Abstract
Aptamers are single-stranded structured oligonucleotides (DNA or RNA) that can bind to a wide range of targets (“apatopes”) with high affinity and specificity. These nucleic acid ligands, generated from pools of random-sequence by an in vitro selection process referred to as systematic evolution of ligands by exponential enrichment (SELEX), have now been identified as excellent tools for chemical biology, therapeutic delivery, diagnosis, research, and monitoring therapy in real-time imaging. Today, aptamers represent an interesting class of modern pharmaceuticals which with their low immunogenic potential mimic extend many of the properties of monoclonal antibodies in diagnostics, research, and therapeutics. More recently, chimeric aptamer approach employing many different possible types of chimerization strategies has generated more stable and efficient chimeric aptamers with aptamer–aptamer, aptamer–nonaptamer biomacromolecules (siRNAs, proteins) and aptamer–nanoparticle chimeras. These chimeric aptamers when conjugated with various biomacromolecules like locked nucleic acid (LNA) to potentiate their stability, biodistribution, and targeting efficiency, have facilitated the accurate targeting in preclinical trials. We developed LNA-aptamer (anti-nucleolin and EpCAM) complexes which were loaded in iron-saturated bovine lactofeerin (Fe-blf)-coated dopamine modified surface of superparamagnetic iron oxide (Fe3O4) nanoparticles (SPIONs). This complex was used to deliver the specific aptamers in tumor cells in a co-culture model of normal and cancer cells. This review focuses on the chimeric aptamers, currently in development that are likely to find future practical applications in concert with other therapeutic molecules and modalities.
Introduction
On the therapeutic front, in the race for developing targeted delivery vehicles, nucleic acid-based aptamers are now regularly in competition with small molecules and antibodies since the inception of aptamer technology more than a decade ago. Aptamers (Latin aptus meaning “to fit”) are the functional nucleic acid ligands generated by a molecular selection process called Systematic Evolution of Ligands by Exponential Enrichment (SELEX) and are also one of only a few classes of molecules that similar to antibodies can be crafted to bind to multiple different targets (Keefe, Citation2008; Kanwar et al., Citation2010c). Aptamers are 20–80 base pair long single-stranded de-oxy ribonucleic acid (DNA) or ribonucleic acid (RNA) oligonucleotides which are folded into unique three-dimensional conformations due to various intramolecular interactions (Kanwar et al., Citation2010c).
The idea of using oligonucleotides as therapeutic agents is not a new one; as in 1990 RNA decoys, which mimic the transactivating responsive (TAR) RNA of human immunodeficiency virus (HIV), were shown to prevent HIV replication by sequestering the Tat protein (Sullenger et al., Citation1990; Bivalkar-Mehla et al., Citation2010). However, among the various nucleic acid-based strategies employed for drug discovery research and therapeutics, aptamers have emerged as the most promising agents for therapy and diagnosis, since the origin of aptamer technology two decades ago (Ellington, Citation1990; Tuerk & Gold, Citation1990). Aptamer-mediated drug delivery enhances the therapeutic efficacy due to their high specificity to the target and thus their affinity reduces the off-target effects or other unwanted side effects, commonly observed with widely used cytotoxic drug therapeutics. In regard to comparison with traditional antibodies, aptamers also known as “chemical antibodies,” have number of advantages such as (i) smaller in size and less complex with low immunogenic potential, (ii) easier to synthesize and modify in vitro, (iii) higher affinity and specificity, (iv) structural flexibility enabling aptamer to bind to hidden epitopes which cannot be targeted by antibodies (Majumder, Citation2009) , and (v) stronger stability and can be stored easily until put to use. As the use of aptamers has been extended from basic biology of cellular processes and gene regulation to therapeutic and diagnostic applications, many patented aptamers are currently being tested in clinical trials and reviewed recently (Majumder, Citation2009). The resulting outcomes will provide important validations of their efficacies and cost saving. The approval of an anti-vascular endothelial growth factor (VEGF) aptamer (Eyetech/Pfizer’s Macugen) six years ago, by the Food and Drug Administration (FDA) for treatment of age-related human macular degeneration has already proved a milestone for the applications of aptamer technology. In this review, we focus on the importance of aptamer and their chimerics in the field of molecular medicine. The review focuses on the various selection procedures along with the established modifications for the aptamers and their chimerics. Various forms of aptamer-chimerics have been discussed with their therapeutic benefits and strategies for targeted delivery. Some of the aptamers in clinical trials are also listed.
