Abstract
Aquaporins are water facilitating proteins embedded in the cellular membranes. Such channels have been identified in almost every living organism – including humans. These proteins are vital molecules and their malfunction can lead to several severe disorders and diseases. Hence, an increased understanding of their structure, function and regulation is of the utmost importance for developing current and future drugs. Heading towards this goal, the first problem to overcome is to acquire the proteins in sufficient amounts to enable functional and structural characterization. Using a suitable host organism, large amounts of target molecules can possibly be produced, but for membrane proteins limitations are frequently encountered. In the work described here, we have produced the 13 human aquaporins (hAQPs) in one of the most successful hosts for recombinant overproduction of eukaryotic proteins; the yeast Pichia pastoris, in order to explore the underlying bottleneck to a successful membrane protein production experiment. Here we present exceptional yield of hAQP1, whereas some other hAQPs were below the threshold needed for scaled up production. In the overproduction process, we have established methods for efficient production screening as well as for accurate determination of the initial production yield. Furthermore, we have optimized the yield of low producing targets, enabling studies of proteins previously out of reach, exemplified with hAQP4 as well as the homologue PfAQP. Taken together, our results. present insight into factors directing high production of eukaryotic membrane proteins together with suggestions on ways to optimize the recombinant production in the yeast P. pastoris.
Introduction
Membrane proteins serve crucial functions in the cell and they constitute the majority of all current drug targets (Lundstrom Citation2006). Thus, detailed understanding of the workings of this class of molecules is of great relevance for both academia and the pharmaceutical industry, encouraging biochemical, functional and structural analysis of this group of molecules. However, membrane proteins are generally poorly understood due to bottlenecks encountered all the way from production to characterization of the isolated protein and they are dramatically underrepresented in structural databases (White Citation2011). Hence, the first hurdle to encounter is the fact that the majority of membrane protein targets are present at very low concentrations in their native membranes (Mus-Veteau Citation2002), requesting novel innovative strategies for recombinant overproduction. Indeed, the main bottleneck for structural determination and characterization of a membrane protein today is the task of overproducing a stable and functional protein in sufficient amounts (Grisshammer and Tate Citation1995, Forstner et al. Citation2007). Membrane protein overproduction is often a matter of a trial-and-error exercise limiting the number of available targets to be studied (Grisshammer Citation2006). Interestingly, it has historically even been considered an art rather than science (Bonander and Bill Citation2009) giving the lack of knowledge and available methods to solve the problems associated with production of the specific protein of interest. Moreover, eukaryotic membrane proteins are known to be even more difficult to produce relative to their prokaryotic counterparts (Tate Citation2001, Grisshammer Citation2006). As a consequence, several ways of circumventing membrane protein overproduction exists, like extracting large quantities of protein from naturally abundant sources (Bill et al. Citation2011). While being quite successful, it limits the selection of targets, especially those of human origin. Consequently, for future progress on eukaryotic membrane proteins, this method has to be replaced by reliable recombinant overproduction, which takes development of effective strategies.
In general, there are reasons to avoid a prokaryotic host for production of eukaryotic membrane proteins; the translation rate, the translocon, and the lipid composition differ, which in combination could have a negative impact on the final yield (Tate Citation2001, Tate et al. Citation2003). The methylotrophic yeast P. pastoris is a commonly used eukaryotic host mainly due to its strong and tightly regulated alcohol oxidase 1 (AOX1) promoter used to drive recombinant protein production. In addition, the preferred respiratory growth of P. pastoris allows growth of high cell density cultures. Moreover, stable transformants from linearized vector DNA can easily be generated by homologous recombination resulting in stable host strains which can grow without selection pressure. Notably, the host P. pastoris has been a vital part of the pipeline leading up to structure determination of eukaryotic membrane proteins; P. pastoris is the most frequently used host-producing protein for structural characterization (Bill et al. Citation2011). Hence, as more researchers are attracted to use this production system in the future, increased understanding in the determinants of high membrane protein production levels is of vital importance.
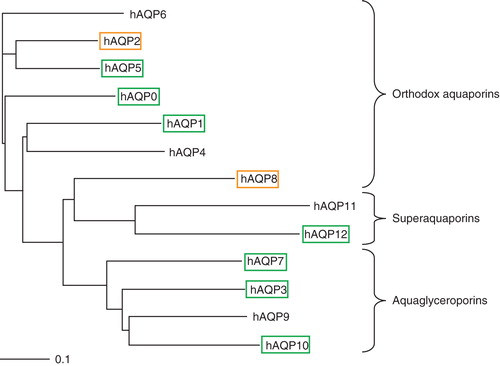
In order to achieve an increased understanding of factors directing a successful membrane protein production experiment we have taken advantage of the family of human aquaporins (hAQPs); in total 13 homologues membrane proteins. The human aquaporins have a high sequence similarity; 63% of the protein sequence is identical between hAQP2 and hAQP5 (). Aquaporins are commonly divided into two subgroups: the orthodox aquaporins (AQP0, AQP1, AQP2, AQP4, AQP5, AQP6, and AQP8), mainly transporting water, and the aquaglyceroporins (AQP3, AQP7, AQP9, and AQP10), transporting water and glycerol. The two remaining aquaporins (AQP11 and AQP12) still have undetermined transport specificity and are usually placed in their own group called superaquaporins. In the work described here, we have produced the hAQPs in P. pastoris with the goal to identify patterns discriminating high and low producers, respectively. In addition, we have evaluated various optimization approaches in order to increase an initially low production level for a certain target. Altogether, reliable comparisons between the production levels of different targets, or modifications thereof, have been possible due to the establishment of a quantitative production screening protocol.
Figure 1. Phylogenetic tree of the 13 human aquaporins. Aquaporins giving a high protein yield are shown in black boxes, poor yield in grey boxes, and proteins with a yield below the detection limit in the quantitative production screen are shown without any boxes. This Figure is reproduced in colour in Molecular Membrane Biology online where high protein yield is shown in green boxes and poor yield in orange boxes, respectively.
Identifying high yielding clones by production screening
In order to conclude if the protein yield is high enough for further production and characterization or if optimization is needed, it is essential to have a fast and reliable method to determine the initial production level of a certain protein target. Thus, we established a protocol for small-scale production screening of novel targets produced in P. pastoris from 2 ml BMMY cultures using a 24-deep-well-block where samples were taken 6, 22 and 54 hours post induction (Fantoni et al. Citation2007). Even though the aeration and agitation are sub-optimal in these small culture vials, they are sufficient for identification of clones having production levels higher than a certain threshold level, as verified by the controls included in each growth experiment.
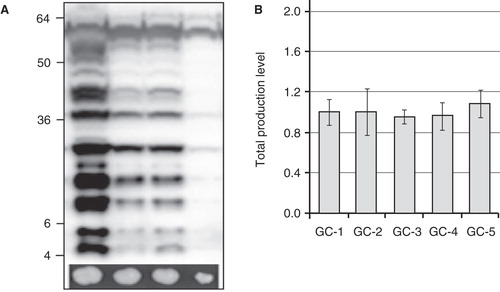
In theory, a higher gene dosage should, in principle, result in a higher level of protein product. While this, intuitively, is more likely to be the case for non trivial production targets like soluble proteins, it could be contra productive for membrane proteins possibly leading to intracellular traffic jam. To shed further light on this question, the correlation between the gene dosage and the aquaporin production level was evaluated providing an alternative small- scale production screen for integral membrane proteins in P. pastoris (Oberg et al. Citation2011a). In this screen we examined the relationship between the aquaporin yield and the ability of the recombinant P. pastoris cells to grow on high concentrations of Zeocin. Cells surviving on high Zeocin concentrations (2000 μg/ml) are likely to have multiple versions of the Zeocin resistance gene, thus, they will also contain multiple copies of the desired gene located on the same expression casette integrated into the P. pastoris genome. Indeed, for the human aquaporins there is a clear positive correlation between large colonies of the recombinant strain on high Zeocin concentrations and the production levels as analyzed by immunoblots (). Thus, increasing the gene dosage can be beneficial also for integral membrane proteins. Consequently, screening for improved growth on high Zeocin concentrations can circumvent the small scale production screening in 2 ml cultures having the additional advantage that more colonies could easily be screened in the search for high producing clones.
