Abstract
Small ankyrin-1 is a splice variant of the ANK1 gene that binds to obscurin A. Previous studies have identified electrostatic interactions that contribute to this interaction. In addition, molecular dynamics (MD) simulations predict four hydrophobic residues in a ‘hot spot’ on the surface of the ankyrin-like repeats of sAnk1, near the charged residues involved in binding. We used site-directed mutagenesis, blot overlays and surface plasmon resonance assays to study the contribution of the hydrophobic residues, V70, F71, I102 and I103, to two different 30-mers of obscurin that bind sAnk1, Obsc6316–6345 and Obsc6231–6260. Alanine mutations of each of the hydrophobic residues disrupted binding to the high affinity binding site, Obsc6316–6345. In contrast, V70A and I102A mutations had no effect on binding to the lower affinity site, Obsc6231–6260. Alanine mutagenesis of the five hydrophobic residues present in Obsc6316–6345 showed that V6328, I6332, and V6334 were critical to sAnk1 binding. Individual alanine mutants of the six hydrophobic residues of Obsc6231–6260 had no effect on binding to sAnk1, although a triple alanine mutant of residues V6233/I6234/I6235 decreased binding. We also examined a model of the Obsc6316–6345-sAnk1 complex in MD simulations and found I102 of sAnk1 to be within 2.2Å of V6334 of Obsc6316–6345. In contrast to the I102A mutation, mutating I102 of sAnk1 to other hydrophobic amino acids such as phenylalanine or leucine did not disrupt binding to obscurin. Our results suggest that hydrophobic interactions contribute to the higher affinity of Obsc6316–6345 for sAnk1 and to the dominant role exhibited by this sequence in binding.
Introduction
Ca2+-dependent contraction in striated muscle depends upon the precise spatial regulation of the activity of the sarcoplasmic reticulum (SR) (Ebashi and Endo Citation1968). The SR stores Ca2+ and the contractile apparatus engages Ca2+ released from the SR to stimulate myosin ATPase and induce muscle shortening (Endo Citation1977). To coordinate activity efficiently, the SR is organized around each contractile unit, or sarcomere. In mammalian skeletal muscle, the two major components of the SR, the network SR (nSR) and junctional SR, surround each sarcomere at distinct locations, but the mechanisms that establish this organization and that stabilize it during the contractile cycle are poorly understood. We have been studying the association of the giant, modular, cytoplasmic protein, obscurin, with a small splice variant of the ankyrin 1 gene, small ankyrin-1 (also known as sAnk1 and Ank1.5), to learn how it contributes to the architecture of the SR (Kontrogianni-Konstantopoulos et al. Citation2003, Citation2006, Borzok et al. Citation2007, Busby et al. Citation2010, Citation2011).
sAnk1 is an integral membrane protein of the nSR, oriented in the membrane with two ankyrin-like repeat (ALR) motifs exposed to the cytoplasm (Porter et al. Citation2005, Borzok et al. Citation2007, Busby et al. Citation2011). In in vitro assays, these motifs bind to two nearby sites within the C-terminal, non-modular domain of obscurin A (Bagnato et al. Citation2003, Kontrogianni-Konstantopoulos et al. Citation2003, Armani et al. Citation2006, Busby et al. Citation2010). Obscurin is a member of the titin superfamily of giant muscle proteins (Kontrogianni-Konstantopoulos et al. Citation2009). The A isoform of obscurin in skeletal muscle is cytoplasmic and localizes to both the Z-disk and M-bands of sarcomeres, where it binds to several different proteins and concentrates at the periphery of the contractile apparatus (Young et al. Citation2001, Kontrogianni-Konstantopoulos et al. Citation2003, Bowman et al. Citation2007, Fukuzawa et al. Citation2008). sAnk1 concentrates in the nSR at these same locations (Zhou et al. Citation1997, Kontrogianni-Konstantopoulos et al. Citation2003). The binding of sAnk1 to obscurin A occurs with affinities in the submicromolar range, and several experiments suggest that they interact in vivo to organize the nSR around each sarcomere (Bagnato et al. Citation2003, Kontrogianni-Konstantopoulos et al. Citation2003, Lange et al. Citation2009).
The molecular mechanism of binding between obscurin A and sAnk1 is an active area of investigation. Recently our laboratory has determined the specific pairs of electrostatic interactions that contribute to the higher affinity (∼ 100 nM) binding site, located within amino acids 6316–6345 of obscurin A, a sequence with significant (∼ 35%) alpha-helical content (Borzok et al. Citation2007, Busby et al. Citation2010). A docked model of this complex was created using Brownian (BD) and Molecular dynamics (MD), and confirmed the experimentally identified pairs of charged residues (Busby et al. Citation2011). A second site, located within residues 6231–6260, binds to sAnk1 (Bagnato et al. Citation2003, Armani et al. Citation2006), but with lower affinity (∼ 350 nM) (Busby et al. Citation2010). This second site contains half the alpha helical content (∼ 17%) of the first (Busby et al. Citation2010). Although both sites on obscurin A use negatively charged amino acids to interact with arginine and lysine side chains exposed on the ALRs of sAnk1, their difference in affinity is unlikely to be explained solely by the difference in electrostatic interactions and secondary structure. We therefore investigated the possibility that hydrophobic interactions mediate binding of Obsc6316–6345 and thus contribute to its higher affinity for sAnk1.
In an earlier study, we described a homology model of sAnk1 that suggested the presence of a hydrophobic ‘hotspot’ flanked by charged residues that mediate binding to obscurin (Borzok et al. Citation2007). We subjected the model to 30ns MD simulations to verify that the conformation of sAnk1 was stable, with the hydrophobic residues exposed on the loops between the α-helices of each of its two ALRs (Busby et al. Citation2011). Although hydrophobic residues within ankyrin repeat (AR) proteins typically stabilize the packing of ARs, several crystal structures of complexes of AR proteins with their ligands, including the structure of IκBα bound to the p65 subunit of NFκB, suggest that hydrophobic contacts also contribute directly to binding (Huxford et al. Citation1998, Jacobs and Harrison Citation1998). Based on these precedents, we hypothesized that the hydrophobic hotspot on the surface of the sAnk1 model contributes to binding to obscurin. We further hypothesized that hydrophobic interactions mediated by these residues are more prominent in the binding of sAnk1 to its high affinity site than to its low affinity site on obscurin A. The data we present here, based on more extensive site-directed mutagenesis studies and MD simulations, verify these predictions and provide insight into ligand recognition by ALRs.