Strategies for aptamer selection
For screening of aptamers, the target and the characteristic of aptamers obtained must be known; a random pool of oligonucleotides both DNA and RNA have to be made for screening; the immobilized targets are used for screening of aptamers from the RNA or DNA pool (Wang, Citation2009). The specificity of the aptamer and its ability to distinguish between the closely related species depends on the type of selection procedure for its isolation. SELEX technique developed by Tuerk, Ellington, and Szostak was the first aptamer selection procedure (Tuerk & Gold, Citation1990; Ellington & Szostak, Citation1992). It involves selection of nucleic acid ligands which interacts with specific target in a repeated binding, selection, and amplification of aptamers from initial library until the desired characteristics have been isolated. First, an oligonucleotide library is created which has sequences that can be amplified by polymerase chain reaction (PCR). The library can be used directly for DNA-based aptamer selection, whereas for selection of an RNA aptamer, the DNA library has to be converted into a RNA library; the known target molecule is then incubated in the oligonucleotide pool along with the target accompanied with heating and cooling to promote formation of stable structures. The oligonucleotides bound to the target are isolated and are put through repeated rounds of selection for obtaining the sequences with the best specificity. These sequences are then cloned in plasmids, amplified, sequenced and known as aptamers (Tuerk & Gold, Citation1990).
In 1992, Ellington and Szostak (Citation1992) discovered another screening method for aptamers with a 10-fold higher affinity called as the negative SELEX, where the nucleotide pool was loaded on a matrix with analog of the target, and the unbound sequences are then used to incubate with the target for normal binding. Thus, aptamers selected by negative SELEX had higher affinity to the target and could discriminate between the closely related targets. Negative SELEX has been used for selection of small molecules (Bassett et al., Citation2004) and proteins (Haller & Sarnow, Citation1997). Jenison and co-workers (Citation1994) established another screening methodology called as counter SELEX, for screening of small molecules’ aptamers where the cycles were similar to negative SELEX; however, here the matrices were exchanged with the analogs of the target molecules. Wang and co-workers (Citation2003) discovered subtractive SELEX which removes single-stranded DNA or RNA sequences that can bind to nonspecific part of a complex target with various binding sites in order to obtain highly specific aptamers in the end. This process has extensively been used to screen aptamers for various cancers.
There are many diagnostic markers and therapeutic targets expressed on the cell surface. The aptamers selected by SELEX may show low or no affinity toward them due to shielding of the aptamer’s binding domain. To overcome this, several groups, as discussed in the review by Xianbin and co-workers (2011), have reported the isolation of aptamers from living cells as targets such as cancer cells (Shangguan et al., Citation2006) by the process called as cell-SELEX. In cell-SELEX, instead of using a purified protein target, whole living cells are used as the targets. Live African and American trypanosomes aptamers (Homann & Göringer, Citation1999; Ulrich et al., Citation2002) are some of the examples of cell-SELEX. The only drawback in this process is that it has to be repeated more than 25 times (Jenison et al., Citation1994; Sooter et al., Citation2001). Capillary electrophoresis (CE-SELEX)-based selection procedure developed by Bowser and Krylov groups is another revolution in field of aptamer selection (Berezovski, Citation2004; Mendonsa & Bowser, Citation2004). Here the oligonucleotide library is mixed with the target molecule and introduced into a free solution CE system. The unbound oligonucleotides migrate at a different rate than the oligonucleotides which are bound to the target. The collected bound sequences are then amplified by PCR and high affinity aptamers can be obtained only after two rounds of selection (Mendonsa & Bowser, Citation2004). An advancement to the CE-SELEX is the micromagnetic selection (M-SELEX), which uses magnetic beads as solid support matrix for linking the target (Bruno & Kiel, Citation2002). More recently, the number of beads can be manipulated to isolate high affinity aptamers. The DNA aptamers against the light chain of botulinum neurotoxin type A has been isolated in a single round of selection by this method (Qian et al., Citation2009).