Figure 2. The screen on high Zeocin concentration. (A) Immunoblot showing the yields from the small scale production screen as compared to growth of the corresponding transformants on high Zeocin concentrations (2000 μg/mL), shown underneath. There is a clear correlation between large colonies grown on high Zeocin and the production yield analyzed by immunoblot. (B) Variation between the growth control (GC), SoPIP2;1, in five different experiments. The same control clone of SoPIP2;1 was cultured in five independent experiments (n = 3), over several years, and the production yield was estimated from independent immunoblots.
Following the observation that a high gene dosage is also beneficial for membrane protein production in P. pastoris, the influence of the transformation method used on the final aquaporin production level was evaluated. For generation of P. pastoris strains, the linearized expression plasmids have routinely been transformed by chemical transformation using the Lithium Chloride Method (Cregg and Barringer Citation1990). However, multiple insertion events occur at higher frequency when electroporation is used (Invitrogen Citation2010). Indeed, from our production trials of the human aquaporins in P. pastoris we found a significant improvement in yield when electroporation was used, which is most likely related to a higher frequency of multiple insertion events. Moreover, the impact by electroporation was mostly pronounced for the low producing aquaporins, suggesting major benefits from an increased number of transcripts for these targets (Oberg et al. Citation2011a).
Independent of the choice of the initial production screen, 2 ml cultures or growth on high Zeocin concentrations, and the transformation method, it is important early on to be able to accurately estimate the relative production level for a certain target. Also, since aquaporins are integral membrane proteins, proper localization to the P. pastoris membrane could be a useful indication of properly folded and functional protein, something that has been confirmed by functional analysis of purified protein reconstituted into liposomes for four different aquaporin targets with varied production levels (see next section below). Hence, after the identification of a clone with a high enough production level from the small scale production screen, a quantitative production screen including a cell fractionation experiment is performed from triplicate 25 ml BMMY cultures in Erlenmeyer flasks 6 hours after induction. Growth in a shaker flask allows better aeration and is less sensitive to measuring errors due to the larger volume as compared to the growth in deep well blocks used in the small-scale production screen. After cell breakage, the crude extract (500 g supernatant) representing the total production and the membrane fraction (100,000 g pellet) are analyzed using immunoblots (Nyblom et al. Citation2007, Oberg et al. Citation2009). As a part of the quality assurance in each individual experiment, a growth control (SoPIP2;1) is always included in the 25 ml quantitative production screen. Its main purpose is to make sure the growth and production experiment progressed as expected, but it also serves as a reference in the estimation of the relative yield of a novel target.
Quantitative immunoblots are a particular challenge where a high variation in the signal strength is commonly observed if particular caution is not taken. Especially, the choice of detection system is critical to ensure that the read out of each signal is within the linear range; some of the commercially available detection kits indicate saturation of the signal while others enhance signals as much as possible on the cost of linearity (unpublished work: F. Öberg and K. Hedfalk). In addition, samples run on different gels at different time points have to be compared in a precise manner which requires that an internal standard is included in the experimental setup. For our study on aquaporin production in P. pastoris, we systematically used a defined amount of purified hAQP1* from one single batch as internal standard in all immunoblot experiments. By using this internal standard a certain signal could be related to a specific protein concentration and hence, we could remove the variation arising from the deviation in the total signal intensity from individual immunoblot experiments. Notably, the level of the growth control (SoPIP2;1) only showed minor variation after scaling to the internal standard () providing an additional quality measurement for our established procedure used for quantitation of relative membrane protein production levels from individual immune blots.
Significant variation in recombinant production levels between homologous aquaporins
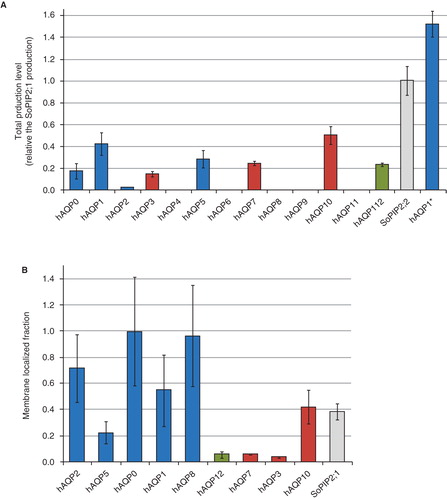
Even though the 13 human aquaporin homologues represent a family of highly-related proteins, there is a substantial variation in their yield when produced recombinantly in P. pastoris (). This is especially pronounced for the two closely-related proteins hAQP1 and hAQP4 where the former one is produced to high levels and the latter one is below the detection limit, as estimated from the quantitative production screen. Interestingly, the two aquaporins produced to the highest levels in P. pastoris, hAQP1 (Nyblom et al. Citation2007) and SoPIP2;1 from spinach leaves (Tornroth-Horsefield et al. Citation2006), also have a high natural abundance in their native plasma membranes. A possible interpretation could be that these proteins have natural intrinsic properties that allow them to be densely packed in the membrane and a concomitant high recombinant yield where aggregation and denaturation can be avoided.
Figure 3. Production of the human aquaporin homologues in P. pastoris. (A) Bar chart showing the total production level of the human aquaporins homologues produced in the host P. pastoris relative the SoPIP2;1 production, for which the production is set to one (shown in grey). The y-axis represents the average from triplicate cultures and error bars show the standard deviation (n = 3). (B) Bar chart showing the membrane localized fraction for the hAQPs produced in the quantitative production screen. The typical membrane insertion is shown for the reference protein SoPIP2;1. The aquaporins are grouped by their position in the phylogenetic three (). This Figure is reproduced in colour in Molecular Membrane Biology online where Orthodox aquaporins have blue bars, Aquaglyceroporins red bars and Superaquaporins green bars, respectively.
Aquaporins have been localized to most organs in the human body. Some of the aquaporins have only been detected in intracellular vesicles whereas others are found in the plasma membrane. At least two of the human aquaporins are known to be trafficked to the plasma membrane (AQP2 and AQP8), thus they reside in intracellular vesicles when trafficking has not been triggered (Garcia et al. Citation2001, Nedvetsky et al. Citation2009). During protein synthesis, signals within the protein sequence itself or external recognition systems can determine the sub-cellular localization for a specific protein. To see whether the endogenous localization of a certain protein correlates with its recombinant yield in the P. pastoris production system, we analysed the estimated yield versus the native localization for each human aquaporin target (). Although not statistically ensured, there is a clear tendency for proteins targeted to the plasma membrane to also have a high yield when recombinantly produced, as compared to the aquaporins found in intracellular vesicles. This observation suggests that yet unidentified signals or sequences within the protein itself are determinants for high or low recombinant yields, respectively.
Table I. Table showing the recombinant yield and sub-cellular localization in the native membrane for the different aquaporins found in mammalians. The localizations have been extracted from the shown references, but they are also stated, with only minor differences, in a review (King et al. Citation2004).
