ER-Golgi intermediate compartment. COS-7 cells expressing wild-type or degron opsin were fixed and stained with an antibody specific for opsin and the ERGIC marker, ERD2. (A–B) Wild-type opsin was evenly distributed throughout the cell (αOpsin, red in merge) whilst ERD2 staining was restricted to a perinuclear location (αERD2, green). In some cells wild-type opsin co-localized with ERD2 (panel A, see merge) whilst in other cells little wild type protein was detected at this location (panel B, merge). (C–D) Degron opsin was distributed in a reticular network (αOpsin) and in many cells no co-localization was apparent (panel C, merge). Occasionally significant localization was apparent (panel D, merge). Scale bars represent 10 μM. This Figure is reproduced in colour in Molecular Membrane Biology online.
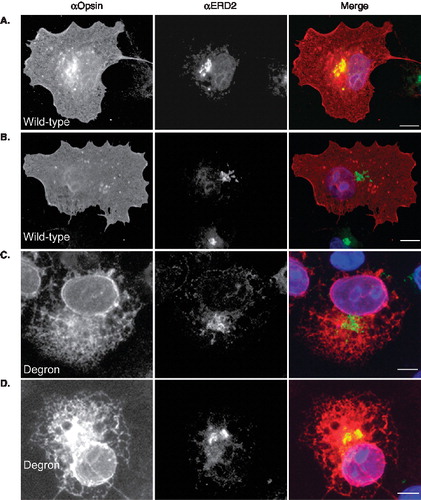
Figure 4. Degron opsin does not reach the cis-Golgi. COS-7 cells were treated as before (see legend to Figure 3) and stained with the anti-opsin antibody and an antibody recognizing the cis-Golgi marker, GM130, followed by appropriate secondary antibodies to visualise the respective proteins. (A–B) Wild-type opsin (αOpsin, red in merge) shows significant overlap with GM130 (green in merge). In contrast, degron opsin (panels C and D) was not apparent in GM130 positive compartments. Scale bars represent 10μM. This Figure is reproduced in colour in Molecular Membrane Biology online.
The opsin degron mutant is rapidly degraded
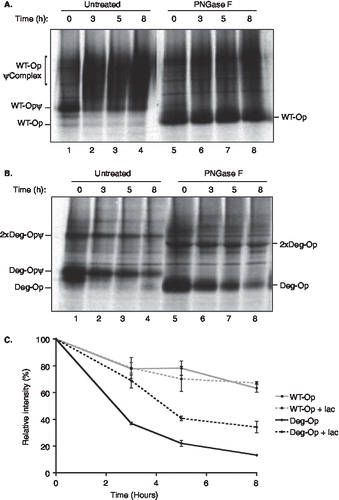
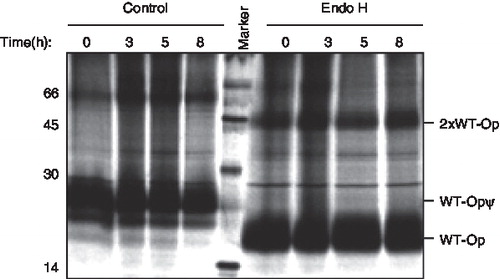
The typical fate of misfolded or unassembled membrane proteins retained at the ER is their rapid removal via a process known as ER-associated degradation (ERAD) [Citation1–3]. We therefore analysed the stability of the wild-type and opsin degron mutant using a pulse chase approach. For the wild-type protein, following metabolic labelling for 1 h, the two high mannose N-linked glycans added in the ER were efficiently converted into complex glycans within three hours and acquired resistance to digestion with Endoglycosidase H (Supplementary Figure 4A, online version only). This is entirely consistent with glycoprotein trafficking through the Golgi complex prior to delivery to the plasma-membrane [Citation36], as deduced from our imaging studies. When the glycans were removed from the wild-type opsin chains and the resulting products were quantified, the wild-type protein was found to be relatively stable over an eight-hour time course (, lanes 5–8; , wtOP). In contrast, the degron opsin chains did not acquire complex N-linked glycans (Supplementary Figure 4B, online version only; lanes 1–4), and the protein was efficiently degraded over the period of an eight-hour chase with a half-life of ∼ 2.5 h (, lanes 5–8; , degOP). Thus, the presence of a degron motif within opsin TM1 results in substantial ER retention and efficient degradation of the resulting opsin mutant. In contrast, the introduction of two basic residues into the TM region of influenza haemagglutinin (HA), or its replacement by the entire TM and short cytosolic tail of TCRα, resulted in the ER retention of only half of the resulting protein with the remainder reaching the lysosome [Citation9]. Hence, the context of the degron motif appears to influence the efficiency with which it exerts an ER retention phenotype.