The other forms of SELEX are covalent SELEX (Jensen et al., Citation1995), which is used to isolate aptamers that bind covalently to the target moiety, Toggle SELEX (White et al., Citation2001) that generates aptamers with the ability of cross reactivity in different species, bead-based SELEX (Yang et al., Citation2002a), Tailored SELEX (Vater et al., Citation2003), which was able to regulate the size of aptamers by removing the fixed nucleotide sequences from the oligonucleotide library, and on chip SELEX (Collett et al., Citation2005). More innovatively, Miyachi et al. used atomic force microscopy based SELEX (AFM-SELEX) which has the capability to detect the force of affinity or adhesion between two molecules, to isolate anti-thrombin aptamers within just three rounds of selection (Miyachi et al., Citation2010).
With time the aptamer selection procedures have evolved due to the developing technology. Better techniques have been devised to select aptamers with high affinity and maximum specificity to the desired target. However, the use of aptamers in diagnosis of biological fluids has been limited. Thus, post-SELEX changes and chemical modifications have made their way into aptamer technology due to a continuous need of stability and efficient activity.
Established aptamer modifications to overcome aptamer instability
The use of modified nucleotides has been the most commonly approached method to overcome aptamer instability. Precise site-specific modifications facilitate engineering of aptamers for delivery into target cells with enhanced specificity. The need for modification is justified by functional optimization, truncation, or multimerization of aptamers which has enhanced the binding efficacy and stability. Various linkage designs, modification strategies, and conjugation approaches are prevalent in aptamer technology. The various modifications of nucleotides, mainly the chemical ones, are compatible with the enzymatic steps of in vitro selection procedure, introduced either at phosphate/ribose backbone or at the nucleobases (Keefe & Cload, Citation2008).
Chemical modifications in aptamers
Replacement of DNA phosphate backbone by phosphorothionate enhanced stability against nucleases and the cell viability of aptamers (Eckstein & Gish, Citation1989). However, most prominent modification of aptamers is derivatization of 2´-ribose, as this position conferred stability of most RNA aptamers (Yang et al., Citation2002b). Several other different approaches have been reviewed for improving aptamer stability in biological fluids due to the sensitivity of both DNA and RNA aptamers to nucleases which limits their therapeutic and diagnostic potential (Kusser, Citation2000). Most of these approaches rely on the incorporation of nucleotides carrying modifications of either sugar residues (2´-F, 2´-NH2 (amino), 2´-O-methyl, 2´-O-methoxyethyl and 2´-O-dimethyl allyl) (Green et al., Citation1995; Ruckman et al., Citation1998) of the phosphate (phosphorothioate and methyl phosphonate) (Eckstein & Gish, Citation1989) or of the nitrogenous base [propenyl, 5-(N aminoalkyl) carbomoyluracil, methyl, trifluoromethyl, phenyl and 2-thiopyrimidine] (Lin et al., Citation1996).