It is noteworthy that there is no apparent correlation between the recombinant yields () and the location of a specific aquaporin in the phylogenetic tree (). In contrast, such a correlation was apparent when analyzing the fraction of the protein localized to the membrane. To get the fraction of membrane localized material, the signal strength for the membrane fraction was divided by the total protein production for each specific target (). Due to error propagation arising from this exercise, the error bars are relatively wide. Nevertheless, our results showed a higher degree of membrane insertion for the orthodox aquaporins as compared to the superaquaporins and the aquaglyceroporins. A two- tailed Fisher's Exact Test comparing the orthodox aquaporins to the non-orthodox gives a statistically significant association (p < 0.05) where the orthodox aquaporins are more prone to be membrane integrated. Thus, this suggests that the substrate specificity also gives rise to protein properties beneficial for proper folding and membrane insertion.
To evaluate the quality of an overproduction experiment, it would be appealing to directly measure the amount of correctly folded aquaporin in a membrane. We have conducted initial tests with GFP tagged AQPs and produced them in the P. pastoris cell. Our results support the production of correctly folded and properly inserted aquaporin for both a high (hAQP1) and low (hAQP8) yielding target in this particular host (Oberg et al. Citation2011a). However, their degree of membrane insertion appears to be different. Without staining, it is hard to distinguish between the different cellular compartments, but nevertheless, there seems to be a higher degree of insertion of the low producing hAQP8 (also see ) as compared to the highly produced hAQP1 which has more pronounced intracellular GFP-signals (Oberg et al. Citation2011a). This implies the presence of aggregates due to saturation and overload of the cellular membrane protein secretion machinery for proteins being produced at exceptionally high yields, as has previously been suggested as a complication for related targets (Bonander et al. Citation2005).
To confirm our statement that proper localization to the P. pastoris membrane could be a useful indication of properly folded and functional protein, a selection of targets varying in production levels have been purified and reconstituted in liposomes. Notably, water transport was confirmed for all four targets, hAQP1 (Nyblom et al. Citation2007), hAQP4 (hAQP4m-N185D) (Oberg et al. Citation2011a), hAQP5 (unpublished work: F. Öberg, J. Sjöhamn, and K. Hedfalk) and hAQP10 (Oberg et al. Citation2011b) (). Moreover, the values for the osmotic water permeability (Pf) did not decrease as the protein yield was enhanced. For example, Pf was higher for hAQP1 than for hAQP5, indicating that the observed difference in functionality was due to the water transport capacity of the different aquaporin channels and not related to the overproduction yield as such. Hence, supported on the functional data for those aquaporins, our general conclusion is that human aquaporins recombinantly produced in the P. pastoris membrane are functional with the assumption that this is valid for the vast majority of the overproduced protein. Taken together, we can conclude that a high gene dosage in general also corresponds to a high total membrane protein production level for a certain target even though the yields of homologue proteins could vary. It is noteworthy that we have not seen any correlation between the production level and the function per se.
Table II. Table showing the osmotic water permeability (Pf) for hAQP4m-N185D (Oberg et al. Citation2011a), hAQP5 (unpublished work: F. Öberg, J. Sjöhamn, and K. Hedfalk), hAQP10 (Oberg et al. Citation2011b and unpublished work: F. Öberg, J. Sjöhamn, and K. Hedfalk), and hAQP1 (Nyblom et al. Citation2007). The Pf for the control liposomes in each experiment is shown just before each protein sample.
Fermentor growth is essential to achieve high yields of stable aquaporins
When a high producing clone has been identified using the quantitative production screen described above, controlled growth and induction is vital to optimize the production and make use of the full capacity of the P. pastoris system. Ideally, the up-scaled growth takes place in a fermentor where the growth conditions are monitored and controllable. Essentially, the oxygen addition can be sufficient due to efficient aeration and agitation while the methanol addition can be maximized without any risk of oxygen limitation. This is especially important for P. pastoris since the protein being responsible for the first oxidation reaction in the methanol utilization pathway, alcohol oxidase 1, has a low affinity for oxygen. Hence, large amounts of oxygen are needed to allow higher methanol concentrations and thereby take the full advantage of the AOX1 promoter (Cregg et al. Citation2000). Furthermore, the controlled regimes accessible in a fermentor also allow fine tuning of the AOX1 promoter by mixed feeding protocol. Consequently, by cultivation in fermentors, we were able to achieve exceptional high yields of hAQP1; 90 mg of pure protein was extracted per litre of culture (Nyblom et al. Citation2007).
Moreover, by using the appropriate sensors, the amount of viable cells in the reactor can easily be monitored providing a sophisticated tool to control active growth as compared to the classical optical density measurements often used to analyze shake flask cultures where all cells are taken into account. Especially for P. pastoris, measuring the fraction of living cells is a useful tool in avoiding addition of excessive, and hence, toxic, amounts of methanol. Interestingly, we observed a difference in growth characteristics between aquaporins from the different sub-families. In general, cell-producing orthodox aquaporins continued to grow upon the switch from glycerol to methanol while clones overproducing aquaglyceroporins had a much slower growth rate on methanol, sometimes with a concomitant decay of living cells. A plausible explanation could be that the slightly wider channels provided by the aquaglyceroporins could allow transportation of the small methanol molecule into the cell where it would be toxic. To evaluate this possibility, three amino acids lining the pore entrance at the ar/R constriction region in hAQP5 were mutated to create a larger pore and thereby changing the pore specificity to not only be selective to water, as has previously been made for AQP1 (Beitz et al. Citation2006). Indeed, the growth on methanol was hampered for this AQP5 mutant indicating that methanol might be taken up by the broader channel (unpublished work: F. Öberg, J. Sjöhamn, and K. Hedfalk).
Following this notion, a mixed feed containing 60% sorbitol and 40% methanol was evaluated for hAQP10 with the intention to lower the concentration of the toxic methanol for aquaglyceroporins (described in Oberg et al. Citation2011b). Sorbitol is selected since it provides an additional carbon source that does not give rise to the gene repression associated with glycerol. In addition, growth on sorbitol has been shown to increase the protein yield by increasing biomass (Jungo et al. Citation2007a, Jungo et al. Citation2007b) as well as by weakening induction to better match the requirements of the metabolism of the cells (Holmes et al. Citation2009). For hAQP10, we observed no significant changes in the total protein yield from the mixed feed. However, a protein degradation product commonly seen in the pure methanol feed disappeared suggesting a reduced cellular stress response under these conditions. Hence, mixed feed could possibly provide a solution for stable production of aquaglyceroporins in general giving homogenous samples suitable for further characterization.
Finally, quantitation of the yield from the fermentor cultures made it possible for us to verify the quality, reliability and scale ability of the quantitative production screen performed in Erlenmeyer flasks. As mentioned above, we produced 90 mg hAQP1 per litre of fermentor culture (Nyblom et al. Citation2007). In comparison, a third of that amount was obtained for hAQP10; 30 mg hAQP10 per litre of fermentor culture (Oberg et al. Citation2011b). The relationship between the yields from the scaled-up cultures were fully consistent with the corresponding quantitated yields for those targets in 25 ml cultures where hAQP1 (denoted hAQP1*) gives exactly three times higher yield than hAQP10 (). In comparison, targets resulting in low yields in , such as hAQP2, hAQP6, and hAQP8, have been problematic to overproduce to sufficient yields for subsequent analysis, even by fermentor growth. Hence, to our satisfaction, the data from the quantitative production screen can be extrapolated to the large-scale production in the fermentor which equips us with a very useful tool early in the process to determine the suitability of a certain target for large scale production and characterization, a tool that saves time and energy as well as money.
