Abstract
A number of studies using chimeric constructs made by fusing endoplasmic/sarcoplasmic reticulum calcium pump (SERCA) sequences with those of the plasma membrane located calcium pump (PMCA) have suggested that the retention/retrieval signal responsible for maintaining SERCA in the endoplasmic reticulum (ER) is located within the N-terminus of these pumps. Because of the difficulties in identifying the presence of constructs at the plasma membrane we have used a trans-Golgi network (TGN) marker to evaluate whether chimeric proteins are retained by the ER or have lost their retention/retrieval sequences and are able to enter the wider endomembrane system and reach the TGN. In this study, attempts to locate this retention/retrieval sequence demonstrate that the retention sequences are located not in the N-terminus, as previously suggested, but in the largely transmembranous C-terminal domain of SERCA. Further attempts to identify the precise retention/retrieval motif using SERCA1/PMCA3 chimeras were unsuccessful. This may be due to the fact that introducing SERCA1 sequences into the C-terminal PMCA3 sequence and vice versa disrupts the organization of the closely packed transmembrane helices leading to retention of such constructs by the quality control mechanisms of the ER. An alternative explanation is that SERCAs have targeting motifs that are non-linear, being made up of several segments of sequence to form a patch that interacts with the retrieval machinery.
Introduction
Proteins required for the functioning of the endoplasmic reticulum (ER) are maintained in this organelle following their synthesis by a process involving retention and/or retrieval. The maintenance of soluble proteins, such as protein disulphide isomerise (PDI), in the ER by the process of retrieval is well characterized. PDI carries a C-terminal KDEL retrieval sequence (Munro and Pelham Citation1987) reviewed in (Pelham Citation1990), and any KDEL-tagged proteins that have escaped the ER and entered the ER-Golgi intermediate compartment (ERGIC) en route for the secretory pathway are recognized by the ERD2 receptors and returned to the ER ( CitationLewis and Pelham Citation1990; Lewis et al. Citation1990; Tang et al. Citation1993). A similar strategy exists for transmembrane proteins carrying a cytoplasmic C-terminal KKXX motif or a cytoplasmic N-terminal double arginine motif (Nilsson et al. Citation1989; Jackson et al. Citation1990; Schutze et al. Citation1994). Binding of the KKXX motif to COPI ensures ER retrieval (Letourneur et al. Citation1994). COPI binding also appears to play a role in the retrieval of double arginine motifs, though the interactions appear more complex (O'Kelly et al. Citation2002; Yuan et al. Citation2003; Brock et al. Citation2005; see review Beck et al. Citation2009). Not all proteins are maintained by a process of retrieval; for example, it appears that the chaperone calnexin does not exit the ER at a significant rate under most circumstances (although see Okazaki et al. Citation2000) and is largely kept in the ER by retention rather than retrieval (Hammond and Helenius Citation1994).
The mechanism of retrieval has not been identified for all ER-retrieved proteins. ER calcium pumps, for example, contain no canonical ER retrieval sequences. Although human SERCA3 has a C-terminal KDEL sequence, homologues from other mammals do not contain this motif. In addition to the SERCAs, retrieval signals have not yet been identified in the Sec61α, β and γ proteins or in the calcium pump regulators phospholamban and sarcolipin, even though these transmembrane proteins are retrieved from the ERGIC (Greenfield and High Citation1999; Newton et al. Citation2003; Butler et al. Citation2007). Attempts have been made to identify potential targeting sequences within SERCA structures, all of them involving the characterization of chimeras built from SERCA sequences and sequences from the plasma membrane located calcium pumps (PMCAs) or of truncated PMCA (Foletti et al. Citation1995; Zvaritch et al. Citation1995; Fresu et al. Citation1999; Guerini and Carafoli Citation1996; Guerini et al. Citation2000; Newton et al. Citation2003). These studies are difficult to interpret because mis-folded proteins are detected by ER quality control mechanisms that lead to ER retrieval and ultimately to protein degradation by the proteosome (see reviews: Ellgaard et al. Citation1999; Cobbold et al. Citation2003).
In this study we have generated 20 SERCA1/PMCA3 chimeras in an attempt to identify the sequence(s) responsible for targeting. Previous studies have had difficulties in identifying constructs as being either plasma membrane or ER located on the basis of visualization of confocal images. Specific SERCA/PMCA chimeras result in apparent ER localization or PM localization in the same cell type within individual experiments (Foletti et al. Citation1995; Guerini et al. Citation2002). It is also difficult to show with certainty that PMCA/SERCA chimeras are located in the plasma membrane. In order to circumvent this difficulty, in this study chimeras that have lost their ER retrieval signals have been identified through their co-localization with a marker of the trans-Golgi network (TGN) (Banting and Ponnambalam Citation1997) indicating that such proteins have escaped the ER/ERGIC and entered the wider endomembrane system en route to the plasma membrane.