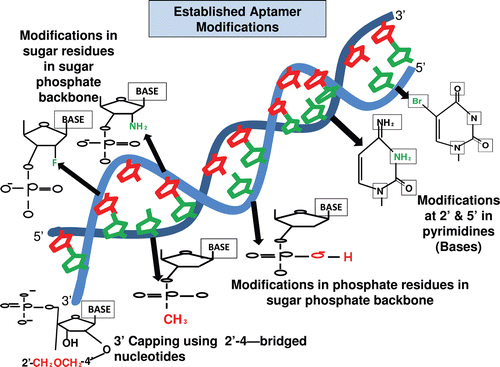
Pyrimidines are modified at fifth position to enhance the stability of the aptamer with iodide (I), bromide (Br), chloride (Cl), amino (NH3), azide (N3) (Kirschenheuter, Citation1997) and 2´ position with amino (NH2), fluoro (F), and methoxy (OCH3) (Barry et al., Citation1996). These modifications also provide nuclease resistance. The 2’ sugar modifications, fifth position pyrimidine modification, substitution of 4-thiouridine, substitution of 5-bromo or 5-iodouracil, backbone modification, methylations and unusual base pairing combinations such as isobases, isocytidine, and isoguanodine (Sampson, Citation2003), and 3’ capping (Kasahara Y, Citation2010) are other forms of modifications which provide thermal stability and nuclease resistance to aptamers (). The SELEX protocol also offers possibility of further diversity during selection and enrichment where modifying the aptamers is much easier when compared to post-SELEX methods (Sampson, Citation2003).
Figure 1. Figure shows the various modifications made in an aptamer to increase its stability and functionality. The common modifications made in the ribose residue of the sugar-phosphate backbone are incorporations of 2´-fluorine (F) and 2´-amino group (NH2), in the phosphate group of the sugar-phosphate backbone are incorporation of phosphorothioate and methyl phosphonate. The modification made in the nucleotide base includes incorporation of 5´- bromide (Br) and amino group (NH2) (Cheung et al., Citation2010). The 3´-capping of the aptamer sequence is also another form of modification, which helps in increasing the stability of aptamer and its prevention from nuclease degradation (Kasahara et al., Citation2010).
Aptamers with photolabile protecting groups offer a possibility for the spatiotemporal regulation of aptamer activity with control over activity of the target molecule. Using this modification the aptamer can be activated or deactivated by specific light intensity (Mayer et al., Citation2005; Heckel et al., Citation2006). Hence aptamers can be modified to fluoresce at their fifth position by chemical or enzymatic synthesis (Sengle, Citation2000). The chemical modifications are induced to increase the half life and stability of the aptamers. The modified aptamers can be obtained by spiegelmer technology, which is also known as mirror image SELEX. In this method, aptamers of chiral target from naturally occurring dextrorotatory oligonucleotide pool are screened and from them the complementary levorotatory oligonucleotide is synthesized called as spiegelmers. These spiegelmers have equivalent capability as the initial aptamer to bind with high affinity to the target (Wlotzka et al., Citation2002) and the chemical modification is induced later as the levorotatory molecule with chemical modifications cannot be recognized by SELEX enzymes; this molecule can recognize the target with more specificity and has better stability (Eulberg & Klussmann, Citation2003).
Chimeric aptamer: an insight
There is an increasing enthusiasm for generating new and more potent cell internalizing, highly specific aptamers with greater stability and nuclease resistance. Recombination is a powerful evolutionary force and it is a known fact that the protein segments are routinely recombined to engineer various chimeric proteins with novel functions. Aptamer diversity is required for the development of more established and better characterized aptamers; thus the need for modification and chimerization arises. Chimerization started with the idea of creating antibodies for any potential target (Riechmann et al., Citation1988; Russell et al., Citation1992). The constant need for higher specificity, stability, and effectiveness motivated better synthesis procedures over time. Similarly to antibodies and proteins’ chimerization, aptamer chimerization has been carried out for characterizing and diversifying them over a long period of time (Goldstein et al., Citation1995; Song et al., Citation2005).