Experimental details
The scaled up growth of P. pastoris was done in 3-litre fermentors (Infors) with an Initial Fermentation Volume (IFV) of 1.5 litres and a start OD600 of about 0.2 according to the Invitrogen Pichia Fermentation Process Guidelines (Invitrogen, Citation2002). During growth, the temperature was set to 30°C, pH adjusted to 5 by NH3 addition, agitation and aeration were varied between 500–1500 rpm and 0.1–1 vvm, respectively, depending on the cell density, dissolved oxygen kept above 20% as verified by frequent oxygen spikes following carbon limitation and the density of living cells continuously monitored. The initial glycerol bath phase typically lasted for 20 hours consuming 60 g glycerol and giving an OD600 of about 25, followed by a glycerol fed-batch phase for 24 hours, just to acclimatize cells to growth during carbon-limited conditions. However, for targets with a limited growth on methanol, the length of the glycerol fed-batch phase could be doubled to allow the formation of more biomass before induction on methanol. The recombinant protein was produced during the methanol fed batch phase (200–400 ml MeOH) which lasted for 24–48 hours resulting in 400–600 g wet cells (OD600 about 200–400).
The construct design has a major impact on the final production level
The family of human aquaporins was used to evaluate several factors directing high production of eukaryotic membrane proteins including alterations around the initiation codon of the mRNA, fusion with Mistic or AQP, chimeric AQP constructs, as well as directing the topological maturation of the aquaporin monomer by directed mutagenesis.
The nucleotide sequence flanking ATG is of major importance
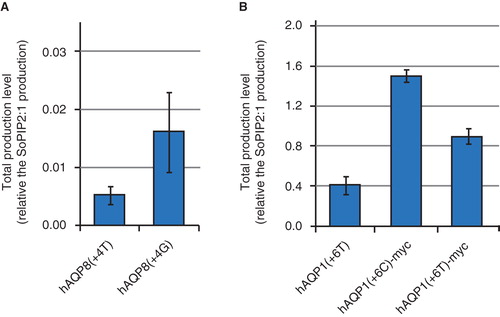
Translation is controlled by the rate of initiation, thus being affected by the 5′ sequence of the mRNA transcript (Romanos et al. Citation1992). For eukaryotic mRNA, the sequence flanking the initiator codon (underlined) was found to be (A/G)NNAUGG by Marilyn Kozak, henceforth called the Kozak sequence (Kozak Citation1981). In yeast, the consensus sequence is different; (A/Y)A(A/U)AAUGUCU (Cigan and Donahue Citation1987). For production of all 13 human aquaporins in P. pastoris (Oberg et al. Citation2009) we adapted the start sequence to the yeast consensus sequence A/YAA/TAATGTCT, as recommended in the EasySelect Pichia Expression Kit manual at the time (Invitrogen Citation2005). Hence, we consistently used Y (C/T) in the −4 position, AAA in the −1 to −3 positions and we aimed to mimic TCT at positions +4 to +6 by just allowing silent mutants in the second codon (). When looking at the difference in production level and the sequence around the initiator ATG, we observed a clear preference of G at the +4 position among the highly produced aqupaorins (5/7 in ). The likely explanation to this is that a G in this position increases the probability for a small non-polar amino acid, like alanine and glycine, which are important for a successful cleavage of the initiator methionine from the nascent polypeptide (Xia Citation2007). To shed further light on the importance of G at the +4 position, we mutated a T to G in this position for an aquaporin produced to moderate yields, hAQP8, which resulted in a mutation of serine to glycine. Notably, the production yield was increased upon mutation from the wild type TCT sequence to GCT (). Another interesting observation was the significant decrease in production level for hAQP1 upon imitation of the yeast consensus sequence (Oberg et al. Citation2009) as compared to a construct with unaltered sequence for the second codon (Nyblom et al. Citation2007) (). A putative determinant for this observation could be the intrinsic cytosine in the +6 position, which upon change to a thymine reduced the yield. Most interestingly, the sequence flanking the initiator codon of the highly produced AOX1 in the wild type P. pastoris cell, (30% of the soluble protein content in methanol grown cells) was found to be ACGATGG (De Schutter et al. Citation2009), perfectly matching the Kozak sequence. This fact combined with our observation, that mimicking TCT for the second codon did not have any positive influence on the aquaporin yield, leads to the conclusion that the eukaryotic consensus sequence is superior to the yeast consensus sequence when overproducing eukaryotic membrane proteins in the yeast P. pastoris. It is worth mentioning that the most recent EasySelect Pichia Expression Kit manual (Invitrogen Citation2010) recommends the (G/A)NNATGG consensus sequence with the notion that the yeast consensus sequence is a less strong alternative to the Kozak sequence showing a 2–3 fold effect in translation initiation efficiency. Interestingly, as compared to higher eukaryotes, translation in yeast is suggested to be more sensitive to secondary structures (Baim and Sherman Citation1988), supporting the importance of optimizing the initiation sequence.
Table III. Table showing protein production yields for all human AQPs overproduced in P. pastori s. The hAQPs are listed based on their production level, starting with the hAQP giving the highest yield. The silent mutations introduced in the second codon are underlined. The original sequence is given in brackets. The two smallest nonpolar residues (alanine and glycine) in the second position are underlined.
Figure 4. The influence of the second triplet on the production yield. Bar chart showing the total production yield for (A) two hAQP8 constructs with mutations in the +4 position, and (B) three hAQP1 constructs with mutations in the +6 position. Variations in the nucleotide sequence for the second codon are shown in brackets. All constructs have a C-terminal 6× histidine tag and some have the additional myc tag in the C-terminus, as shown in the figure. Production is relative to the SoPIP2;1 production, for which the production is set to one. The y-axis represents the average from triplicate cultures and error bars show the standard deviation (n = 3). This Figure is reproduced in colour in Molecular Membrane Biology online.
Evaluation of fusion proteins in P. pastoris
Apart from the flanking sequences of the initiator ATG codon, other aspects of the construct design were also evaluated for the human aquaporins produced in a simple eukaryotic host. The intention was to test whether the fusion to a stable and highly-produced protein, or peptide sequence, would enhance the production of the membrane protein of interest, analogous to similar approaches previously shown to be successful for soluble proteins in both E. coli and S. cerevisiae using maltose binding protein as well as other fusion partners (Wang et al. Citation2003, Hennig and Schafer Citation1998, Perez-Martin et al. Citation1997, Lian et al. Citation2009). In comparison, Mistic (acronym for ‘membrane-integrating sequence for translation of integral membrane protein constructs'), a membrane anchored protein found in the bacteria Bacillus subtilis, has been applied as fusion partner in bacteria (Roosild et al. Citation2005) where it has been able to increase the production of G-protein coupled receptors among others (Petrovskaya et al. Citation2010). To evaluate the possibility of transferring this approach to a eukaryotic host, the Mistic sequence was codon optimized (Opt-Mistic) for P. pastoris. Notably, our results showed a remarkably stable and high level of production of Opt-Mistic alone, in the same range as hAQP1 (Oberg et al. Citation2011a). When fusing Opt-Mistic to either a high or a low yielding AQP, the Mistic-AQP fusions resulted in a lower yield than for the AQP alone. Hence, these data imply that the concept of Mistic fusions to increase eukaryotic membrane protein yields cannot be directly transferred from E. coli to a eukaryotic host like P. pastoris. Consequently, we evaluated the use of an aquaporin with an intrinsically high and stable yield in this particular host, hAQP1, as fusion partner. However, both full length hAQP1 and parts thereof failed to increase the moderate production level of hAQP8. A set of chimeric constructs, where either the amino-terminus, transmembrane domain 1 (TMD1), TMD1-2 or TMD1-3 of hAQP8 were substituted for the corresponding protein sequence of hAQP1, did not lead to any improvements in recombinant production of hAQP8 either, indirectly indicating an importance of the carboxyl terminal half of hAQP1 as determinant for high production. Indeed, increased production was also observed for SoPIP2;1, having a high and stable production in itself, having its C-terminus swapped for the one of hAQP1* further supporting the importance of the hAQP1 C-terminus for high recombinant production levels (Oberg et al. Citation2011a).