Figure 5. The degron motif from TCRα destabilizes opsin. COS-7 cells were transiently transfected with wild-type opsin (A) or the degron mutant (B), metabolically labelled for 1 hour and then incubated for the chase periods 0, 3, 5 and 8 hours. Opsin chains were recovered by immunoprecipitation and analysed directly (lanes 1 to 4) or after deglycosylation with PNGase F (lanes 5–8) using SDS PAGE. The location of wild-type opsin bearing Golgi modified N-linked glycans (WT-OpψComplex) and a presumptive dimer of the degron mutant (2 × Deg-Op) are indicated (see legend to ). (C) Opsin chains were analysed as before except that 50 µM lactacystin was present (+ lac) or absent during the metabolic labelling and chase period as indicated. After immunoprecipitation and deglycosylation, samples were resolved by SDS-PAGE and quantified by phoshphorimaging. Duplicate experiments were performed and the results are expressed graphically including the standard error of the mean.
Opsin degron removal is proteasome mediated
A key feature of classical ERAD pathways is that substrate removal occurs via degradation at cytosolic proteasomes following retrotranslocation from the ER [Citation3]. Hence, any perturbation of proteasome function will inhibit substrate removal during the course of a pulse-chase analysis [Citation9]. When a pulse chase analysis of degron opsin stability was performed in the presence of the proteasome inhibitor lactacystin, the mutant protein was partially stabilized and the estimated half-life increased to over 4 h (, degOP + lac). A similar stabilizing effect was obtained using proteasome inhibitor I (Supplementary Figure 4C), and we conclude that the opsin degron mutant acts as a bona fide ERAD substrate being retained at the ER and degraded by cytosolic proteasomes, presumably after retro-translocation out of the ER membrane.
An additional feature of the opsin degron mutant was the presence of what appear to be SDS-stable dimers of the mutant chains, as deduced from their mobility and sensitivity to glycosidase treatment (, 2 × Deg-Op). Dimers of wild-type opsin have been previously reported in COS1 cells, and the dimerization interface has been mapped to the C-terminal half of the polypeptide [Citation37]. The introduction of the degron motif into opsin TM1 does not affect the capacity of the polypeptide to form dimers indicating that this region of the polypeptide most likely assumes a native like fold. The dimeric forms of wild-type opsin are much more readily apparent when the protein is prevented from acquiring complex carbohydrates in the Golgi (see below).
ER retention is not sufficient for opsin degradation
If the degron motif serves to simply retain opsin in the ER, and its subsequent degradation is by ‘default’ then retaining wild-type opsin at the ER via another strategy might also lead to its degradation. To this end, we used the mannosidase I inhibitor kifunensine to prevent mannose-trimming and thereby perturb the export of wild-type opsin from the ER in line with previous studies of angiotensin II type 1 receptor trafficking in COS-7 cells [Citation38]. In the presence of kifunensine, conversion from the high mannose form of N-linked glycans initially added to the opsin polypeptides, to the complex N-linked glycans typically acquired in the Golgi, was prevented throughout an eight-hour period (, lanes 1–4). The resulting glycoproteins remained sensitive to digestion with endoglycosidase H (, lanes 5–8, cf. Supplementary Figure 4A, online version only), confirming the absence of any Golgi mediated glycan processing and suggesting that the bulk of the wild-type protein was retained at the ER and/or ERGIC (cf. [Citation38]). Despite this efficient ER retention, no obvious effect upon the stability of the wild-type protein was observed (, lanes 1–8). It remains a formal possibility that in addition to causing the ER retention of the wild-type opsin, kifunensine treatment also inhibits its potential degradation by preventing entry into the EDEM pathway of glycoprotein removal for which mannose trimming is typically a prerequisite. However, Fagioli and Sitia [Citation17] have shown that kifunensine treatment does not inhibit that degradation of a chimera of the Ig μ chain containing the transmembrane region of TCRα, and including its degron motif, concluding that this chimeric molecule follows a distinct pathway for ER degradation that is independent of ER mannosidase I activity.
Figure 6. Kifunensine treatment prevents wild-type opsin acquiring complex N-linked glycans. COS-7 cells expressing wild-type opsin were pulse-labelled and then chased for up to 8 hours in the presence of 5 μg/ml of kifunensine. Samples were recovered by immunoprecipitation, deglycosylated with Endo H where indicated, and resolved by SDS-PAGE. The location of a presumptive opsin dimer is shown (2 × WT-Op). All other symbols are as described in the legend to .