Chimerization of aptamers refers to its combination with another aptamer, biomacromolecule(s) and/or compounds where the functional capability of the chimera is a combined factor of aptamer as well as the other biomacromolecules and/or compounds used in combination (Burke & Willis, Citation1998).
Chimeric aptamer approach aims to combine two aptamers or an aptamer with another non-aptamer moiety (biomacromolecules, drug, or dyes), where one molecule engages with the target and the other has a functional effect on the target molecule. When two aptamers or an apatmer with another biomacromolecule are added together, either through natural recombination or chemical engineering, it is difficult to know beforehand whether the joining may diminish the activity of one or both of the recombining partners; hence, chimerization was considered as a challenging procedure (Burke & Gold, Citation1997a; Burke & Willis, Citation1998). However, with the progress of research in this field as discussed here, it is very clear that chimeric aptamers are not only highly stable and efficient in activity but are also able to deliver drug loads.
Selection of the aptamer chimeras
The selection of chimeric aptamer over the parent type aptamer needs efficient comparison between them in terms of specificity of binding and the binding affinity. Within a chimeric SELEX, two or more different libraries are used for the production of chimeric aptamers with more than one wanted feature or which are able to function in different ways. Each of the parent libraries will be selected first for a distinct feature. The selected biomolecules are then fused in such a way that they acquire several properties (Burke & Willis, Citation1998). Chimeric SELEX method simulates random recombination among functional aptamers derived from nucleotide population with 70 to 80 positions of random sequences as a mean to generate aptamers with novel functions. Another method to obtain multifunctional aptamers is multiple SELEX; two aptamers with specificity toward coenzyme-A, chloramplenicol or adenosine could be fused. This selection strategy was applied to obtain a suitable chimera capable of binding both the targets. This will have several applications such as catalysis, therapeutics, structure studies, etc. (Burke et al., Citation1997b; Burke & Willis, Citation1998) In another work, two nucleic acid pools of 40 and 60 units were used to screen aptamers for cibacron blue and cholic acid. After five to six rounds of selection, the aptamers were fused and the ones with the ability to bind both cibacron blue and cholic acid were obtained. It was observed that the chimeric aptamer could bind either to cibacron blue and could be eluted by cholic acid or could bind to cholic acid and be eluted by cibacron blue; these results highlighted the importance of allosteric interactions in mechanism of chimerism (Wu & Curran, Citation1999).
Zinc fingers have diverse functions in a cell, mainly dealing with interactions with DNA, RNA, or proteins and play a major role in the regulation of gene expression (Laity et al., Citation2001). Work done in this field compared interaction of several different zinc finger proteins with RNA1 (a high affinity aptamer for zinc finger) where, site-directed mutagenesis in RNA1 was used to identify key and structural elements in RNA1 involved in the formation of high affinity interactions with a broad range of zinc fingers (Lu et al., Citation2003). The modified forms, RNA1, RNA21, and RNA 22, were compared to characterize RNA molecules with high affinity for a set of zinc fingers (Weiss et al., Citation2010). Such studies help in identifying the most specific aptamer for a target which has several applications such as coordinated release of protein on a specific time scale or release in a specific cellular location. A similar work describes isolation of 2´-fluoro (2´-F)-substituted RNA aptamers that bind to streptavidin with high affinity. After selection of aptamer SA19 (specific for streptavidin), it was mutated