Application of protein engineering to influence folding and target stability
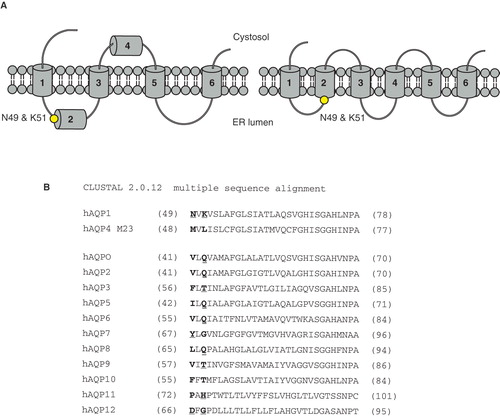
Yet another method of construct design was evaluated for the protein family of human aquaporins, this time based on detailed information available on the alternative topology maturation for AQP1 and AQP4, respectively. During folding of AQP1 in the ER, a four spanning intermediate is initially observed (Skach et al. Citation1994), which subsequently matures into the final six transmembrane fold (Lu et al. Citation2000) (). In contrast, AQP4 folds sequentially into the six spanning topology (Shi et al. Citation1995, Sadlish et al. Citation2005). Recent studies identified two amino acids near TMD2 as responsible for the difference in maturation between the two homologues aquaporins (Foster et al. Citation2000): Asn49 and Lys51 for AQP1 corresponding to Met48 and Leu50 in AQP4. Interestingly, engineering these specific amino acids from hAQP1 on hAQP4 (hAQP4m) lead to a swap in folding pathway (Buck et al. Citation2007). Hence, it was tempting to test whether the settings for topology maturation could also have a positive influence on the aquaporin production level in P. pastoris. Indeed, a significant yield increase was observed for hAQP4m, as compared to hAQP4, allowing scaled up production for this target (Oberg et al. Citation2009). When carefully comparing all the human aquaporin sequences in this specific region, a unique presence of positively charged amino acids is observed in hAQP1, something that is completely lacking in hAQP4 (). In correlation, the hydrophobicity of the amino acids in and in close proximity of a given amino acid segment is a known determinant for the insertion of that TMD into the membrane (Hessa et al. Citation2005, Hessa et al. Citation2007). Hence, a plausible explanation for the observed difference in the aquaporin folding pathway could be the variation in hydrophobicity in the TMD2 segment, where hAQP1 is less hydrophobic due to the two positively charged residues before TMD2. Hence, the concept of increasing the production yield by redirecting the folding pathway is unlikely to be transferable to other family members since no other family members possess two hydrophobic amino acids in these specific positions () and the folding pathway is yet to be established for those aquaporins. Nonetheless, the improvement of the hAQP4 yield by two point mutations exemplifies the great value of biochemical insight in the design of a successful overproduction experiment for a desired target.
Figure 5. The folding of human aquaporin 1. (A) Topology maturation for hAQP1 with a four spanning intermediate (left) and the mature fold with six TMD (right) (Lu et al. Citation2000). Asn49 and Lys51 in the N-terminal part of TMD2 (their position is marked with a black dot) are determinants for this fold (Foster et al. Citation2000). (B) Sequence alignment of all hAQPs around TMD2, with the amino acids corresponding to Asn49 and Lys51 in hAQP1 highlighted. For hAQP4 M23 the corresponding amino acids are Met48 and Leu50. Nonpolar amino acids (hydrophobic) are marked in bold and polar and electrically charged (hydrophilic) are marked in bold and underlined. This Figure is reproduced in colour in Molecular Membrane Biology online where specific positions in are marked with a yellow dot.
Gene optimization to increases yield
An intuitive and attractive strategy to improve the production level of a certain target is to optimize its gene sequence to perfectly match the codon usage of the selected host system and the ease of creating synthetic genes has improved. Services offering gene optimization take various parameters into account: codon usage, GC content, splicing signals, stable mRNA secondary structures, DNA repeats and, where applicable, the most efficient signal peptide is identified for the gene of interest (GeneArt Citation2011, GenScript Citation2011). Among those parameters, the main focus has been the optimizing of the translation process based on the codon usage and host adaptation presented almost 25 years ago (Sharp and Li Citation1987). However, to produce high yields of functional proteins, the nascent polypeptide must also be correctly folded, properly translocated, and undergo any necessary posttranslational modifications.
The genetic code provides 64 possible ways of combining a triplet of nucleotides, codons, to code for either one of the 20 amino acids or the three stop codons giving that most amino acids can be encoded by several codons, methionine and tryptophan being the only exceptions having only one triplet coding for them. Moreover, the frequency of a given combination of a nucleotide codon and tRNA anticodon shows a great variation between different organisms (). Therefore, as a consequence of recombinant gene expression, the difference in the preferred codon usage in the native gene and the tRNA pools of the expression system may cause inefficient translation hampering the protein production. One solution is to increase the intracellular tRNA pool by overexpressing genes coding for the rare tRNAs. This strategy, commonly used for E. coli, was evaluated for the malaria parasite aquaporin (PfAQP), a putative drug target in the search for more efficient anti malarial treatment, without any successful outcome (Hedfalk et al. Citation2008). In comparison, moving this concept to higher organisms has been found to be impractical (Gustafsson et al. Citation2004). Hence, the alternative approach of changing the gene is left as a plausible solution. By codon optimization it is possible to change the codons for the encoded amino acids using silent mutations and thereby make use of the more favourable codons of the host. Codon Adaptation Index (CAI) is a measurement of how well the codon usage in a protein coding DNA sequence matches the bias of a certain host. CAI can also be used as a numerical estimator of gene expressivity where a high value would indicate a highly expressed gene (Wu et al. Citation2005). shows CAI values for all hAQPs and PfAQP in their native host and in P. pastoris, respectively, using three different calculators; CAIcal (Puigbo et al. Citation2008), EMBOSS:cai (Bleasby Citation2001) and JCat (Grote et al. Citation2005), respectively. Two constructs optimized for production in P. pastoris have been included in and they are both named ‘Opt-'. As seen here, these genes have exceptionally high CAI values in P. pastoris as compared to their native gene sequences and would thus be expected to give a higher protein yield. Indeed, the gene optimization of PfAQP is an illustrative example of production improvements in P. pastoris giving a functional and membrane localized protein product (Hedfalk et al. Citation2008). We anticipated that the explanation to this was mainly related to the unusually low GC content: only 31% in the PfAQP gene and 24% in the coding region of the whole genome ( and ). Whether the reason for an increased production of PfAQP in P. pastoris is found in the codon adaptation, in the alterations of the GC content or in the combination thereof is not obvious. It has been reported that the change in GC content is the major contributor to the increased translational efficiency in P. pastoris (Sinclair and Choy Citation2002) and considering the large deviation in GC content for P. falciparum it seems to be a probable explanation. Interestingly, the wild type PfAQP gene has a higher CAI in P. pastoris than all the human aquaporins suggesting it could be produced to high levels without any optimization. This, however, was not observed in our study. Consequently, the usage of CAI alone for prediction of membrane protein production levels is limited.
Table IV. Table showing the codon usage for P. pastoris (Pp), H. sapiens (Hs) and P. falciparum (Pf), respectively. The genetic code includes 64 possible ways of making a triplet, of these, three are stop codons in most organisms and are here marked with a star (*). The codon usage is given as fractions where 1 equals ‘always used' and the most commonly used codon is highlighted in bold. The GC content of the coded DNA is also given for each species. Gene frequencies are adapted from Codon Usage Database (Kazusa Citation2007) for P. pastoris, H. sapiens and P. falciparum where the species had the numbers 4922, 9606 and 36329 respectively.