Materials and methods
Construction of chimeric calcium pumps from SERCA1 and PMCA3
Calcium pump chimeras were constructed from cDNAs encoding rabbit SERCA1 and rat PMCA3, provided by Dr P. Adams and Prof. G. Shull as in Newton et al. (Citation2003). The human SERCA2b construct was a gift from Prof. F. Wuytack (Eggermont et al. Citation1989). All constructs were built in pcDNA3.1 (+) (Invitrogen) and were positioned upstream of the EGFP gene, producing a C-terminal fluorescent tag. Where possible, sections of SERCA1 or PMCA3 were amplified using the polymerase chain reaction (PCR) and flanked by restriction sites, allowing ligation of the inserts into either SERCA1 or PMCA3 sequences. Multi-step PCR was employed to build chimeras in which smaller sections of sequence were exchanged (Grandori et al. Citation1997). Using this approach, inserts were built from two or three PCR products using further rounds of PCR in which products were combined. Inserts were then cut using the appropriate restriction enzymes and cloned into SERCA1 or PMCA3.
CD8 reporter constructs
The original CD8 construct was a gift from Dr M. N. J. Seaman (Seaman Citation2004). The CD8 gene was amplified from this construct using PCR and inserted into the pcDNA3.1 (+) vector upstream of EGFP, resulting in a C-terminally tagged CD8-EGFP construct. CD8 SERCA M10 and CD8 PMCA M10 were then built, in which the transmembrane domain of CD8 was replaced by the SERCA1 or PMCA3 M10 sequence. Multi-step PCR was used to insert the short sequence encoding SERCA1 or PMCA3 transmembrane domains into the CD8 sequence.
COS-7 cell culture and transfection
COS-7 cells were harvested at 80% confluency and seeded onto cover-slips in 24-well plates. Cells were then transfected with vector DNA encoding the appropriate construct using FuGENE-6 (Roche) according to the manufacturer's instructions. Cells were left for two days before cover-slips were removed and viewed.
Localization of the trans-Golgi by immunofluorescence
Following transfection and incubation for two days, COS-7 cells were treated with 5 μg/ml brefeldin A (Sigma-Aldrich) for 1 h, to improve separation and visualization of the trans-Golgi network from the ERGIC (see Banting and Ponnambalam Citation1997; Greenfield and High Citation1999). Cells were fixed for 15 min in ice-cold methanol and pre-incubated in PBS Triton X-100 (0.01%) supplemented with 2% low fat dried milk for 30 min. Primary antibody (sheep anti-human TGN46; Serotech) was added at a 1:50 dilution and cells incubated for 1 h. Cells were then washed and then treated with secondary antibody (rabbit anti-sheep IgG conjugated to Texas Red; Abcam) diluted 1:100, for 1 h. Cover-slips were washed and mounted in mowiol (0.1% citifluor).
Permeabilization of COS-7 cells
Characterization of constructs containing the C-terminus of SERCA2b was carried out by a selective permeabilization protocol modified from (Butler et al. Citation2007). Two days after transfection in 24-well plates, COS-7 cells expressing the appropriate constructs were washed in PBS and fixed with 4% formaldehyde (in PBS) for 15 min. Cells were washed with PBS alone (no membrane permeabilization), or supplemented with 0.01 mg/ml saponin (for plasma membrane permeabilization) or 0.1% Triton X-100 (for entire membrane permeabilization). Blocking was carried out with PBS supplemented with 2% low fat dried milk and either no detergent, saponin or Triton X-100 (buffer P) for 30 min. Mouse anti-GFP antibodies (Roche) were added at a 1:100 dilution in the appropriate buffer P for 1 h at 37°C. Antibody was removed, and cells washed in buffer P. Anti-mouse Texas Red conjugated secondary antibody (GE healthcare) was added at 1:50 in Triton X-100 buffer P and incubated for 1 h at 37°C. Secondary antibody was removed and cells washed in Triton X-100 buffer P and once in PBS before being mounted with mowiol.
Confocal microscopy
Samples were viewed with a Leica TCS SP2 confocal microscope under oil with a 40× objective and pinhole diameter of Airy 1. Leica LCS software was used for image acquisition and analysis. EGFP was excited at a wavelength of 488 nm and emission measured between 500–600 nm and Texas Red was excited at 594 nm and emission measured between 605 and 700 nm. All emission bandwidths were freely adjustable with the acousto-optical beam splitter (AOBS™). Ar458, 476, 488, 496, 514, and HeNe 594 lasers were used to excite fluorescence. Confocal images were analysed using ImageJ software from NIH (http://rsbweb.nih.gov/ij/)
Results and discussion
Previous studies by a number of investigators have suggested that the ER retrieval sequence of SERCA1 is located in the N-terminal segment of SERCA1 (Foletti et al. Citation1995; Guerini and Carafoli Citation1996; Guerini et al. Citation1998; Newton et al. Citation2003). Using PMCA3/SERCA1 chimeras the requirement of the N-terminal 211 amino acids of SERCA for ER retrieval was reported (Foletti et al. Citation1995; Newton et al. Citation2003) and using a similar approach showed that the first 85 amino acid residues of SERCA were sufficient to retain a PMCA/SERCA chimera in the ER. Guerini et al. (Citation1998) suggested that a retention/retrieval sequence was located in the first 28 residues of SERCA; however, these authors pointed out that additional retrieval signals might occur elsewhere since ER retention was demonstrated only in 50% of the transfected cells; in the others the construct appears to be located in the plasma membrane.