Methods
Generation of GST and MBP-fusion constructs
GST constructs were previously generated with the pGEX4T-1 (Amersham Biosciences, http://www.gelifesciences.com) vector for Rattus norvegicus Obsc6316–6435 and Obsc6231–6260 (Busby et al. Citation2010). PCR was used to amplify the sequence of interest, which was enzymatically digested with BamH1 and EcoR1 and ligated into the PGEX4T-1 vector. Fused constructs were transformed into DH5α competent cells (Invitrogen, Carlsbad, CA, http://www.invitrogen.com). After DNA was extracted, the sequence was verified by the UMB Biopolymer Core Facility.
sAnk1-MBP constructs were created with the pMal-c2x vector (New England Biolabs, Beverly, MA, USA). PCR was used to amplify the sequence of interest, encoding Rattus norvegicus sAnk1 residues 29–155, which was enzymatically digested with SalI and EcoR1 and ligated into the pMal-c2x vector which contains the maltose-binding tag at the C-terminus of the fusion protein. Constructs were transformed into DH5α competent cells and DNA was extracted and sequence verified.
Site-directed mutagenesis
The Quik-Change II mutagenesis kit (Stratagene, Cedar Creek, TX, USA, http://www.stratagene.com/) was used to generate mutations, following the manufacturer's instructions. Briefly, primers were made to cover the site of interest and mutations were generated via PCR. Template DNA was removed using Dpn-1 and the mutated plasmid was transformed into XL-1 competent cells. Mutagenesis was verified by sequencing.
Production of proteins
DNA was transformed into competent BL21* pLysS cells (Promega, Madison, WI, USA) to reduce proteolytic degradation. Cells were grown in sequentially diluted cultures, induced with 1 mM Isopropyl-β-D-thio-galactoside and allowed to produce protein for 4 h. Soluble fusion constructs were extracted from sonicated supernatants and purified by gravity flow affinity chromatography, following procedures recommended by the manufacturers. Multiple batches of all constructs were expressed and purified to repeat each of the experiments reported here.
Far-Western blots
Blot overlays were performed as previously described (Borzok et al. Citation2007) with one minor modification: to obtain the data for blot overlays, we used a goat anti-mouse 800 IRDye secondary antibody at 1:15,000 (LI-COR Biosciences, Lincoln, NE, USA, http://www.licor.com/) and we detected bound antibody with a LI-COR Odyssey Infrared Imager.
Surface plasmon resonance
Quantitative binding studies were performed by surface plasmon resonance (SPR) with a Biacore 3000 (GE Healthcare, http://www.gelifesciences.com), as described (Busby et al. Citation2010). To compare the ability of sAnk1-MBP wild type and mutants to bind to GST-Obsc6316–6345 and GST-Obsc6231–6260, we captured the GST constructs of both sequences with antibodies to GST (GE Healthcare) bound to a Biacore CM5 chip (GE Healthcare). We then applied sAnk1-MBP over a wide range of concentrations to determine the dissociation constant, KD. We eliminated non-specific binding by subtracting the signal generated by binding of the MBP fusion proteins to chips bound to GST alone, as well as the signal generated by exposing the chip charged with GST-obscurin to a solution blank. We fitted the data with a 1:1 binding model using Biaeval software to determine the kinetic rate constants and KD for the binding of each of the GST-obscurin constructs with all sAnk1-MBP constructs.
Reagents
Unless otherwise noted, all reagents were from Sigma Chemical Co. (St Louis, MO, USA) and were of the highest grade available. All primers for site-directed mutagenesis and cloning were made by the Biopolymer Core Facility, UMB.
Molecular dynamics simulations
Empirical force field calculations were performed with the programs CHARMM (Brooks et al. Citation2009) and NAMD (Phillips et al. Citation2005). Calculations used the CHARMM all-atom protein force field (MacKerell et al. Citation1998) including the CMAP backbone energy correction (MacKerell et al. Citation2004). The structure of the sAnk1-obscurin complex was taken from the 30ns snapshot from our previous MD simulation (Busby et al. Citation2011) and used as the initial structure for the wild type simulation and to model mutants of the sAnk1-obscurin complex. Images were generated using the VMD package. VMD is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign. The initial structures of mutant sAnk1-obscurin complexes (I102A and I102F) were prepared by replacing I102 with either alanine or phenylalanine in sAnk1, with the orientation of the side chains based on the internal coordinates in the CHARMM force field. Monomer sAnk1 structures were taken from each sAnk1-obscurin complex and used as initial structures.
Preparation for the MD simulations involved overlaying the structures of wild type (I102) and two mutant sAnk1-obscurin complexes (I102A and I102F) and the corresponding sAnk1 monomers with 76 and 62 Å pre-equilibrated cubic boxes of TIP3P water (Jorgensen et al. Citation1983) that contained 150 mM NaCl, for the complexes and monomers, respectively. Solvent molecules with non-hydrogen atoms within 2.8 Å of protein non-hydrogen atoms were deleted. Each system was minimized and heated to 298 K at a rate of 10 K/ps and equilibrated in the NPT ensemble (1 atm, 298 K) (Feller et al. Citation1995) for 5 ns. Simulations were performed with a 2 fs integration timestep using the SAE algorithm (Ryckaert et al. Citation1977) to constrain covalent bonds to hydrogens. Electrostatic forces were calculated with the particle mesh Ewald method (Darden et al. Citation1993) using a real space cutoff of 12 Å with a kappa value of 0.4 Å-1 and a 4th order spline interpolation. Van der Waals (VDW) forces were truncated with a cut-off distance of 12 Å with smoothing performed using a force switching function (Steinbach and Brooks Citation1994) starting at 10 Å. Production runs were performed in the NPT (1 atm, 298 K) ensemble for 30 ns each. Time frames from the trajectories were saved every 5 ps for subsequent analysis. The solvent accessibility was calculated using the method developed by Lee and Richards (Citation1971) with a probe radius of 1.4 Å. Block averages of the solvent accessibility were calculated for six 5 ns blocks, from which average and standard error were calculated.
Results
Our laboratory previously published a molecular model of sAnk1, based on the similar pattern of its charged and hydrophobic residues to the ARD of human Notch1 (Borzok et al. Citation2007). This model predicted that the side chains of residues Val-70, Ile-102, and Ile-103 cluster at the surface of sAnk1, where they are surrounded by the positively charged residues that mediate binding to obscurin. We subsequently modified this model, based on the observation that 30 ns MD simulation in aqueous solution resulted in a more relaxed confirmation (Busby et al. Citation2011). Inspection of the resulting structure () shows the two ALRs to be properly folded with a fourth, surface-exposed, hydrophobic side chain, Phe-71 (yellow), clustered together with the three original residues in the hydrophobic ‘hotspot’. We studied this cluster of hydrophobic residues to learn if it is involved in mediating binding to obscurin.