Table V. Table showing the codon adaptation index (CAI) for aquaporins in their native host and P. pastori s, respectively, where the optimized genes are highlighted in bold. CAI calculations using CAIcal (Puigbo et al. Citation2008) and EMBOSS:cai (Bleasby Citation2001) were based on the Codon Usage Database (Kazusa Citation2007) for P. pastoris (Pp), H. sapiens (Hs) and P. falciparum (Pf) where the species had the numbers 4922, 9606 and 36329 respectively. CAI calculations from JCat (Grote et al. Citation2005) use its own codon tables and since it does not contain data for Pp or Pf, these columns have been omitted. For clarity, the values obtained from the calculators are separated by a dotted line. The GC content and the Nc value is also given. Nc represents the extent of the gene's codon bias; 20: strong bias, 61: no bias.
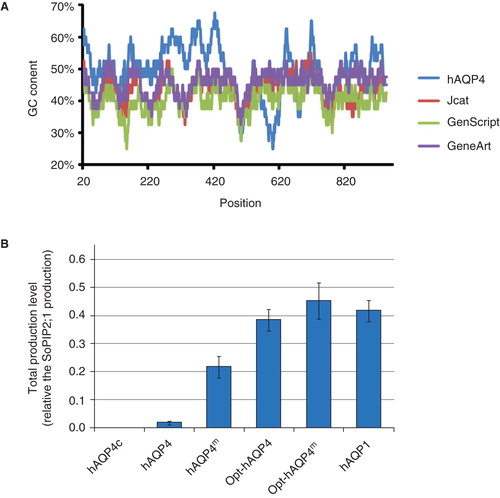
In comparison, the difference in GC content of the coding genome is not always as distinct as for P. falciparum and P. pastoris. In the case of human AQP4, the difference between H. sapiens and P. pastoris is significantly smaller and no obvious production problems can be predicted based on the GC content. The CAI values are also similar for hAQP4 in the native host and in P. pastoris, neither giving any indication of production problems related to differences in codon usage in the two hosts. Nevertheless, the wild type hAQP4 sequence was submitted to two large companies specialized in gene engineering: GenScript and GeneArt. As a comparison, a manual codon optimized gene was made using JCat (Grote et al. Citation2005). The result is presented in . The ‘effective number of codons used', Nc, is a measurement of the extent of codon bias in a gene; its value range from 20, corresponding to one codon being exclusively used for an amino acid, to 61 where the probability is equal for all possible codons (Wright Citation1990). Independent of the method used, the major changes are seen in nucleotide no3 of each codon, as expected, since substitutions at this site more seldom change the amino acid. Interestingly, the variation between the optimizations performed by the companies is substantial. Moreover, it appears that human genes have a relatively high GC content, 52%, as compared to P. pastoris, 43% (). Commonly, a decrease in GC content is observed for all the hAQP4 genes adapted to P. pastoris () due to the more AT biased codon choice for this lower eukaryote. To further analyze the importance of the GC content, a plot was made for all the optimized genes in and compared to the wild type hAQP4 (). From this GC-plot large variations in GC content are observed for the native protein, with some regions spiking over 67% and some dipping to 25%. In contrast, the general theme for the optimized genes is to stay within a much more narrow range of variation. To analyze the effect of the codon adaptation in practice, we used the GenScript optimized version of hAQP4 (Opt-hAQP4) for production in P. pastoris (Oberg et al. Citation2011a) which resulted in a significant increase in yield (). The precise explanation for this observation remains to be evaluated. Anyhow, we intuitively believe that the combinatorial effect of enhancing both the translation and translocation of a protein would result in the highest possible level of production, hence, we introduced the mutants directing the folding pathway of hAQP4 (hAQP4m) in the codon optimized hAQP4, resulting in Opt-hAQP4m. Certainly, the yield of hAQP4 was increased even further giving a yield comparable to the highly produced hAQP1 (). To summarize, the optimization strategies used for hAQP4 illustrates how detailed knowledge of the protein target provides the tools for designing a successful overproduction experiment. However, such knowledge is rarely available which emphasizes the importance of further studies of various aspects of novel protein targets. On the other hand, when information of this sort exists, combining strategies is likely to have a great impact on the final yield of functional membrane protein.
Table VI. Table summarizing the different results obtained when optimizing hAQP4 using JCat, GenScript or GeneArt. Total GC content is shown as well as the GC content of the first, second and third position in the codon. The number of bases changed is also presented as a percentage of the 969 nucleotides in the gene. Nc is a value representing the extent of the codon bias; 20: strong bias, 61: no bias.
Figure 6. Various strategies to optimize the hAQP4 production in P. pastoris. (A) Plot showing the GC content in a 40 bp window centred at the indicated nucleotide position. Data were acquired using the EMBOSS:isochore (Rice Citation2011). (B) Bar chart showing the yield of four different hAQP4 versions. The genes have been transformed by electroporation except hAQP4c where chemical transformation has been used. Constructs having mutations causing an AQP1 like topology maturation have been indicated as hAQP4m. The prefix Opt symbolizes constructs where the gene has been codon optimized to better match the P. pastoris host system. This Figure is reproduced in colour in Molecular Membrane Biology online.
Conclusions
We have used the homologues proteins from the human aquaporin family in order to establish and validate quantitative screening protocols in the methylotrophic yeast P. Pastoris, generic for eukaryotic membrane proteins. By systematically using these protocols we can early in the process estimate our final yield from scaled up fermentor cultures. The main gain is the possibility of evaluating whether the production level of a novel target will be sufficient for downstream purification and characterization or if optimization of its production needs to be undertaken. In brief, we recommend an initial small scale screen on high concentrations of Zeocin to identify high producing clones since increasing the gene dosage was found to be beneficial also for membrane protein production experiments. As a consequence, for the final protein yield, electroporation is superior to chemical transformation, something that was especially pronounced for targets produced at low to moderate initial levels.
For the 13 human aquaporins, we observed a significant variation in the total production level even between proteins having the highest degree of sequence identity. The underlying reason for this variation is not fully unravelled, but specific properties of the proteins are a plausible explanation. On the contrary, a relationship between the transport specificities and the degree of membrane localization was observed, indicating that the pure water channels are more likely to be densely packed in the membrane. In general, the highest yields are achieved when P. pastoris cells are grown in tightly controlled fermentor cultures, conditions allowing maximal use of the methanol-induced AOX1 promoter in high cell density cultures. This was particularly pronounced for one of our most highly produced human aquaporin homologues, hAQP1*, where fermentor growth resulted in nearly five times higher yields (90 mg of pure protein per litre) as compared to shaker flasks.
Further, using the recombinant hAQP clones we have evaluated the importance of the construct design for recovery of high membrane protein levels in P. pastoris. Obviously, the sequence flanking the initiator methionine should mimic the Kozak sequence to improve the processing of the nascent polypeptide chain. Moreover, we found that the use of fusion partners is a non-trivial approach for eukaryotic membrane proteins in a eukaryotic host system. On the contrary, for the few cases where biochemical data is available for the target of interest, alteration of the topology maturation in the translation and translocation process might prove to be useful. A more generic approach is to apply gene optimization in order to increase the production level for membrane proteins. In this process, codons will be adapted to use the host tRNA pool in an efficient way, the GC content will be changed to suit the requirements of the host, and the mRNA stability will be enhanced. However, while shown to often be beneficial, the various algorithms used for codon adaptation and the precise reasons behind its actual effect remain elusive. PfAQP is an example where the production level was significantly improved when the GC content of the gene was closer to the GC content of the host's coding DNA. On the contrary, several of the wild type human aquaporins giving high yields, like hAQP1 and hAQP5, have a very high average GC content while some of the low producers, like hAQP4 and hAQP9, have a lower GC content resembling that of P. pastoris. Consequently, the reasons behind high production yields cannot simply be explained by the GC content of a certain gene. Rather, the explanation lies in the combination of different, possibly unknown, factors. However, when applicable, combining multiple strategies, such as alteration of the topology maturation and codon optimization, it is likely to be successful in the production experiment.