by DPTP [6-(2-deoxy-β-D-erythropentofuranosyl)-3, 4-dihydro-8H-pyrimido-(4, 5C) (1, 2) oxamine-7-one-5´-triphosphate], a nucleotide analog. The 2´-F-pyrimidine containing RNA transcripts from various mutagenesis cycles were analyzed by surface Plasmon resonance (SPR) to verify the abolition of binding to streptavidin. Metal transporting ATPases CopA and CopT sequences were inserted downstream of SA19 aptamer to form chimeric SA19-CopA or SA19-CopT. Their properties were then compared with SA19 unmodified aptamer for characterization of binding affinity and specificity (Alaoui et al., Citation2002). In the study done by McNamara and co-workers, even though the first generation prostrate specific membrane antigen (PSMA) aptamer/ polo-like kinase 1(Plk1)-siRNA (A10-Plk1) chimera inhibited tumor growth, it lacked the ability to be systemically administered (McNamara et al., Citation2006). Thus, in continuation of their work, a second generation of PSMA-Plk1 chimeras was designed to facilitate chemical synthesis, to enhance silencing activity, specificity, and to enable modifications to optimize in vivo kinetics. The changes made to the first generation chimeras to facilitate chemical synthesis were to reduce the aptamer size from 71 nucleotides to 39 nucleotides and a 2´-F was added to the longer strand and the shorter strand was unmodified. To increase the silencing activity, several chimeras were engineered, ones with an overhang at 3´ end of siRNA duplex, ones with a wobble base at 5´ end of guiding strand of siRNA, ones where the passenger and the guide strand were swapped, and ones with a stem loop. Then, the binding of optimized chimeras to PSMA expressing cells was tested; all chimeras had retained the binding ability confirming that modifications made to first generation did not alter binding or specificity. To determine the enhancement of silencing activity of the chimeras for gene-specific silencing, a quantitative real time polymerase chain reaction (qRT PCR) was used and it was confirmed that siRNA portion of chimera enhanced Plk1 silencing, most active were swap and stem-loop chimeras. Finally, the effect of chimeras on growth and survival of prostate cancer cells was tested and it was found that second generation chimeras inhibited cell growth and proliferation at a lower concentration than the first generation chimeras (Justin et al., Citation2009). Even though they are engineered well and have more capability, the chimeric aptamers are as prone to degradation as an unmodified aptamer. However, there are some molecules such as locked nucleic acids (LNA’s) (Lebars et al., Citation2007), which when incorporated into the chimeras provide a higher grade of stability and specificity and thus help in the selection of the chimeras by giving them an additional advantage over the parental types.
LNA-based modifications
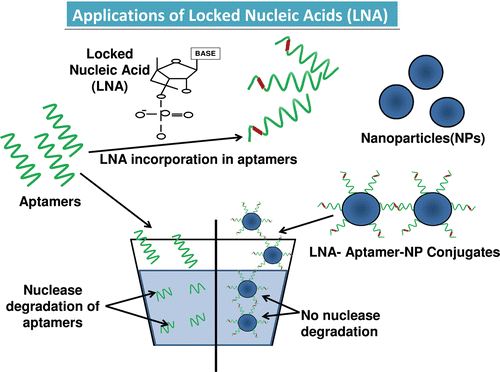
Both DNA and RNA oligonucleotides are highly susceptible to degradation by nucleases. Thus, the potential applications and the usage of aptamers in drug delivery are limited by the susceptibility of aptamers to degradation by intracellular nucleases. To further improve the stability against nucleases and to overcome this limitation, the use of non-natural bases, such as LNA, were used (Orum & Wengel, Citation2001; Petersen & Wengel, Citation2003; Hernandez et al., Citation2009;). The LNA comprises of a new class of bicyclic high affinity analogs (2´-O, 4´-C-methylene-β-D-ribofuranosyl nucleotide) in which furanose ring of ribose sugar is chemically locked in an RNA mimicking conformation by introduction of 2´-O,4´-C-methylene bridge. These impart high degree of thermal stability when hybridized with their DNA and RNA target molecule (). A specific feature of LNA is that they induce the neighboring DNA to adopt an A-type conformation in a DNA–RNA duplex. Since LNA hybrids are not compatible with standard enzymes that are used in SELEX, only post-SELEX is followed for their selection (Barciszewski et al., Citation2009). The