Figure 1. Images of sAnk-1 alone and the sAnk-157–122:Obsc6322–6339 complex. (A) Top and (B) side views of the hydrophobic hotspot within the binding region of sAnk1, from the 30 ns time frame of the sAnk-1 monomer MD simulations. The backbone is shown in red cartoon representation while the four surface-exposed hydrophobic residues: Val-70 (green), Phe-71 (yellow), Ile-102 (blue), and Ile-103 (red) are shown as surface representations, (C) Front and (D) rear views of the sAnk-157–122:Obsc6322–6339 complex. sAnk-1 is shown in a red cartoon representation along with the four residues shown in panels A and B; obscurin is shown in a blue cartoon representation with L6326 (green), V6328 (yellow), I6332 (blue), V6334 (red) and V6336 (cyan) shown as a surface representation. Full solvent accessible surfaces representations of both proteins are included as white transparent surfaces. Structures for panels C and D were from the 7.7 ns time frame of the 30 ns MD simulation of wild type sAnk1 in complex with Obsc6322–6339, which has an average VDW interaction energy of 1.62 kcal/mol between I102 of sAnk1 and V6334 of obscurin.
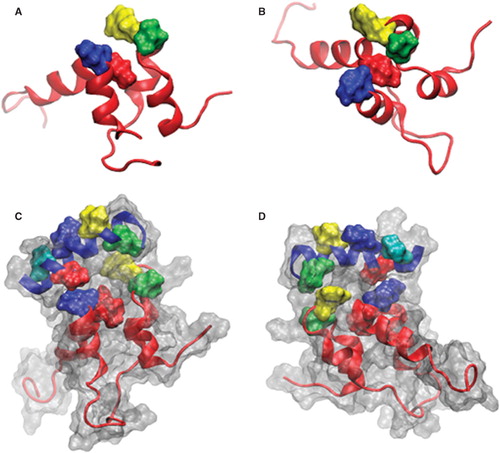
The potential interactions of the sAnk1 hydrophobic residues with obscurin are illustrated in . The structure, from the 7.7 ns time frame of the sAnk1-obscurin MD simulation, shows the extensive interface between these regions of the two proteins. Notable is the orientation of selected hydrophobic residues on the individual proteins that allow them to interact with the partner protein, thereby potentially contributing to binding.
To test the individual contribution of each of these hydrophobic residues to binding to the high and low affinity sites on obscurin A experimentally, we created MBP fusion constructs of sAnk1 with each of these residues mutated to alanine, and then assayed binding to GST fusion proteins of the high and low affinity binding sites on obscurin Obsc6316–6345 and Obsc6231–6260, respectively.
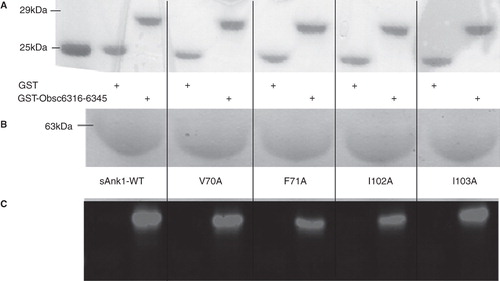
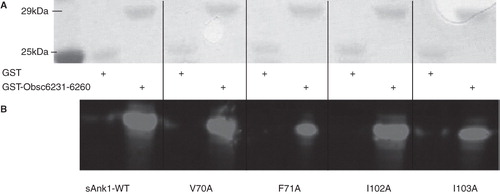
We first assayed binding of sAnk1 and each of its site-directed mutants to the high affinity site, Obsc6316–6345, in blot overlays. Proteins were quantified before SDS-PAGE with a Bradford assay (Bio-Rad, Hercules, CA, USA); staining with Ponceau Red confirmed equal loading and transfer (), as well as the proper molecular weight of each of the constructs. shows Coomassie Blue staining of a gel for the wild type and mutant sAnk1 variants also used in subsequent studies. Overlay of blots of GST-Obsc6316–6345 with the wild type and mutant sAnk1-MBP constructs shows that each of the four alanine mutations decreased specific binding to Obsc6316–6345 (). These results suggest that, in addition to electrostatic interactions (Busby et al. Citation2011), Obsc6316–6345 requires the entire surface-exposed cluster of hydrophobic amino acid side chains of sAnk1 to form a stable complex with ∼ 130 nM binding affinity. These results are consistent with our previously published blot overlays, that identified other key residues of sAnk1-obscurin binding, and that showed that binding could be completely inhibited by serial alanine mutagenesis (Borzok et al. Citation2007, Busby et al. Citation2011).
Figure 2. Alanine mutations within the hydrophobic hotspot of sAnk1-MBP reduce binding to GST-Obsc6316–6345. (A) The nitrocellulose membrane following transfer from the SDS-PAGE gel was stained with Ponceau Red confirmed equal loading and transfer of GST and GST-Obsc6316–6345. (B) Site-directed mutagenesis was used to create the four alanine-substituted MBP fusion protein mutants of sAnk1. Coomassie Blue stain shows the affinity-purified proteins, after analysis at equal load by SDS-PAGE. (C) For the blot overlay, a ladder of protein standards was run in lane 1 and the 25 kDa marker is shown. The remaining even lanes were loaded with GST and odd lanes with GST-Obsc6316–6345, at equal protein loads. The resulting gel was transferred to nitrocellulose. Each pair of lanes was then overlaid individually with each of the five sAnk1-MBP fusion proteins from B and probed with anti-MBP, followed by a fluorescent secondary antibody. The results show that each of the four alanine mutants of sAnk1 reduces binding to GST-Obsc6316–6345. Binding to each construct is specific, as none of the sAnk1 constructs bind to GST alone.
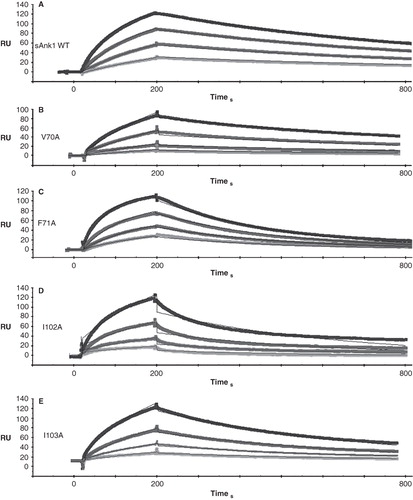
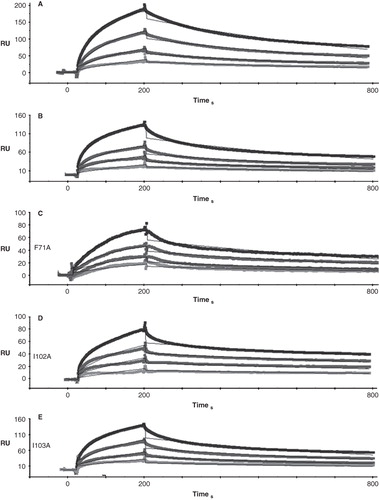
shows quantitative results of experiments in which we used SPR to measure the kinetic constants of binding of GST-Obsc6316–6345 to wild type sAnk1 and to each of the alanine-substituted mutants. Examples of SPR traces for GST-Obsc6316–6345 binding to wild type sAnk1 and all four sAnk1 mutants are shown with a 1:1 Langmuir fit in .