In conclusion, taking advantage of the 13 hAQP homologues, we have investigated and analyzed several factors of importance for a successful design of an overproduction experiment specifically for eukaryotic membrane proteins in the eukaryotic host P. pastoris where the transformation method, the effects of specific mutations, the protein stability, the sequence flanking the initiator methionine, the fusion partners, the GC content, and the codon adaptation have been taken into account. Our established screening protocol for production has made it possible to quantitate the membrane protein yield and carefully evaluate the impact of a certain modification on the final protein yield. Thus, we could efficiently identify the clone with the highest production level for each homologue, as well as quantitate the effect of any optimization experiment with great accuracy. Taken together, our results establish the possibility to take overproduction of recombinant eukaryotic membrane proteins to a quantitative level, which is an essential step towards revealing the complex factors influencing the final yield for this group of highly interesting and relevant protein targets.
Acknowledgements
We would like to acknowledge Jennifer Carbrey (Duke University Medical Center) who kindly provided the cDNA for all hAQPs, William Skach (Oregon Health Sciences University) for providing rAQP4 cDNA and for sharing useful perspectives, the Proteomics Centre at Sahlgrenska Academy in Göteborg for perfoming the LC-MS/MS analysis and the Wallenberg Foundation for their support of the Membrane Protein Centre; Lundberg Laboratory, Göteborg, Sweden. Financial support: This work was supported by Swedish Science Research Council (VR), Carnegiestiftelsen, SWEGENE and financial support from the European Commission by contracts LSHG-CT-2004-504601 (E-MeP), LSHG-CT-2006-037793 (OptiCryst), HEALTH-F4-2007-201924 (EDICT), QLG2-CT-2002-00988 (SPINE) and LSHP-CT2004-012189 (MalariaPorin project) as well as the Marie Curie Training Network Aquaglyceroporins.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Baim SB, Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol Cell Biol 1988;8:1591–1601.
- Beitz E, Wu B, Holm LM, Schultz JE, Zeuthen T. 2006. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc Natl Acad Sci USA 103:269–274.
- Bill RM, Henderson PJ, Iwata S, Kunji ER, Michel H, Neutze R, 2011. Overcoming barriers to membrane protein structure determination. Nat Biotechnol 29:335–340.
- Bleasby A. 2001. EMBOSS: cai [Online] Accessed 25 March 2011 from. http://emboss.bioinformatics.nl/cgi-bin/emboss/cai.
- Bonander N, Bill RM. 2009. Relieving the first bottleneck in the drug discovery pipeline: Using array technologies to rationalize membrane protein production. Expert Rev Proteomics 6:501–505.
- Bonander N, Hedfalk K, Larsson C, Mostad P, Chang C, Gustafsson L, 2005. Design of improved membrane protein production experiments: quantitation of the host response. Protein Sci 14:1729–1740.
- Buck TM, Wagner J, Grund S, Skach WR. 2007. A novel tripartite motif involved in aquaporin topogenesis, monomer folding and tetramerization. Nat Struct Mol Biol 14:762–769.
- Chepelinsky AB. 2009. Structural function of MIP/aquaporin 0 in the eye lens; genetic defects lead to congenital inherited cataracts. Handb Exp Pharmacol 190:265–97.
- Cigan AM, Donahue TF. 1987. Sequence and structural features associated with translational initiator regions in yeast – a review. Gene 59:1–18.
- Cregg JM, Barringer KJ. 1990. Pichia transformation. United States patent application 07/110148.
- Cregg JM, Cereghino JL, Shi J, Higgins DR. 2000. Recombinant protein expression in Pichia pastoris. Mol Biotechnol 16:23–52.
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, 2009. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol 27:561–566.
- Elkjaer M, Vajda Z, Nejsum LN, Kwon T, Jensen UB, Amiry-Moghaddam M, Frokiaer J, Nielsen S. 2000. Immunolocalization of AQP9 in liver, epididymis, testis, spleen, and brain. Biochem Biophys Res Commun 276:1118–1128.
- Fantoni A, Bill RM, Gustafsson L, Hedfalk K. 2007. Improved yields of full-length functional human FGF1 can be achieved using the methylotrophic yeast Pichia pastoris. Protein Expr Purif 52:31–39.
- Forstner M, Leder L, Mayr LM. 2007. Optimization of protein expression systems for modern drug discovery. Expert Rev Proteomics 4:67–78.
- Foster W, Helm A, Turnbull I, Gulati H, Yang B, Verkman AS, 2000. Identification of sequence determinants that direct different intracellular folding pathways for aquaporin-1 and aquaporin-4. J Biol Chem 275:34157–34165.
- Garcia F, Kierbel A, Larocca MC, Gradilone SA, Splinter P, LaRusso NF, 2001. The water channel aquaporin-8 is mainly intracellular in rat hepatocytes, and its plasma membrane insertion is stimulated by cyclic AMP. J Biol Chem 276:12147–12152.
- GeneArt. 2011. Gene Synthesis [Online] Accessed 25 March 2011 from. http://www.geneart.com/english/products-services/gene-synthesis/index.html.
- GenScript. 2011. Gene Synthesis [Online] Accessed 25 March 2011 from. http://www.genscript.com/gene_synthesis.html.
- Grisshammer R. 2006. Understanding recombinant expression of membrane proteins. Curr Opin Biotechnol 17:337–340.
- Grisshammer R, Tate CG. 1995. Overexpression of integral membrane proteins for structural studies. Q Rev Biophys 28:315–422.
- Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, 2005. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33:W526–W531.
- Gustafsson C, Govindarajan S, Minshull J. 2004. Codon bias and heterologous protein expression. Trends Biotechnol 22:346–353.
- Hedfalk K, Pettersson N, Oberg F, Hohmann S, Gordon E. 2008. Production, characterization and crystallization of the Plasmodium falciparum aquaporin. Protein Expr Purif 59:69–78.
- Hennig L, Schafer E. 1998. Protein purification with C-terminal fusion of maltose binding protein. Protein Expr Purif 14:367–370.
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433:377–381.
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030.
- Holmes WJ, Darby RA, Wilks MD, Smith R, Bill RM. 2009. Developing a scalable model of recombinant protein yield from Pichia pastoris: the influence of culture conditions, biomass and induction regime. Microb Cell Fact 8:35.
- Invitrogen. 2002. Pichia Fermentation Process Guidelines [Online] Accessed 5 February 2008 from. http://tools.invitrogen.com/content/sfs/manuals/pichiaferm_prot.pdf.
- Invitrogen. 2005. EasySelect Pichia Expression Kit, version G [Online] Accessed 14 February 2005 from. http://www.invitrogen.com/content/sfs/manuals/easyselect_man.pdf.
- Invitrogen. 2010. EasySelect Pichia Expression Kit, Version MAN0000042 [Online] Accessed 18 June 2010 from. http://www.invitrogen.com/content/sfs/manuals/easyselect_man.pdf.
- Itoh T, Rai T, Kuwahara M, Ko SB, Uchida S, Sasaki S, Ishibashi K. 2005. Identification of a novel aquaporin, AQP12, expressed in pancreatic acinar cells. Biochem Biophys Res Commun 330:832–838.