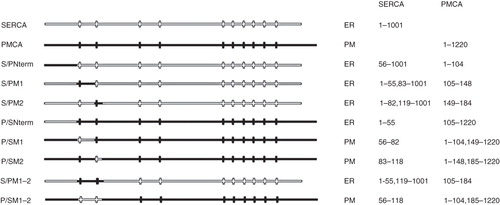
In this study, to clarify the location of a potential N-terminal ER retrieval sequence, eight chimeras were constructed in which N-terminal segments of SERCA1 were replaced with the cognate sequences of PMCA3 and vice versa (see ). Since it is not possible to discern from a protein's location in the ER whether it is there because it contains an ER retrieval/retention sequence or because it is aberrantly folded and has been detected and retained by the quality control apparatus of the ER the most informative chimeras are those that escape the ERGIC en route for the plasma membrane. These chimeras must lack retention signals and thus help to identify where retention signals are located.
Figure 1. Summary of chimeras used to clarify the role of the N-terminus of SERCA1 in ER retrieval. All of the chimeras were tagged with EGFP at their C-termini. The designated names are provided in the left column and their proposed locations, from analysis of co-localization studies, are indicated as well the precise sequences making up the chimeras. The filled horizontal lines represent PMCA3 sequence and the unfilled lines SERCA1 sequence. The vertical lines indicate the approximate locations of the transmembrane sequences.
The classical technique for the detection of surface-exposed proteins relies on labelling cell surface exposed residues with biotin and detecting them using labelled streptavidin (Sargiacomo et al. Citation1989). In the context of these chimeras, using such an approach to demonstrate that chimeras are located in the plasma membrane is problematic; this is because insufficient sequence is exposed on the extracellular surface. To provide an alterative method of discriminating between ER-retained constructs and constructs bound for the plasma membrane, we have used co-localization of constructs with a TGN marker to indicate whether the chimeras have been retained by the ER by a process of retention/retrieval as occurs for SERCA1, or whether they are en route for the plasma membrane via the TGN, as occurs for PMCA3 (Newton et al. Citation2003). This has led us to recategorize the location of three of our constructs that were studied previously (Newton et al. Citation2003) (see below for further details). However, because it is possible to say with certainty whether a construct has moved into the TGN, this method of categorizing location is a more objective way of discriminating between constructs maintained in the ER and those which escape from the ERGIC to enter the wider endomembrane system than a method simply based on a comparison with confocal images of fluorescently tagged SERCA1 and PMCA3.
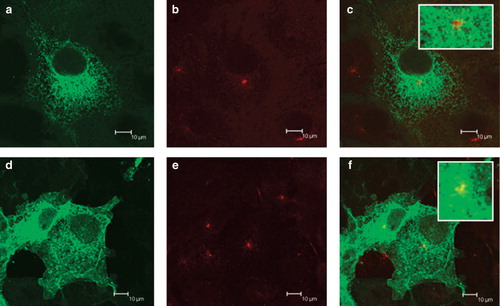
The expression patterns of SERCA1-EGFP and PMCA3-EGFP in COS-7 cells show what would be expected of a protein retained in the ER and a protein that has escaped the retention machinery and is en route for the plasma membrane respectively (see ). The SERCA-EGFP construct shows the reticular pattern () demonstrated by other studies for this particular construct (Newton et al. Citation2003; Butler et al. Citation2007). The TGN, revealed by anti-TGN46 antibodies demonstrated a perinuclear location () that does not co-localize with SERCA-EGFP (). This shows, as has been demonstrated previously (Newton et al. Citation2003) that although SERCA-EGFP can enter the ERGIC it does not progress to the TGN and is retrieved. By contrast PMCA3-EGFP, as well as showing a reticular pattern in the cytoplasm, appeared at the cell boundary indicating that it was targeted to the plasma membrane (). That the construct had escaped the retrieval mechanisms of the ERGIC and cis-Golgi was demonstrated by its co-localization with the TGN marker, TGN46, (, ).
Figure 2. Expression of EGFP-tagged SERCA1 and PMCA3 by COS-7 cells. COS-7 cells were transfected with DNA encoding for SERCA1-EGFP (a–c) and PMCA3-EGFP (d–f) in the expression vector pcDNA3.1(+). After two days the cells were treated with brefeldin A, fixed in methanol, permeabilized with triton X100, before treating with antibodies directed against TGN46, followed by a Texas Red conjugated secondary antibody. Laser scanning confocal images of EGFP are shown in panels a and d, Texas Red in b and e and overlaid images in c and f. Scale bars are indicated.