A dominant negative mutant of HRD1 inhibits degradation of the opsin degron mutant
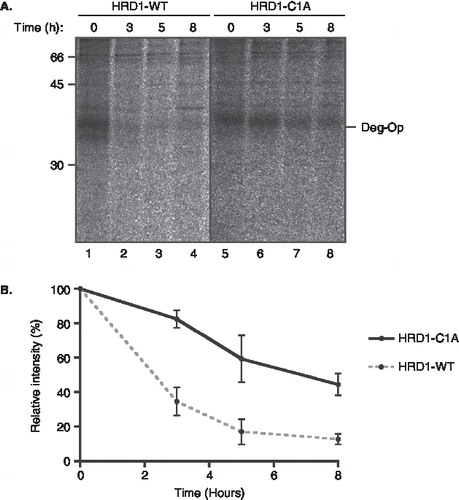
It has recently been established that actions of the yeast E3 ubiquitin ligase Hrd1p in promoting the degradation of misfolded membrane proteins most likely require interactions between the transmembrane domain of Hrd1p and its substrate [Citation39]. Given that the degron motif was introduced into the first TM of opsin, we speculated that HRD1, the mammalian equivalent of Hrd1p, may play some role in its rapid degradation. To test this hypothesis, we exploited a well-characterized dominant negative version of human HRD1 (HRD1-C1A) that has previously been shown to delay the degradation of TCRα when expressed in trans [Citation13]. When the opsin degron mutant was co-expressed with the wild-type form of HRD1, the protein continued to be rapidly degraded as previously observed (, lanes 1–4). In contrast, co-expression of the dominant negative C1A form of HRD1 resulted in a pronounced stabilization of the opsin degron mutant, with its apparent half-life increasing from ∼ 2.5 h to over 6 h (, lanes 5–8 and ). These data strongly support our conclusion that the degradation of the opsin degron mutant is proteasome mediated, and suggest that the HRD1 E3 ligase most likely contributes to the ERAD process for this substrate. Although we have not directly addressed the issue during the course of this study, we assume that the proteasomal degradation of the opsin mutant involves the generation of a ubiquitinated intermediate, most likely in concert with the extraction of the polypeptide from the ER membrane [Citation3].
Figure 7. A dominant negative HRD1 mutant stabilizes the opsin degron mutant. (A) The degradation of the opsin degron mutant was analysed by pulse-chase using COS-7 cells expressing the opsin degron mutant together with either wild-type human HRD1 (lanes 1–4) or the HRD1-C1A mutant (lanes 5–8) as indicated. Samples were recovered by immunoprecipitation between 0 and 8 hours after labelling, deglycosylated, and resolved by SDS-PAGE. (B) Products recovered by immunoprecipitation at different times were quantified by phosphorimaging. Three independent experiments were performed and the results are expressed graphically including the standard error of the mean.
Taken together, our data show that the two basic residues derived from the degron motif are highly effective when introduced into opsin, and suggest the motif is responsible for both ER retention and degradation, most likely via a process that requires the recruitment of specific chaperones or quality control components such as BiP [Citation40] and Hrd1 [Citation39]. This study defines a new model membrane protein for the analysis of the ERAD pathway and shows that the ER degron motif is functional even in the context of a complex polytopic membrane protein. The effect of the relative location of the degron motif within a polytopic protein upon its capacity to mediate ERAD has not yet been established, and we conclude that opsin provides an excellent model to examine this facet of membrane protein quality control and explore the cellular components that mediate this process.
Supplementary Material 1
Download JPEG Image (312.7 KB)Supplementary Material 2
Download JPEG Image (173.6 KB)Supplementary Material 3
Download JPEG Image (584.9 KB)Supplementary Material 4
Download JPEG Image (216.6 KB)Acknowledgements
We thank Samuel Crawshaw for valuable discussions during this project and Paul Hargrave, Martin Lowe, Hans-Dieter Söling, Sjaak van Voorden and Marjolein Kikkert for providing reagents. This work was supported by a Wellcome Trust PhD award (AR-S) and funding from the BBSRC (BCSC and SH) grant number BBSRC, BB/G000948/1, and MRC (SH) grant number G0501725.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Anelli T, Sitia R. 2008. Protein quality control in the early secretory pathway. EMBO J 27:315–327.
- Hebert DN, Molinari M. 2007. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev 87:1377–1408.
- Vembar SS, Brodsky JL. 2008. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9:944–957.
- Lilley BN, Ploegh HL. 2005. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci USA 102:14296–14301.
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. 2005. Inaugural article: Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA 102:14132–14138.
- Scott DC, Schekman R. 2008. Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J Cell Biol 181:1095–1105.
- Bonifacino JS, Cosson P, Shah N, Klausner RD. 1991. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J 10:2783–2793.