- Jungo C, Marison I, von Stockar U. 2007a. Mixed feeds of glycerol and methanol can improve the performance of Pichia pastoris cultures: A quantitative study based on concentration gradients in transient continuous cultures. J Biotechnol 128:824–837.
- Jungo C, Schenk J, Pasquier M, Marison IW, von Stockar U. 2007b. A quantitative analysis of the benefits of mixed feeds of sorbitol and methanol for the production of recombinant avidin with Pichia pastoris. J Biotechnol 131:57–66.
- Karabasil MR, Hasegawa T, Azlina A, Purevjav J, Purwanti N, Yao C, Akamatsu T, Hosoi K. 2009. Trafficking of GFP-AQP5 chimeric proteins conferred with unphosphorylated amino acids at their PKA-target motif ((152)SRRTS) in MDCK-II cells. J Med Invest 56:55–63.
- King LS, Kozono D, Agre P. 2004. From structure to disease: the evolving tale of aquaporin biology. Nat Rev Mol Cell Biol 5:687–698.
- Kazusa. 2007. Avilable from: http://www.kazusa.or.jp/codon
- Kozak M. 1981. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res 9:5233–5252.
- Lian J, Ding S, Cai J, Zhang D, Xu Z, Wang X. 2009. Improving aquaporin Z expression in Escherichia coli by fusion partners and subsequent condition optimization. Appl Microbiol Biotechnol 82:463–470.
- Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. 2000. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol Biol Cell 11:2973–2985.
- Lundstrom K. 2006. Structural genomics: The ultimate approach for rational drug design. Mol Biotechnol 34:205–212.
- Mobasheri A, Shakibaei M, Marples D. 2004. Immunohistochemical localization of aquaporin 10 in the apical membranes of the human ileum: a potential pathway for luminal water and small solute absorption. Histochem Cell Biol 121:463–471.
- Morishita Y, Matsuzaki T, Hara-chikuma M, Andoo A, Shimono M, Matsuki A, Kobayashi K, Ikeda M, Disruption of aquaporin-11 produces polycystic kidneys following vacuolization of the proximal tubule. Mol Cell Biol 2005;25:7770–7779.
- Mus-Veteau I. 2002. Heterologous expression and purification systems for structural proteomics of mammalian membrane proteins. Comp Funct Genomics 3:511–517.
- Nedvetsky PI, Tamma G, Beulshausen S, Valenti G, Rosenthal W, Klussmann E. 2009. Regulation of aquaporin-2 trafficking. Handb Exp Pharmacol 190:133–157.
- Neely JD, Amiry-Moghaddam M, Ottersen OP, Froehner SC, Agre P, Adams ME. 2001. Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proc Natl Acad Sci SA 98:14108–14113.
- Nielsen S, Nagelhus EA, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen OP. 1997. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J Neurosci 17:171–180.
- Nielsen S, Smith BL, Christensen EI, Agre P. 1993. Distribution of the aquaporin CHIP in secretory and resorptive epithelia and capillary endothelia. Proc Natl Acad Sci USA 90:7275–7279.
- Nyblom M, Oberg F, Lindkvist-Petersson K, Hallgren K, Findlay H, Wikstrom J, 2007. Exceptional overproduction of a functional human membrane protein. Protein Expr Purif 56:110–120.
- Oberg F, Ekvall M, Nyblom M, Backmark A, Neutze R, Hedfalk K. 2009. Insight into factors directing high production of eukaryotic membrane proteins; production of 13 human AQPs in Pichia pastoris. Mol Membr Biol 26:215–227.
- Oberg F, Sjohamn J, Conner MT, Bill RM, Hedfalk K. 2011a. Improving recombinant eukaryotic membrane protein yields in Pichia pastoris: the importance of codon optimization and clone selection. Mol Membr Biol 28:398–411.
- Oberg F, Sjohamn J, Fischer G, Moberg A, Pedersen A, Neutze R, 2011b. Glycosylation increases the thermostability of human aquaporin 10. J Biol Chem 286:31915–31923.
- Perez-Martin J, Cases I, de Lorenzo V. 1997. Design of a solubilization pathway for recombinant polypeptides in vivo through processing of a bi-protein with a viral protease. Protein Eng 10:725–730.
- Petrovskaya LE, Shulga AA, Bocharova OV, Ermolyuk YS, Kryukova EA, Chupin VV, 2010. Expression of G-protein coupled receptors in Escherichia coli for structural studies. Biochemistry (Mosc) 75:881–891.
- Puigbo P, Bravo IG, Garcia-Vallve S. 2008. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct 3:38.
- Rai T, Sasaki S, Uchida S. 2006. Polarized trafficking of the aquaporin-3 water channel is mediated by an NH2-terminal sorting signal. American journal of physiology. Cell physiology 290:C298–C304.
- Rice P. 2011. EMBOSS:isochore [Online] Accessed 28 March 2011 from. http://emboss.bioinformatics.nl/cgi-bin/emboss/isochore.
- Romanos MA, Scorer CA, Clare JJ. 1992. Foreign gene expression in yeast: A review. Yeast 8:423–488.
- Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. 2005. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science 307:1317–1321
- Sadlish H, Pitonzo D, Johnson AE, Skach WR. 2005. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat Struct Mol Biol 12:870–878.
- Sharp PM, Li WH. 1987. The codon Adaptation Index – a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15:1281–1295.
- Shi LB, Skach WR, Ma T, Verkman AS. 1995. Distinct biogenesis mechanisms for the water channels MIWC and CHIP28 at the endoplasmic reticulum. Biochemistry 34:8250–8256.
- Sinclair G, Choy FY. 2002. Synonymous codon usage bias and the expression of human glucocerebrosidase in the methylotrophic yeast, Pichia pastoris. Protein Expr Purif 26:96–105.
- Skach WR, Shi LB, Calayag MC, Frigeri A, Lingappa VR, Verkman AS. 1994. Biogenesis and transmembrane topology of the CHIP28 water channel at the endoplasmic reticulum. J Cell Biol 125:803–815.
- Skowronski MT, Lebeck J, Rojek A, Praetorius J, Fuchtbauer EM, Frokiaer J, Nielsen S. 2007. AQP7 is localized in capillaries of adipose tissue, cardiac and striated muscle: implications in glycerol metabolism. Am J Physiol Renal Physiol 292:F956–F965.
- Yasui M, Hazama A, Kwon TH, Nielsen S, Guggino WB, Agre P. 1999. Rapid gating and anion permeability of an intracellular aquaporin. Nature 402:184–187.
- Tate CG. 2001. Overexpression of mammalian integral membrane proteins for structural studies. FEBS Lett 504:94–98.
- Tate CG, Haase J, Baker C, Boorsma M, Magnani F, Vallis Y, 2003. Comparison of seven different heterologous protein expression systems for the production of the serotonin transporter. Biochim Biophys Acta 1610:141–153.
- Tornroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, 2006. Structural mechanism of plant aquaporin gating. Nature 439:688–694.
- Wang A, Clapper J, Guderian JA, Foy TM, Fanger GR, Retter MW, 2003. A novel method for increasing the expression level of recombinant proteins. Protein Expr Purif 30:124–133.
- White S. 2011. Database of Membrane Proteins of Known Structure [Online] Accessed 8 April 2011 from. http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- Wright F. 1990. The 'effective number of codons' used in a gene. Gene 87:23–29.
- Wu G, Culley DE, Zhang W. 2005. Predicted highly expressed genes in the genomes of Streptomyces coelicolor and Streptomyces avermitilis and the implications for their metabolism. Microbiology 151:2175–2187.
- Xia X. 2007. The +4G site in Kozak consensus is not related to the efficiency of translation initiation. PLoS One 2:e188.