Figure 3. Representative SPR traces of GST-Obsc6316–6345 binding to wild type and mutant sAnk1. Four serial dilutions of sAnk1 (A) and the mutants (B-E), at concentrations of 1 μM and lower, were prepared and flowed over chip surfaces bound with GST-Obsc6316–6345 for 3 min, before dissociation. 1 μM is shown in black as the top curve followed by 500 nM, 250 nM and the lightest shade of grey at the bottom is 125 nM. The overall fit of the binding curves was based on the 1:1 Langmuir model provided by Biaeval software.
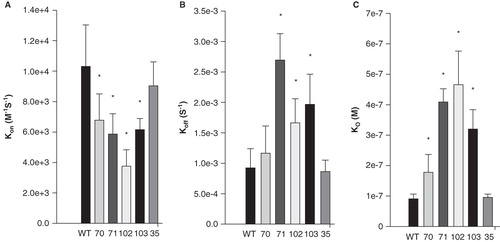
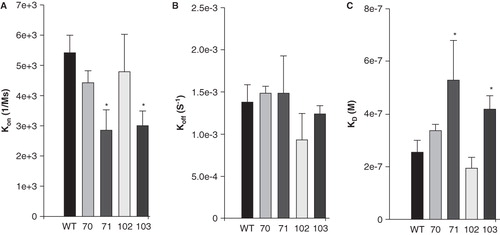
The ‘on’ rate, kon, for all four mutants was significantly decreased compared to wild type (); the greatest change was a ∼ 3-fold reduction in kon for I102A. The ‘off’ rate, koff, was significantly increased in each mutant except V70A (). These mutations of the hydrophobic residues in sAnk1 therefore caused a significant increase in the dissociation constant, KD, calculated as koff/kon, for binding to Obsc6316–6345. By contrast, we observed no significant change in the kon and koff for a control mutant, in which we exchanged a hydrophobic residue outside of the hydrophobic cluster, Phe-35, for alanine (sAnk1-F35A). Thus, the hydrophobic residues, predicted by modeling and MD simulation to be exposed in a ‘hotspot’ on the surface of sAnk1, contribute specifically to binding to Obsc6316–6345.
Figure 4. Quantitation of the kinetics for binding of hydrophobic mutants of sAnk1 to Obsc6316–6345. Surface plasmon resonance (SPR) was used to measure the binding kinetics of the high affinity binding site Obsc6316–6345 to wild type sAnk1 and mutants in which the hydrophobic residues were converted to alanines. (A) The on rate, kon, for all mutants was significantly decreased compared to wild type. (B) The dissociation rate, koff, for all of the mutants except V70A was significantly increased, compared to wild type. (C) The calculated KD's, based on the results in A and B, show that all mutants result in a decreased overall binding affinity compared to wild-type sAnk1. The F35A mutant protein was used as a control to show the alanine mutations are specific to the hydrophobic hotspot. n = 12 for WT, n = 9 for V70A, n = 5 for F71A, I102A, and I103A, and n = 4 for F35A. * = p < 0.05.
We next compared the binding of the same sAnk1 mutants to the more N-terminal, lower affinity binding site for sAnk1, Obsc6231–6260. Blot overlays indicated that only two of the four mutations, F71A and I103A, reduced binding of sAnk1 to Obsc6231–6260 ().
Figure 5. sAnk1-MBP mutations of two hydrophobic residues in hotspot reduce binding to GST-Obsc6231–6260. Methods were the same as in , except site-directed mutants of Obsc6231–6260 were assayed. As in , even lanes were loaded with GST and odd lanes with GST-Obsc6231–6260, at equal protein loads, except lane 1, which shows the standard proteins. Qualitatively, a reduction in binding is observed in lanes 7 and 11 corresponding to decreased binding affinity of F71A and I103A for Obsc6231–6260.
We again used SPR to quantitate the kinetics of binding between Obsc6231–6260 and the five sAnk1 constructs. Traces are shown, with a 1:1 Langmuir fit, in .
Figure 6. Representative SPR traces of GST-Obsc6231–6260 binding to wild type and mutant sAnk1. Experiments were performed as stated in , with GST-Obsc6231–6260 attached to the chip surface and sAnk1 analytes applied at serial dilutions from 1 μM (top curve in black) to 125 nM (bottom curve light grey).
SPR assays confirmed this and showed that these two mutants, but not V70A or I102A, significantly decreased kon () and increased the overall values of KD (). None of the mutants showed significant changes in koff (). The role of the cluster of hydrophobic residues at the surface of the ALR domain of sAnk1 therefore differs significantly, depending on the binding site on obscurin with which it interacts.
Figure 7. Quantitative evaluation of the binding kinetics for sAnk1 constructs binding to Obsc6231–6260. SPR was used to measure the binding kinetics of this region of obscurin to the wild type sAnk1 and its hydrophobic alanine mutants, as in , but the Obsc6231–6260 sequence was assayed. (A) The association rate, kon, for the F71A and I103A mutants of sAnk1, but not for the V70A or I102A mutants, was significantly decreased compared to wild type. (B) The dissociation rate, koff, for the mutants was not significantly different for binding to Obsc6231–6260 compared to wild type (p = 0.44 for V70A, 0.69 for F71A, 0.18 for I102A and 0.31 for I103A). (C) The calculated overall binding constants, KD, based on the results in A and B, show only the F71A and I103A mutants of sAnk1 with decreased binding affinities, compared to wild type. n = 4 for all panels, * = p < 0.05.