Eight constructs were produced swapping the transmembrane domains TM1 or TM2 of SERCA1 or PMCA3 with those from its cognate partner (). Of these, five (S/PNterm, S/PM1, S/PM2, P/SNterm and S/PM1–2) were retrieved to the ER. The location of S/PM1–2 (which is typical of this group) is shown in where the location of this construct is similar to that of SERCA-EGFP in and additionally, the construct was not co-localized with the TGN marker () indicating that it has been retrieved from the ERGIC. In contrast the three constructs (P/SM1, P/SM2 and P/SM1–2) have a similar localization to PMCA3 ( panels a, d and j) and show co-localization with the TGN marker ( panels c, f and l) indicating that they have escaped from the ERGIC. These three constructs that have evaded retrieval from the ERGIC and entered the endomembrane system en route for the plasma membrane demonstrate that the retrieval sequence cannot be located in either of the first two transmembrane domains of SERCA, because replacement of PMCA3 sequences with either TM1 or TM2 or TM1–2 from SERCA did not lead to retrieval of the construct, indicating that the retrieval sequence must have been absent from these SERCA sequences.
Figure 3. Location of chimeras used to examine the role of the N-terminus of SERCA1 in ER retrieval. COS-7 cells were transfected with the chimeric constructs all tagged at their C-terminus EGFP: P/SM1 (panels a–c); P/SM2 (panels d–f); S/PM1-2 (panels g–i); P/SM1–2 (panels j–l). After two days the cells were treated with brefeldin A, fixed in cold methanol, and permeabilized with triton X100 before incubating with antibodies against TGN46, visualized with Texas Red conjugated to a secondary antibody. Confocal images of EGFP fluorescence are shown in panels a, d, g, j, m, p, s and v. Texas Red images are shown in panels b, e, h, k, n, q, t and w. The merged images are shown in panels c, f, i, l, o, r, u and x.
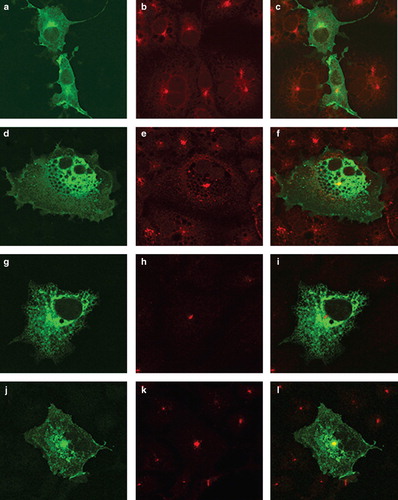
It is not possible to say whether the retention signal is located in the N-terminal 55 residues of SERCA before TM1 because although replacing the first 104 N-terminal residues of PMCA3 with the 55 N-terminal residues of SERCA (construct P/SNterm) resulted in retention by the ER (showing a similar pattern to that shown for S/PM1–2 in panels g–i), so did replacement of the N-terminus of SERCA with the N-terminus of PMCA3 (S/PNterm). This suggests that joining SERCA1 and PMCA3 at this point leads to a construct that is recognized by the ER quality control machinery, leading to retention. Thus it is not possible to assert with any certainty, based on this information, whether a retrieval signal is located in this N-terminal segment of SERCA.
Interestingly, Foletti et al. (Citation1995) assigned the SERCA retrieval signal to the first 85 residues of the protein based on the observation that replacing the cognate sequence of PMCA with the first 85 residues of SERCA resulted in a construct that was located in the ER. However, they did not rule out that this was the result of retention of mis-folded protein. Similarly Guerini et al. (Citation1998) reported that grafting the first 28 residues of SERCA onto the complete sequence of PMCA also resulted in ER retention. Again however, it is not possible to argue definitively that the ER retention signal is located at the N-terminus of SERCA because mis-folding of the construct cannot be ruled out. Zvaritch et al. (Citation1995) identified what they referred to as a possible masked retention signal at the C-terminus of PMCA which was revealed when PMCA was truncated at residue Arg1087, but as with all the other constructs, it is not possible to exclude retention due to mis-folding. In addition, this sequence is unlikely to be of relevance to SERCA targeting because there is no analogous sequence in SERCA; it is located in the cytoplasmic C-terminus of PMCA, a domain which is absent from SERCA.