introduction of LNA into aptamers renders the molecule highly resistant toward nuclease degradation. Significant stabilization of aptamers has been achieved by introduction of LNA at various positions of oligonucleotides. There are mainly three types of LNA: antigenic LNA, antisense LNA, and LNA aptamers. Immense work has been done with LNA hybrid aptamers, such as in a 39-mer oligonucleotide TTA1 (RNA aptamer); LNA was introduced which exhibited significant stem stabilization and improved plasma stability of the aptamer while maintaining high binding affinity to the target (Barciszewski et al., Citation2009). Another study showed that LNA was used for modification of R06 aptamer to generate stable hybrid with improved binding affinity to form kissing complexes with the target with higher nuclease resistance (Lebars et al., Citation2007). Recently invented LNA nucleotides have been shown to be effective both in and without combination of taxol for inhibiting expression of survivin thereby leading to tumor growth inhibition when compared to LNA nucleotides alone (Kanwar et al., Citation2010a). Survivin is a member of inhibitor of apoptosis (IAP) family of proteins that has a dual role in cell division (mitosis) and apoptosis (Kanwar et al., Citation2001; Altieri, Citation2003; Baratchi et al., Citation2010; Kanwar RK et al., Citation2010; Kanwar, Citation2010a). It is known to be over-expressed in most of the tumors and has become recently an attractive target for novel anticancer therapies (Kanwar et al., Citation2001; Altieri, Citation2003; Baratchi et al., Citation2010; Kanwar RK et al., Citation2010; Kanwar et al., Citation2010a). A set of researchers prepared locked nucleic acid nanoparticle conjugate (LNP’s) by adding thiol-terminated LNA oligonucleotides to a solution containing gold nanoparticles (AuNPs) in phosphate buffer and also prepared DNA-modified nanoparticle analogs for comparative study by using antisense DNA and LNA, and they found out that LNP’s were more stable and induced a better relative decrease in survivin expression (Seferos et al., Citation2007). In another work, chimeras were synthesized by surface plasmon resonance (SPR) from 64 LNA/2´-o-methyl sequence to all possible combination in a kissing RNA-aptamer loop complementary to epsilon-nucleotide loop of TAR element of HIV-1. Following which a chimera of higher specificity than the parent RNA aptamer was obtained and one of the chimeras also showed inhibition of TAR-dependant luciferase expression in a cell assay (DI Primo et al., Citation2007). The LNA incorporations have successfully changed the scenario of aptamer instability and their effectiveness has been well established with time.
Figure 2. The structure of the locked nucleic acid molecule is shown in . The reason for its stability is due to the presence of an additional bond between the hydrogen on the 4th carbon and the oxygen atom on the 2nd carbon, which also gives the molecule a locked structure and makes it more resistant to thermal degradation. The incorporation of the LNA molecules into the aptamer is also shown which stabilizes the structure, and finally the LNA-incorporated aptamers are loaded onto a nanoparticle for target delivery (DI Primo et al., Citation2007; Lebars et al., Citation2007; Seferos et al., Citation2007; Barciszewski et al., Citation2009).
In spite of being able to incorporate several beneficial properties into the chimeras, their delivery is yet another necessary stage which plays a key role in their action.
Various types of aptamer chimeras and their functional applications in therapeutics
There are many possible types of chimerization such as aptamer–siRNA chimeras, aptamer–enzyme chimeras, and aptamer–aptamer chimeras ( and ). Aptamer technology has evolved in a multidirectional fashion finding importance and playing essential roles in fields of biosensing, imaging, drug delivery, and modern molecular medicine for therapy for various diseases and disorders.
Table 1. Chimeric aptamers and their action.