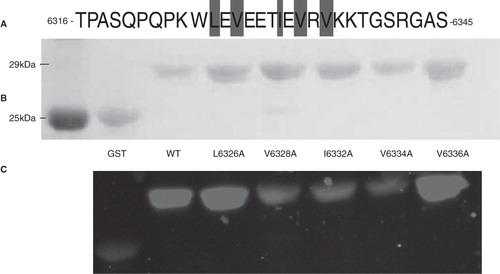
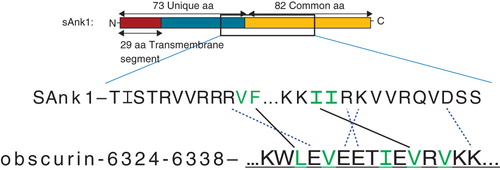
We next assayed the role of the hydrophobic residues in Obsc6316–6345 in binding to wild type sAnk129–155. This 30-mer has five hydrophobic amino acids (), Leu-6326, Val-6328, Ile-6332, Val-6334 and Val-6336, all of which are located near the middle of the sequence (). Alanine scanning mutagenesis of these residues, followed by blot overlays of binding to fusion proteins of the wild type sAnk1 fusion protein, showed that mutants of the middle three residues, V6328A, I6332A and V6334A, but not the two flanking residues, L6326A and V6336A, inhibited binding to sAnk1. This suggests that these hydrophobic residues in the central helical region of the high affinity binding site of obscurin participate specifically in binding to sAnk1.
Figure 8. Mutations of three hydrophobic residues within GST-Obsc6316–6345 reduce binding to sAnk1-MBP. (A) The primary sequence of this region of obscurin contains five hydrophobic residues within the binding site (Leu-6326, Val-6328, Ile-6332, Val-6334 and Val-6336), all highlighted. (B) Blot overlay was used to assay the binding of each of the alanine mutants to wild type sAnk1-MBP. Lanes: (1) Standard; (2) GST; (3) GST-Obsc6316–6345; (4) GST-Obsc6316–6345-L6326A; (5) GST-Obsc6316–6345-V6328A; (6) GST-Obsc6316–6345-I6332A; (7) GST-Obsc6316–6345-V6334A; and (8) GST-Obsc6316–6345-V6336A.
Similar studies of each of the six hydrophobic residues in the lower affinity binding site, Obsc6231–6260, showed no detectable differences in the binding of any mutant (V6233A, I6234A, I6235A, I6239A, V6242A, V6243A) to sAnk1, suggesting that none of them play as prominent a role in binding to sAnk1 as V6328, I6332 or V6334 (). We reported earlier that the three C-terminal hydrophobic residues, V6233, I6234 and I6235, together are necessary for binding to sAnk1 (Busby et al. Citation2010). As the F71A and I103A mutants of sAnk1 inhibit binding to Obsc6231–6260, we hypothesized that the general hydrophobic character within the N-terminal region of Obsc6231–6260 is necessary for binding to sAnk1. We tested this by comparing the binding of the triple alanine mutant, Obsc6231–6260-V6233A/I6234A/I6235A, with mutations at the N-terminus of Obsc6231–6260, to a double mutant of the two more C-terminal hydrophobic residues, Obsc6231–6260-V6242A/V6243A. The triple mutant but not the double mutant reduced binding to sAnk1 (), indicating that the N-terminal hydrophobic residues of Obsc6231–6260 are more likely to interact with the hydrophobic cluster on sAnk1, in particular with residues F71 and I103.
Figure 9. Multiple but not single alanine mutants of GST-Obsc6231–6260 reduce binding to sAnk1-MBP. (A) This region of obscurin has six hydrophobic residues located toward the N-terminus (Val-6233, Ile-6234, Ile-6235, Ile-6239, Val-6242 and Val-6243), all highlighted. (B) Blot overlays were used to assay the ability of each alanine mutant within Obsc6231–6260 to bind to wild type sAnk1-MBP. In addition, a triple mutant of the three N-terminal hydrophobic residues, as well as a double mutant of the last two neighboring hydrophobic residues, were tested. Lanes: (1) Standards; (2) GST; (3) GST-Obsc6231–6260; (4) GST-Obsc6231–6260-V6233A; (5) GST-Obsc6231–6260-I6234A; (6) GST-Obsc6231–6260-I6235A; (7) GST-Obsc6231–6260-I6239A; (8) GST-Obsc6231–6260-V6242A; (9) GST-Obsc6231–6260-V6243A; (10) GST-Obsc6231–6260-V6233A/I6234A/I6235A; and (11) GST-Obsc6231–6260-V6242A/V6243A.

We performed MD simulations for the ALR domain of wild type sAnk1 as a monomer and in complex with obscurin, to examine the nonpolar interactions involved in the binding of the high affinity site, Obsc6316–6345, to sAnk1. To identify the residues making hydrophobic contributions, we calculated the average per-residue solvent accessibilities in sAnk1, and compared them with those in the corresponding sAnk1-obscurin complexes for the four targeted hydrophobic residues. In addition, we estimated the contribution of direct residue-residue interactions to binding between obscurin and sAnk1 (I102) as the average Van der Waals (VDW) interaction energies for the targeted residues.
presents the results of our calculations of solvent accessibility and shows that significant changes occur with F71 and I102 upon complex formation. These results are consistent with our observation that mutation of those residues leads to the largest decrease in binding to Obsc6316–6345 in blot overlay () and SPR ( and ) experiments. Further analysis of the MD simulation based on the VDW interaction energies () indicate that F71 is in direct contact with L6326 of obscurin, while I102 is in direct contact with V6334. These findings are summarized in which also shows previously reported charged interactions.
Table I. Per-residue solvent accessibility (SA) of wild type and mutant sAnk1 as a monomer and in complex with obscurin. ΔSA = SA(Complex) − SA(Monomer).
Table II. VDW interaction energies between hydrophobic residues in sAnk1 and obscurin tested experimentally.
The importance of the interaction between I102 of sAnk1 and V6334 of obscurin is consistent with the blot overlay results for the obscurin mutants discussed above (). By contrast, the predicted interaction between F71 and L6326 is inconsistent with the results of our blot overlay experiments, which suggest that the L6326A mutation does not inhibit binding. Although Ala substitution of L6326 may facilitate binding by retaining some of the hydrophobic character of the side chain, we did not study this potential interaction further. We focused instead on sAnk1-I102 and its hydrophobic interaction with Obsc-V6334, for which all our results are consistent.
As our experiments and MD simulations both suggest that I102 makes direct contact with a hydrophobic side chain on obscurin, we undertook additional studies of its role in the binding of sAnk1 to Obsc6316–6345. These involved mutation of the Ile to Phe or Leu, to determine how subtle changes in the hydrophobic character of that residue could affect binding. The binding of the mutants was assayed in blot overlays and SPR experiments. We also generated the I102D mutant of sAnk1 and studied it in blot overlays to verify the importance of hydrophobicity at residue 102. In blot overlay assays, all three sAnk1 constructs with hydrophobic character at position 102 (, lanes 3, 5, 7) showed significant binding to Obsc6316–6345, whereas binding of sAnk1-I102D (lane 9) was considerably decreased. In the SPR experiments, the association curves for the two hydrophobic mutants were similar to the WT (). Although the dissociation curves for I102L matched those of wild type sAnk1, the I102F mutant formed an even stronger complex that dissociated ∼ 3 times slower than wild type (). This slow dissociation phase contributed to tighter overall binding (and a lower KD) for sAnk1-I102F (), whereas sAnk1-I102L showed a binding comparable to wild type sAnk1. By contrast, sAnk1-I102D showed no appreciable binding in SPR assays (data not shown).