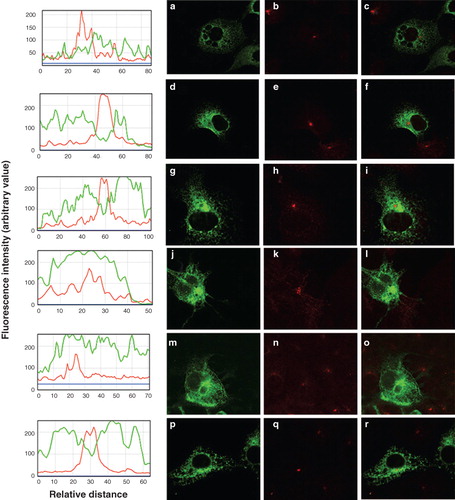
Six further chimeras were produced to investigate the possible location of a retrieval sequence in the C-terminal half of SERCA (). Of the six constructs four (P/SNtermM1–2, S/PNtermM1–2, S/PM3–4 and P/SM5–10) were located in the ER ( panels a, d, g and p, respectively) and failed to reach the TGN ( panels c, f, i and r, respectively) indicating that these constructs were retrieved from the ERGIC. To further demonstrate the absence of these constructs from the TGN the data were evaluated using ImageJ software (http://rsbweb.nih.gov/ij/). The profile of fluorescence intensity across the TGN, shown by the line scan, falls dramatically for the EGFP signals from constructs P/SNtermM1–2, S/PNtermM1–2, S/PM3–4 and P/SM5–10, consistent with the failure of these constructs to reach the TGN ( plots adjacent to panels a, d, g and p, respectively). Constructs P/SM3–4 and S/PM5–10 appeared to be located in the plasma membrane ( j and m, respectively) and were co-localized with the TGN marker ( l and o, respectively) indicating that these constructs have escaped from the ERGIC into the wider endomembrane system en route for the plasma membrane. In this case the profile for the EGFP fluorescence is maintained across the TGN indicating that it is not excluded from this organelle ( plots adjacent to panels j and m, respectively).
Figure 4. Summary of chimeras used to examine the role of SERCA1 C-terminus in ER retrieval. All of the chimeras were tagged with EGFP at their C-termini. The designated names are provided in the left column and their proposed locations, from analysis of co-localization studies, are indicated as well the precise sequences making up the chimeras. The filled horizontal lines represent PMCA3 sequence and the unfilled lines SERCA1 sequence. The vertical lines indicate the approximate locations of the transmembrane sequences.
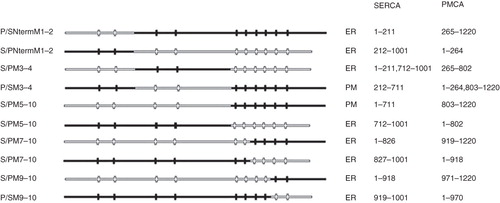
Figure 5. Location of chimeras used to examine the role of the C-terminus of SERCA1 in ER retrieval. (a) COS-7 cells were transfected with the chimeric constructs all tagged at their C-terminus with EGFP: P/SNtermM1–2 (panels a–c); S/PNtermM1–2 (panels d–f); S/PM3–4 (panels g–i); P/SM3–4 (panels j–l); S/PM5–10 (panels m–o); P/SM5–10 (panels p–r). After two days the cells were treated with brefeldin A, fixed in cold methanol, and permeabilized with triton X100 before incubating with antibodies against TGN46, visualized with Texas Red conjugated to a secondary antibody. Confocal images of EGFP fluorescence are shown in panels a, d, g, j, m and p. Texas Red images are shown in panels b, e, h, k, n and q. The merged images are shown in panels c, f, i, l, o, and r. The images from panels 5 c, f, i, l, o and r were analyzed using ImageJ software from NIH (http://rsbweb.nih.gov/ij/). The profiles of fluorescence intensity across the TGN for EGFP and Texas Red fluorescence are plotted in green and red respectively.
These data indicate that the retrieval sequence cannot be located in the N-terminal segment of SERCA as suggested previously as S/PM5–10 which contains the first 711 residues of SERCA1 escapes retrieval from the ERGIC to reach the TGN (). The proposal that residues 212-711 of SERCA1 do not contain the retrieval sequence is supported by the observation that P/SM3–4 has the same cellular distribution as S/PM5–10 () and PMCA (). These two observations suggest that the retrieval sequence has to be located in the C-terminus of SERCA1 (residues 712-1001). The ER location of constructs in this group containing the PMCA3 C-terminus can be explained by the retrieval of mis-folded proteins. For example, if the C-terminus of SERCA1 contains a retrieval signal, construct P/SNtermM1–2 would be expected to be located in the plasma membrane, since it contains the C-terminus of PMCA3, but it is found in the ER. Presumably mis-folding of the construct has led to its retention by the quality control apparatus of the cell. In a previous paper (Newton et al. Citation2003) S/PNtermM1–2 and P/SM5–10 were assigned to the PM and S/PM5–10 to the ER, but here S/PNtermM1–2 and P/SM5–10 are re-assigned to the ER and S/PM5–10 to the PM. The reason for the differences between these studies is unclear. As discussed above, there are examples in the literature of specific SERCA/PMCA chimeras that result in apparent ER or PM localization in the same cell type within individual experiments (Foletti et al. Citation1995; Guerini et al. Citation1998, Citation2002). We have found the identification of chimeras at the PM challenging. To overcome this problem we have utilized the TGN marker to determine whether chimeric proteins have escaped from the ERGIC into the wider endombrane system. This provides a more reliable method for detecting ER retention of chimeric proteins rather relying on the resemblance of the localization to that demonstrated by unmodified SERCA or PMCA. In view of this we believe that the current study provides a more accurate evaluation of the targeting of these chimeras. We cannot however exclude the possibility that the constructs used previously (Newton et al. Citation2003) may have contained spurious mutations that may have led to apparent mis-targeting.