- Bonifacino JS, Suzuki CK, Klausner RD. 1990. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science 247:79–82.
- Fayadat L, Kopito RR. 2003. Recognition of a single transmembrane degron by sequential quality control checkpoints. Mol Biol Cell 14:1268–1278.
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. 2000. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289:739–745.
- Adamus G, Zam ZS, Arendt A, Palczewski K, McDowell JH, Hargrave PA. 1991. Anti-rhodopsin monoclonal antibodies of defined specificity: Characterization and application. Vision Res 31:17–31.
- Laird V, High S. 1997. Discrete cross-linking products identified during membrane protein biosynthesis. J Biol Chem 272:1983–1989.
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. 2004. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem 279:3525–3534.
- Cross BC, High S. 2009. Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J Cell Sci 122:1768–1777.
- Ismail N, Crawshaw SG, Cross BC, Haagsma AC, High S. 2008. Specific transmembrane segments are selectively delayed at the ER translocon during opsin biogenesis. Biochem J 411:495–506.
- Wilson CM, Roebuck Q, High S. 2008. Ribophorin I regulates substrate delivery to the oligosaccharyltransferase core. Proc Natl Acad Sci USA 105:9534–9539.
- Fagioli C, Sitia R. 2001. Glycoprotein quality control in the endoplasmic reticulum. Mannose trimming by endoplasmic reticulum mannosidase I times the proteasomal degradation of unassembled immunoglobulin subunits. J Biol Chem 276:12885–12892.
- Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. 2008. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454:183–187.
- Friedlander M, Blobel G. 1985. Bovine opsin has more than one signal sequence. Nature 318:338–343.
- Kanner EM, Klein IK, Friedlander M, Simon SM. 2002. The amino terminus of opsin translocates ‘posttranslationally’ as efficiently as cotranslationally. Biochemistry 41:7707–7715.
- Pool MR. 2005. Signal recognition particles in chloroplasts, bacteria, yeast and mammals (review). Mol Membr Biol 22:3–15.
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030.
- Lundin C, Kim H, Nilsson I, White SH, von Heijne G. 2008. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc Natl Acad Sci USA 105:15702–15707.
- Abell BM, Pool MR, Schlenker O, Sinning I, High S. 2004. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J 23:2755–2764.
- Cross BC, Sinning I, Luirink J, High S. 2009. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol 10:255–264.
- Audigier Y, Friedlander M, Blobel G. 1987. Multiple topogenic sequences in bovine opsin. Proc Natl Acad Sci USA 84:5783–5787.
- Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. 2008. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133:440–451.
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433:377–381.
- Ismail N, Crawshaw SG, High S. 2006. Active and passive displacement of transmembrane domains both occur during opsin biogenesis at the Sec61 translocon. J Cell Sci 119:2826–2836.
- Kaushal S, Khorana HG. 1994. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry 33:6121–6128.
- Kaushal S, Ridge KD, Khorana HG. 1994. Structure and function in rhodopsin: The role of asparagine-linked glycosylation. Proc Natl Acad Sci USA 91:4024–4028.
- Saliba RS, Munro PM, Luthert PJ, Cheetham ME. 2002. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci 115:2907–2918.
- Nagahama M, Suzuki M, Hamada Y, Hatsuzawa K, Tani K, Yamamoto A, Tagaya M. 2003. SVIP is a novel VCP/p97-interacting protein whose expression causes cell vacuolation. Mol Biol Cell 14:262–273.
- Zuber C, Fan JY, Guhl B, Parodi A, Fessler JH, Parker C, Roth J. 2001. Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc Natl Acad Sci USA 98:10710–10715.
- Appenzeller-Herzog C, Hauri HP. 2006. The ER-Golgi intermediate compartment (ERGIC): In search of its identity and function. J Cell Sci 119:2173–2183.
- Kornfeld R, Kornfeld S. 1985. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem 54:631–664.
- Kota P, Reeves PJ, Rajbhandary UL, Khorana HG. 2006. Opsin is present as dimers in COS1 cells: Identification of amino acids at the dimeric interface. Proc Natl Acad Sci USA 103:3054–3059.
- Lanctot PM, Leclerc PC, Escher E, Guillemette G, Leduc R. 2006. Role of N-glycan-dependent quality control in the cell-surface expression of the AT1 receptor. Biochem Biophys Res Commun 340:395–402.
- Sato BK, Schulz D, Do PH, Hampton RY. 2009. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell 34:212–222.
- Lass A, Kujawa M, McConnell E, Paton AW, Paton JC, Wojcik C. 2008. Decreased ER-associated degradation of alpha-TCR induced by Grp78 depletion with the SubAB cytotoxin. Int J Biochem Cell Biol 40:2865–2879.