Figure 10. Hydrophobic character at position 102 of sAnk1 is necessary for high affinity binding to Obsc6316–6345. SPR measured the binding kinetics for sAnk1 constructs with different amino acid residues at position 102. (A) The on rate, kon, for wild type sAnk1, sAnk1-I102F and -I102L show no differences in association profiles for Obsc6316–6345. (B) The dissociation rate, koff, for the mutants are not significantly increased compared to wild type. Rather, the dissociation rate of sAnk1-I102F for binding to Obsc6316–6345 is significantly decreased. (C) The calculated overall binding constant, KD, based on the results in A and B. Both I102F and I102L retain high affinity binding for Obsc6316–6345 while sAnk1-I102F forms a stronger complex than wild type. sAnk1-I102D did not produce curves that could be analyzed, due to poor association and dissociation. n = 5 for WT sAnk1, sAnk1-I102F and sAnk1-I102L, n = 3 for sAnk1-I102D, * = p < 0.05. (D) Blot overlay assays of GST (lanes 2, 4, 6, 8) and GST-Obsc6316–6345 (lanes 3, 5, 7, 9). Lane 1 shows standard proteins. Lanes 2–3 were probed with wild-type sAnk1, lanes 4–5 with sAnk1-I102F, lanes 6–7 with I102L, and lanes 8–9 with sAnk1-I102D.
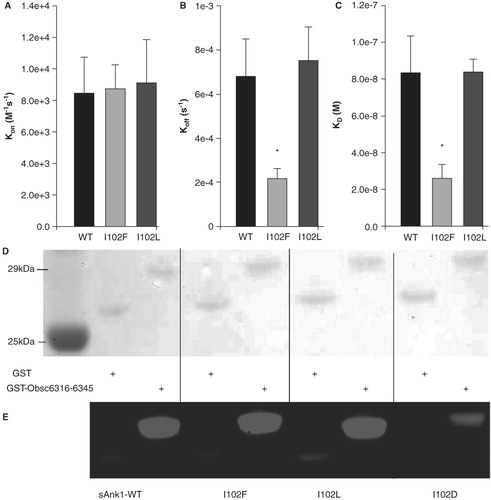
Figure 11. Residues involved in binding between the high affinity binding site of obscurin and sAnk1. Our lab has previously identified charged pairs of amino acids involved in binding between these proteins. The binding residues are matched to their partner with dotted lines. The hydrophobic residues necessary to preserve high affinity binding are highlighted in green. In addition, the favorable interactions between I102/V6334 and F71/L6326 as observed during MD simulations and I102/V6334 as confirmed biochemically, are shown with solid lines to highlight how high affinity binding is achieved.
We carried out MD simulations of the sAnk1-I102F mutant, both alone and in complex with Obsc6316–6345, to learn how this conservative substitution could improve binding. Notably, solvent accessibility of F71 decreased to a greater extent upon complex formation in the I102F mutant compared to wild type (). In addition, the VDW interaction energy of the Phe at position 102 was more favorable that Ile in the wild type (), and the VDW interaction energy of F71 with L6326 also became more favorable. These results are consistent with the increased binding of sAnk1-I102F to Obsc6316–6345 seen in the SPR experiments.
Discussion
We and others have proposed that the interaction between obscurin, a peripheral protein of the sarcomere, and sAnk1, an integral protein of the SR membrane, organizes the SR around the contractile apparatus in striated muscle (Zhou et al. Citation1997, Bagnato et al. Citation2003, Kontrogianni-Konstantopoulos et al. Citation2003, Armani et al. Citation2006, Giacomello and Sorrentino Citation2009, Lange et al. Citation2009). Previous studies revealed a high affinity interaction between these two proteins and showed that the absence of obscurin results in the disorganization of the SR or its network compartment (Bagnato et al. Citation2003, Kontrogianni-Konstantopoulos et al. Citation2003, Citation2006, Borzok et al. Citation2007, Lange et al. Citation2009, Busby et al. Citation2010). Binding is mediated by the interaction of tandem ALRs of sAnk1 with two possible binding sites on obscurin, located near each other in its C-terminal non-modular region and differing in affinity by ∼ 3-fold (Bagnato et al. Citation2003, Kontrogianni-Konstantopoulos et al. Citation2003, Armani et al. Citation2006, Borzok et al. Citation2007, Busby et al. Citation2010). We have been studying the interactions of these binding regions in obscurin in detail, with two aims: to learn how to disrupt the formation of the sAnk1-obscurin complex in situ without eliminating either protein from the sarcoplasm, and to understand the mechanism by which ARD proteins, including those with ALRs such as sAnk1, mediate protein-protein interactions. We focus on the latter question here. In particular, we show specific hydrophobic contacts between the hydrophobic ‘hotspot’ on the surface of sAnk1 and obscurin's high affinity binding site. We also show that general hydrophobic interactions, that cannot be ascribed to particular amino acid residues, contribute to the binding of obscurin's lower affinity site to sAnk1, representing a different mechanism of ALR recognition.
We previously reported that electrostatic interactions between specific charged residues on obscurin and the ALRs of sAnk1 contribute significantly to binding (Borzok et al. Citation2007, Busby et al. Citation2011). Site-directed mutagenesis, which created reciprocal changes in the charged residues on obscurin and sAnk1, identified particular pairs of interacting residues. We supported our findings with molecular modeling studies, which predicted an additional electrostatic interaction that we verified experimentally (Busby et al. Citation2011). Typically however, high affinity binding between proteins involves the burying of exposed hydrophobic residues in the bound complex, and our molecular models show sAnk1 to have a large hydrophobic ‘hotspot’ on its surface (). Here we describe additional site-directed mutagenesis and molecular modeling studies to identify the contributions of these hydrophobic residues to the binding between sAnk1 and two sAnk1-binding regions of obscurin. Studies of hydrophobic interactions are not compatible with approaches involving reciprocal mutagenesis, which would require significant changes in the nature of the residues under study (e.g., conversion to amino acids with polar or charged side chains). Despite this limitation, we successfully identified a particular pair of amino acid residues, I102 of sAnk1 and V6334 of obscurin that make direct hydrophobic contact in the complex.