A further series of constructs were produced to attempt to identify the location of the retrieval sequence within the C-terminus of SERCA1 (). In these constructs the transmembrane domains of the SERCA1 and PMCA3 C-terminus (TMs 5–10) were swapped. However swapping TM 7–10 and TM 9–10 (S/PM7–10, P/SM7–10, S/PM9–10 and P/SM9–10) resulted in constructs that were all located in the ER (see Supplementary Figure 1, panels a, d, g and j, respectively, available online) and did not co-localize with the TGN marker (see Supplementary Figure 1, panels c, f, i and l, respectively, available online) indicating that they were all retrieved from the ERGIC. Since none of the constructs escaped from the ERGIC to reach the TGN it is not possible to say from these data where the retrieval sequence is located. Presumably some of these constructs are in the ER because they are mis-folded and not because they contain an ER retention signal.
To determine whether the C-termini of SERCA1 and PMCA3 in isolation could act as retention signals, the N-terminal domain of SERCA1, including transmembrane domains TM1 and TM2, was attached via a flexible amino acid linker (Gly-Gly-Gly-Gly-Ser)2 to the C-terminus (starting at TM9) of SERCA1 and PMCA3 (constructs SM1–2M9–10 and SM1–2/PM9–10, ). Domain M1 was included to ensure that the construct was targeted to the ER and M2 was added to ensure the correct topology of M9 and M10. Both constructs were located in the ER and neither co-localized with the TGN marker (see Supplementary Figure 2a–c and Figure 2d–f, respectively, available online) and thus provide no further information about the location of the ER retrieval sequence. In addition, the single transmembrane domain of CD8 (a plasma membrane located protein (see Supplementary Figure 3a–c, available online) was exchanged with M10 of SERCA1 or PMCA3 ( and Supplementary Figures 3d–f and Figure 3g–I, respectively, available online) to determine whether M10 of SERCA1 would result in the ER retention of this construct. Since substituting M10 of PMCA3 or SERCA1 for the CD8 TM domain resulted in location in the ER, the most likely explanation is that this is the result of the ER quality control mechanism retaining an aberrant protein.
Figure 6. Summary of other constructs used to identify the location of the C-terminal ER retrieval sequence of SERCA1. All of the constructs were tagged with EGFP at their C-termini. The designated names are provided in the left column and their proposed locations, from analysis of co-localization studies, are indicated as well the precise sequences making up the chimeras. The filled horizontal lines represent PMCA3 sequence and the unfilled lines SERCA1 sequence. The vertical lines indicate the approximate locations of the transmembrane sequences. The exceptions are construct CD8, which contains the human CD8 sequence and CD8-SERCA M10 and CD8-PMCA M10 in which the transmembranous domain of CD8 is replaced by M10 of SERCA1 and PMCA3, respectively. The dashed lines in constructs SM1–2M9–10 and SM1–2/PM9–10 represent flexible linkers between the SERCA1 and PMCA3 sequences.
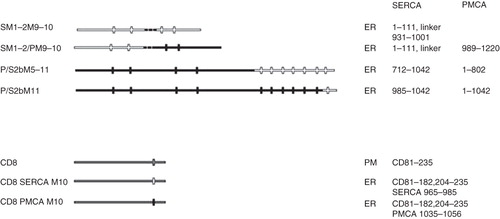
SERCA2b contains an additional transmembrane helix, designated M11 (Campbell et al. Citation1992). M11 was added to the C-terminus of PMCA3 to explore the possibility of producing constructs that would have their EGFP tag located in the extra-cellular environment and hence provide an additional means to demonstrate plasma membrane location. For example anti-EGFP antibodies should bind to non-permeabilized cells expressing such constructs if they were located in the plasma membrane. Two constructs were made; one in which M5 to M11 of SERCA2b was used to replace the cognate sequence of PMCA3 (P/S2bM5–11) and another where M11 of SERCA2b was added directly after PMCA3 M10 (P/S2bM11) (). Permeabilization studies revealed that, as expected, the EGFP tagged PMCA3 was detected by immunofluorescence microscopy only after the plasma membrane had been permeabilized with saponin allowing access of the anti-EGFP antibodies to the cytoplasm (see Supplementary Figure 4, panels a–h, available online). However, neither P/S2bM5–11 nor P/S2bM11 was detectable using anti-EGFP antibodies with intact cells, demonstrating that the EGFP tag of these constructs was not expressed on the cell surface. Neither was permeabilization of the plasma membrane by saponin sufficient to allow anti-EGFP antibodies access to the EGFP tag. The EGFP tag was only accessible after permeabilization of the ER by Triton X-100 (Supplementary Figure 4, panels i–o and p–v, available online). This shows that both P/S2bM5-11 and P/S2bM11 were located in the ER with the EGFP tag in the ER lumen, which is the topology expected for SERCA2b. P/S2bM11 failed to reach the plasma membrane and thus was unsuitable for the purpose for which it was designed. The addition of SERCA2b M11 to PMCA3 clearly leads to retention of the sequence though it is not evident whether this is the result of protein mis-folding or whether M11 contains retrieval information.