The previous 3D model of sAnk1 reported by Borzok et al. (Citation2007) showed that it contains a ‘hotspot’ consisting of three hydrophobic residues, Val-70, Ile-102 and Ile-103, the side chains of which are clustered on the protein surface. These residues are surrounded by the positive charges that confer specificity to the negative residues on obscurin's high affinity binding site, Obsc6316–6345. As the majority of the charged residues around these hydrophobic residues have previously been implicated in binding to Obsc6316–6345, it seemed likely that the hydrophobic cluster was also involved. The binding interface between protein 4.1 and the erythrocyte anion exchanger is an example of hydrophobic side chains flanked by charged residues that are important for binding (Jöns and Drenckhahn Citation1992). Hydrophobic as well as electrostatic interactions participate in high affinity binding of spectrin and ankyrin (Kolondra et al. Citation2010) and of NFκB and IκBα (Bergqvist et al. Citation2006), which we used to model the high affinity binding site of obscurin (Busby et al. Citation2010). Consistent with these earlier studies, we show here that hydrophobic interactions contribute significantly to the binding of obscurin to the ALRs of sAnk1.
In the course of these experiments, we tested our earlier homology model of sAnk1 by subjecting it to 30 ns MD simulation. The results supported the general features of the original model, although deviations by about 8 Å (root mean square deviation of Cα atoms) from the original model occurred. This deviation from a characteristic AR has been previously observed with solved solution structures of AR proteins containing terminal ALRs, such as myotrophin and p19INK4d, in which the terminal ALRs project outward (Baumgartner et al. Citation1998, Yang et al. Citation1998). Both of their structures closely resemble the ALRs in our model of sAnk1, shown in .
Although the second helix in the second ALR of sAnk1 points away from the more N-terminal helical structures of the ALRs, the hydrophobic hotspot we originally predicted by homology modeling remains intact. Indeed, in addition to Val-70, Ile-102 and Ile-103, our MD model also predicts the presence of a fourth hydrophobic residue, Phe-71, in the cluster. All four of the surface exposed hydrophobic residues are highly conserved among species, as well as within other small splice variants of ankyrins (Hopitzan et al. Citation2005), consistent with their having an important role in the structure and function of sAnk1. A role for a set of four hydrophobic residues in binding to a scaffolding protein has been documented for NFκB and its interaction with IκBα (Huxford et al. Citation1998). Consistent with these examples, the hotspot formed by the four hydrophobic residues at the surface of sAnk1 form a docking site for the two binding sites of obscurin.
Previously we reported that the 3-fold difference in affinity between Obsc6316–6345 and Obsc6231–6260 is due to the 3-fold increase in the rate of dissociation of the latter sequence (Busby et al. Citation2010). Our data suggest that sAnk1's cluster of hydrophobic residues contributes to the slower dissociation of obscurin's high affinity site, and that each of the four residues in the hydrophobic hotspot of sAnk1 are involved ( and ). The kinetic constants change consistently, as the kon rate was reduced from 2- to 4.5-fold, whereas koff was increased (with the exception of V70A) by 2- to 3-fold, to decrease the overall binding affinity of all four alanine mutants. These differences are specific to sAnk1, as previous work from our laboratory has shown that the binding is not significantly altered by the exchange of the fusion protein partners (Kontrogianni-Konstantopoulos et al. 2003, Borzok et al. Citation2007). Mutagenesis of Obsc6316–6345 further suggested that three central hydrophobic side chains were involved in binding. MD simulations revealed two pairs of hydrophobic interactions, between sAnk1-F71 and Obsc-L6326, and sAnk1-I102 and Obsc-V6334 ( and ). The latter interaction, though not the former, is consistent with our biochemical studies. These results suggest that each of the side chains in the hydrophobic hotspot of sAnk1 is involved in binding to the high affinity site of obscurin, Obsc6316–6345, and that Obsc6316–6345 provides at least one, and likely more, side chains that interact with the hydrophobic hotspot of sAnk1.
Remarkably, however, this was not the case for the binding of sAnk1 to the low affinity site, Obsc6231–6260. Rather, our results suggest that only F71 and I103 are involved in binding to this sequence. Kinetic results show that the kon rate for both mutants decreased ∼ 2-fold compared to the wild type. Once the complex is formed, dissociation rates for all four mutants were similar to the wild type construct. Considered together with our observation that a cluster of hydrophobic residues near the N-terminus of Obsc6231–6260, but no individual residue, is required for binding to sAnk1, these results suggest that general, rather than specific, hydrophobic interactions are involved in low affinity binding of Obsc6231–6260 to sAnk1. As the affinity of sAnk1 for Obsc6316–6345 decreased to values comparable to those for Obsc6231–6260 when we eliminated specific hydrophobic contacts, these distinct hydrophobic interactions are likely to contribute to the different values of KD of these two sequences.
It is significant that the I102A mutant of sAnk1 is the most potent in inhibiting binding to Obsc6316–6345, but that it has no effect on binding to Obsc6231–6260, consistent with its involvement in specific binding of the former to sAnk1. In agreement with this, mutating I102 to Asp inhibited binding but retaining its hydrophobic character maintained high affinity binding, with the I102F mutant even enhancing binding. MD simulations indicate that this increase is due to an increase in the VDW interactions between I102F and Obsc-V6334 and a greater decrease in the solvent accessible surface area of I102F in the complex. Although we recognize that mutations of sAnk1 could result in altered folding that could affect the binding surfaces indirectly, our earlier studies of sAnk1 mutants showed no significant changes in circular dichroism or light scattering measurements (Borzok et al. Citation2007). We therefore favor the idea that the mutants alter the residues involved directly in binding.
The simulations further suggested more favorable interactions between F71 and L6326 of obscurin upon complex formation by the I102F mutant. We have summarized the favorable hydrophobic interactions between sAnk1 and Obsc6316–6345 in , which also shows previously identified charged interactions. F71 of sAnk1 is involved in binding to both regions of obscurin and may interact specifically with L6326 of obscurin. However, without the results of our molecular modeling studies, we would neither have identified this residue as a component of the hydrophobic hotspot nor assayed its role in binding. As the bulkiest hydrophobic residue, it is not surprising that F71 contributes to binding by promoting the formation of general or specific hydrophobic contacts for Obsc6231–6260 and Obsc6316–6345, respectively. I103 may also play a more general role in establishing hydrophobic interactions with Obsc6231–6260 than I102 because of its closer proximity to F71A (see ).
In summary, our studies indicate that the two short sequences at the end of obscurin, Obsc6316–6345 and Obsc6231–6260, use their hydrophobic side chains in different ways to promote binding to sAnk1. Our results are consistent with the idea that hydrophobic interactions contribute to a limited extent to the interaction of each obscurin binding site with sAnk1, and suggest that they are more important, as well as more specific, for the higher affinity binding site, Obsc6316–6345.
Although we have focused on the surface-exposed, hydrophobic hotspot on the convex surface of sAnk1, hydrophobic clusters on the concave surface of AR domains, in the beta loop region, can also mediate binding to ligands (Mosavi et al. Citation2004). This raises the possibility that sAnk1 can bind other proteins simultaneously with obscurin. Proteins that bind to the concave surface of sAnk1 remain to be identified.
Acknowledgements
The authors acknowledge a generous grant of computer time from the Pittsburgh Supercomputing Center and National Center for Supercomputing Applications. This work was supported by stipends to CW and BB from two training grants, T32 GM08181 (RJB, PI) and T32 AR07592 (Dr M. Schneider, PI), by grants from the NIH (RO1 AR056330, to RJB; CA120215 and GM051501, to ADM) and the Muscular Dystrophy Association (to RJB), and by the University of Maryland Computer-Aided Drug Design Center.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Armani A, Galli S, Giacomello E, Bagnato V, Rossi D, Sorrentino V. 2006. Molecular interactions with obscurin are involved in the localization of muscle-specific small ankyrin1 isoforms to subcompartments of the sarcoplasmic reticulum. Exp Cell Res 312:3546–3558.
- Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. 2003. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol 160:245–253.
- Baumgartner R, Fernandez-Catalan C, Winoto A, Huber R, Engh RA, Holak TA. 1998. Structure of the human cyclin-dependent kinase inhibitor p19INK4d: Comparison to known ankyrin-repeat containing structures and implications for the dysfunction of tumor suppressor p16INK4d. Structure 6:1279–1290.
- Bergqvist S, Croy CH, Kjaergaard M, Huxford T, Ghosh G, Komives EA. 2006. Thermodynamics reveal that helix four in the NLS of NFκB p65 anchors IκBα, forming a very stable complex. J Mol Biol 360:421–434.
- Borzok MA, Catino DH, Nicholson JD, Kontrogianni-Konstantopoulos A, Bloch RJ. 2007. Mapping the binding site on small ankyrin 1 for obscurin. J Biol Chem 282:32384–32396.
- Bowman AL, Kontrogianni-Konstantopoulos A, Hirsch SS, Geisler SB, Gonzalez-Serratos H, Russell MW, 2007. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett 581:1549–1554.
- Brooks BR, Brooks CL 3rd, MacKerell AD JR, Nilsson L, Petrella RJ, Roux B, 2009. Charmm: The biomolecular simulation program. J Comput Chem 30:1545–1614.
- Busby B, Willis CD, Ackermann MA, Kontrogianni-Konstantopoulos A, Bloch RJ. 2010. Characterization and comparison of two binding sites on obscurin for small ankyrin-1. Biochemistry 49:9948–9956.
- Busby B, Oashi T, Willis CD, Ackermann MA, Kontrogianni-Konstantopoulos A, MacKerell AD, 2011. Electrostatic interactions mediate binding of obscurin to small ankyrin 1: Biochemical and molecular modeling studies. J Mol Biol 408:321–334.
- Darden T, York D, Pedersen L. 1993. Particle mesh ewald-an N.log(N) method for ewald sums in large systems. J Chem Phys 98:10089–10092.
- Ebashi S, Endo M. 1968. Calcium ion and muscle contraction. Prog Biophys Mol Biol 18:123–183.
- Endo M. 1977. Calcium release from the sarcoplasmic reticulum. Physiol Rev 57:71–108.
- Feller SE, Zhang YH, Pastor RW, Brooks BR. 1995. Constant pressure molecular dynamics simulations: The langevin piston method. J Chem Phys 103:4613–4621.
- Fukuzawa A, Lange S, Holt M, Vihola A, Ferreiro A, Udd B, 2008. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: Implications for hereditary myopathies. J Cell Sci 121:1841–1851.
- Giacomello E, Sorrentino V. 2009. Localization of ank1.5 in the sarcoplasmic reticulum precedes that of SERCA and Ryr: Relationship with the organization of obscurin in developing sarcomeres. Histochem Cell Biol 131:371–382.
- Hopitzan AA, Baines AJ, Ludosky MA, Recouvreur M, Kordeli E. 2005. Ankyrin-G in skeletal muscle: Tissue-specific alternative splicing contributes to the complexity of the sarcolemmal cytoskeleton. Exp Cell Res 309:86–98.
- Huxford T, Huang DB, Malek S, Ghosh G. 1998. The crystal structure of the IκBα/NFκB complex reveals mechanisms of NFκB interaction. Cell 95:759–770.
- Jacobs MD, Harrison SC. 1998. Structure of an IκBα/NFκB complex. Cell 95:749–758.
- Jöns T, Drenckhahn D. 1992. Identification of the binding interface involved in linkage of cytoskeletal protein 4.1 to the erythrocyte anion exchanger. EMBO J 11:2863–2867.
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935.
- Kolondra A, Lenoir M, Wolny M, Czogalla A, Sikorski AF, Grzbek M. 2010. The role of hydrophobic interactions in ankyrin-spectrin complex formation. Biochim Biophys Acta 1798:2084–2089.
- Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. 2003. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell 14:1138–1148.
- Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, 2006. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J 20:2102–2111.
- Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. 2009. Muscle giants: Molecular scaffolds in sarcomerogenesis. Physiol Rev 89:1217–1267.
- Lange S, Ouyang K, Meyer K, Cui L, Cheng H, Lieber RL, 2009. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J Cell Sci 122:2640–2650.
- Lee B, Richards FM. 1971. The interpretation of protein structures: Estimation of static accessibility. J Mol Biol 55:379–400.
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field M J, 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616.
- MacKerell AD Jr, Feig M, Brooks CL 3rd. 2004. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25:1400–1415.
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. 2004. The ankyrin repeat as molecular architecture for protein recognition. Prot Sci 13:1435–1448.
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, 2005. Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802.
- Porter NC, Resneck WG, O'Neill A, Van Rossum DB, Stone MR, Bloch RJ. 2005. Association of small ankyrin 1 with the sarcoplasmic reticulum. Mol Membr Biol 22:421–432.
- Ryckaert JP, Ciccotti G, Berendsen HJC. 1977. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys 23:327–341.
- Steinbach PJ, Brooks BR. 1994. New spherical-cutoff methods for long-range forces in macromolecular simulation. J Comput Chem 15:667–683.
- Yang Y, Nanduri S, Sen S, Qin J. 1998. The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin. Structure 6:619–626.
- Young P, Ehler E, Gautel M. 2001. Obscurin, a giant sarcomeric Rho guananine nucleotide exchange protein involved in sarcomere assembly. J Cell Biol 154:123–126.
- Zhou D, Birkenmeier CS, Williams MW, Sharp JJ, Barker JE, Bloch RJ. 1997. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J Cell Biol 136:621–631.