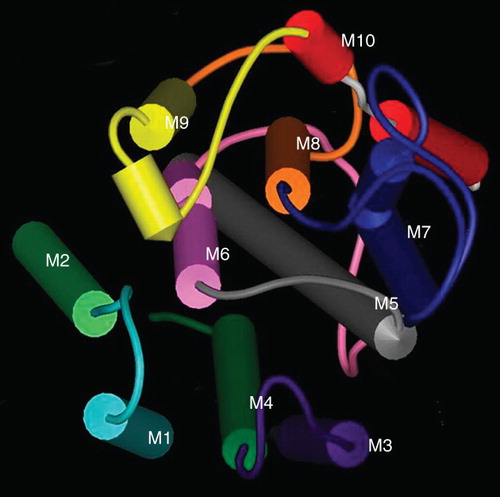
Overall, this study demonstrates that the ER retrieval sequence of SERCA1 is located in M5–M10 or the short cytoplasmic C-terminus. However, it seems that the helices M5–M10 of SERCA1 and PMCA3 are sufficiently dissimilar to lead to mis-folding when they are reorganized into chimeras. For example in construct S/PM7–10 () where SERCA1 M5 and M6 are substituted into the PMCA3 C-terminus, M5 of SERCA1 can interact with M7 and M8 of PMCA3 and M6 will interact with M9 and M8 of PMCA3 as shown by scrutinizing the X-ray structure of SERCA1 (). There are numerous possibilities in such interactions that might lead to non-native folding of chimeras involving helices M5–10 resulting in retention by the quality control machinery. A similar effect on protein folding may explain why S/PM7–10 and S/PM9–10 are also located in the ER.
Figure 7. The organization of the transmembrane helices of SERCA1. A space fill model of the transmembrane domains of SERCA1 produced using the coordinates deposited in the Protein Data Base (ID 1SU4). The helices have been colour-coded M1 light blue, M2 light green, M3 purple, M4 dark green, M5 grey, M6 pink, M7 blue, M8 orange, M9 yellow, M10 red. The view is from the ER lumen.
It is possible that binding of SERCA to proteins such as scaffolds or chaperones in the ER may contribute to the maintenance of the calcium pump in this organelle. Such binding may be disrupted in some of the chimeras in this study, resulting in detection of the protein at the plasma membrane. In this study it has been assumed that proteins localized to the plasma membrane have lost the ER retrieval signal of SERCA. Whether a chimera is at the plasma membrane due to its inability to bind a scaffold, a chaperone, or a retrieval receptor, its maintenance in the correct organelle is still compromised and therefore these chimeras can be considered to have lost at least a part of the sequence required for the ER localization of SERCA.
Of course the fact that SERCA, as well as other ER located proteins such as Sec61α have none of the canonical ER localization signals may indicate that their targeting motifs are non-linear and instead are made up of several segments of sequence, forming a patch on the surface of the protein. These signals therefore may only become apparent once the binding partner involved in the retrieval mechanism is characterized. Considering the findings presented here, it seems unlikely that more information about the SERCA retrieval signal will be obtained from studies with SERCA/PMCA chimeras, and instead focus should be shifted to the elucidation of the mechanism of retrieval and the protein machinery involved.
Supplementary Figures 1–4
Download MS Word (1,009 KB)Declaration of interest: We gratefully acknowledge the British Heart Foundation for funding this work. HW was a British Heart Foundation student (FS/06/018) and JB was funded by the British Heart Foundation (PG/04/057) during the undertaking of the research presented here. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Banting G, Ponnambalam S. 1997. TGN38 and its orthologues: Roles in post-TGN vesicle formation and maintenance of TGN morphology. Biochimica et Biophysica Acta-Molec Cell Res 1355:209–217.
- Beck R, Rawet M, Wieland FT, Cassel D. 2009. The COPI system: Molecular mechanisms and function. FEBS Letters 58:2701–2709.
- Brock C, Boudier L, Maurel D, Blahos J, Pin JP. 2005. Assembly-dependent surface targeting of the heterodimeric GABA(B) receptor is controlled by COPI but not 14-3-3. Molec Biol Cell 16:5572–5578.
- Butler J, Lee AG, Wilson DI, Spalluto C, Hanley NA, East JM. 2007. Phospholamban and sarcolipin are maintained in the endoplasmic reticulum by retrieval from the ER-Golgi intermediate compartment. Cardiovasc Res 74:114–123.
- Campbell AM, Kessler PD, Famborough DM. 1992. The alternative carboxyl termini of avian cardiac and brain sarcoplasmic-reticulum endoplasmic-reticulum Ca2+-ATPases are on opposite sides of the membrane. J Biolog Chem 267:9321–9325.
- Cobbold C, Monaco AP, Sivaprasadarao A, Ponnambalam S. 2003. Aberrant trafficking of transmembrane proteins in human disease. Trends Cell Biol 13:639–647.
- Eggermont JA, Wuytack F, Dejaegere S, Nelles L, Casteels R. 1989. Evidence for 2 isoforms of the endoplasmic-reticulum Ca-2+ pump in pig smooth-muscle. Biochem J 260:757–761.
- Ellgaard L, Molinari M, Helenius A. 1999. Setting the standards: Quality control in the secretory pathway. Science 286:1882–1888.
- Foletti D, Guerini D, Carafoli E. 1995. Subcellular targeting of the endoplasmic reticulum and plasma membrane Ca2+ pumps – a study using recombinant chimeras. FASEB J 9:670–680.
- Fresu L, Dehpour A, Genazzani AA, Carafoli E, Guerini D. 1999. Plasma membrane calcium ATPase isoforms in astrocytes. Glia 28:150–155.
- Grandori R, Struck K, Giovanielli K, Carey J. 1997. A three-step PCR protocol for construction of chimeric proteins. Protein Engineer 10:1099–1100.
- Greenfield JJA, High S. 1999. The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J Cell Sci 112:1477–1486.
- Guerini D, Carafoli E. 1996. The targeting of the plasma membrane calcium pump in the cell. Biosci Rep 16:129–137.
- Guerini D, Guidi F, Carafoli E. 2002. Differential membrane targeting of the SERCA and PMCA calcium pumps: Experiments with recombinant chimeras. FASEB J 16:519–528.
- Guerini D, Garcia-Martin E, Zecca A, Guidi F, Carafoli E. 1998. The calcium pump of the plasma membrane: Membrane targeting, calcium binding sites, tissue-specific isoform expression. Acta Physiolog Scan 163:265–273.
- Guerini D, Zecca-Mazza A, Carafoli E. 2000. Single amino acid mutations in transmembrane domain 5 confer to the plasma membrane Ca2+ pump properties typical of the Ca2+ pump of endo(sarco)plasmic reticulum. J Biolog Chem 275:31361–31368.
- Hammond C, Helenius A. 1994. Quality-control in the secretory pathway – retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, Golgi-apparatus. J Cell Biol 126:41–52.
- Jackson MR, Nilsson T, Peterson PA. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic-reticulum. EMBO J 9:3153–3162.
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic-reticulum. Cell 79:1199–1207.
- Lewis MJ, Pelham HRB. 1990. A human homolog of the yeast Hdel receptor. Nature 348:162–163.
- Lewis MJ, Sweet DJ, Pelham HRB. 1990. The Erd2 gene determines the specificity of the luminal ER protein retention system. Cell 61:1359–1363.
- Munro S, Pelham HRB. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899–907.
- Newton T, Black JPJ, Butler J, Chad J, Lee AG, East JM. 2003. Sarco/endoplasmic-reticulum calcium ATPase SERCA1 is maintained in the endoplasmic reticulum by a retrieval signal located between residues 1 and 211. Biochem J 371:775–782.
- Nilsson T, Jackson M, Peterson PA. 1989. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic-reticulum. Cell 58:707–718.
- O'Kelly I, Butler MH, Zilberberg N, Goldstein SAN. 2002. Forward transport: 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell 111:577–588.
- Okazaki Y, Ohno H, Takase K, Ochiai T, Saito T. 2000. Cell surface expression of calnexin, a molecular chaperone in the endoplasmic reticulum. J Biolog Chem 275:35751–35758.
- Pelham HRB. 1990. The retention signal for soluble-proteins of the endoplasmic-reticulum. Trends Biochem Sci 15:483–486.
- Sargiacomo M, Lisanti M, Graeve L, Lebivic A, Rodriguezboulan E. 1989. Integral and peripheral protein-composition of the apical and basolateral membrane domains in MDCK cells. J Membrane Biol 107:277–286.
- Schutze MP, Peterson PA., Jackson MR. 1994. An N-terminal double-arginine motif maintains type-II membrane-proteins in the endoplasmic-reticulum. EMBO J 13:1696–1705.
- Seaman MNJ. 2004. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol 165:111–122.
- Tang BL, Wong SH, Qi XL, Low SH, Hong WJ. 1993. Molecular-cloning, characterisation, subcellular-localisation and dynamics of p23, the mammalian KDEL receptor. J Cell Biol 120:325–338.
- Yuan HB, Michelsen K, Schwappach B. 2003. 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Current Biol 13:638–646.
- Zvaritch E, Vellani F, Guerini D, Carafoli E. 1995. A signal for endoplasmic-reticulum retention located at the carboxyl-terminus of the plasma-membrane Ca2+-atpase isoform 4CI. J Biolog Chem 270:2679–2688.