Figure 3. Illustrations of the various types of chimeric forms of aptamers established, such as the aptamer–antibody (Ohk et al., Citation2010), aptamer–protein (Zhou & Rossi, Citation2010), aptamer–siRNA (Kawata et al., Citation2010) (Pastor et al., Citation2010), aptamer–miRNA (Lunse, Citation2010), and the aptamer–aptamer (Held et al., Citation2006) chimeras. The use of sticky bridges is also depicted which helps in binding of the aptamer moiety to the siRNA and the miRNA’s due to presence of complementary regions on the sticky loop for both siRNA/miRNA and the aptamer. The aptamer conjugates are then shown to conjugate with the nanoparticles binding to the target cells by the interaction of aptamer–receptor interaction, and finally the nanoparticles are internalized inside the target cells and results in release of siRNA, miRNA, protein, or the aptamer molecule, respectively (Lundberg et al., Citation2007; Farokhzad et al., Citation2004; Khaled et al., Citation2005; Alonso et al., Citation1994; Molpeceres et al., Citation1999).
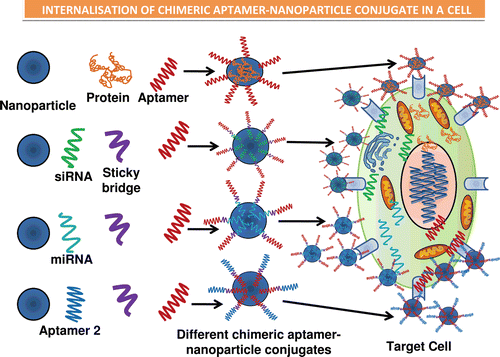
Aptamer–biomolecule conjugate
A recent work done on aptamer–antibody chimera provides an effective methodology for sensitive and specific identification of a pathogenic bacteria Listeria monocytogenes in food. Aptamer A-8, specific for internalin A, an invasion protein of L. monocytogenes was used in fiber optic sensor together with biotinylated P66 antibody in a sandwich format for the detection of L. monocytogenes from food samples (Ohk et al., Citation2010). Many bacterial strains have become drug resistant with the overexposure to antibiotics worldwide. Bacterial infection is a major health concern and demands more advanced treatments, such as use of nanotechnology or nanomedicine (Mathews et al., Citation2010). Chimeric aptamers can play potential role in antibacterial diagnostics and therapeutics due to obvious benefits over traditional antibacterial medicines. An aptamer–aptamer chimera has been developed recently, containing adenosine DNA aptamer and malachite green (MG) RNA aptamer joined by bridging strands that has complementary sequences to both aptamers. In the presence of adenosine, aptamer strand binds to adenosine leaving fewer numbers of complementary base pairs between aptamer and bridge strand, which is less stable, and results in release of bridge strand and observance of fluorescence of malachite green (XU & LU, Citation2010). The fluorescence can be detected by eye and no sophisticated instrument is required for this. Such chimeras can be used very efficiently in real time detection and quantification of small analytes. Work done by another group shows an example of aptamer–enzyme chimeras, the aptamer against transferrin receptor was isolated and coupled to a dephosphorylated α-L-iduronidase enzyme. It was shown that the aptamer–enzyme chimera was successfully taken up by receptor-mediated endocytosis and restored activity in cultured fibroblasts lacking this enzyme (Chi-Hong et al., Citation2008).
Aptamer–biomacromolecule conjugate
The most established and best-characterized chimeras are the aptamer–siRNA chimeras. This interesting and recent innovative approach aims to use the aptamers in siRNA therapeutics. Though work has been done with these chimeras (Eli, Citation2010; Zhou et al., Citation2011), yet most prevalent one targeted for therapy is against PSMA, a cell-surface receptor over-expressed in prostate cancer cells and tumor vascular endothelium (McNamara et al., Citation2006). Some of the work done on PSMA is summarized here. An aptamer–siRNA chimera was developed where the aptamer portion of the chimeras mediated binding to PSMA, whereas the siRNA portion targeted the expression of tumor survival genes polo-like kinase 1 (Plk1)17 and Bcl-2. The chimera effectively delivered the associated siRNA specifically to LNCaP prostate cancer expressing PSMA cells and triggered apoptosis that resulted in cell death both in culture and in a prostrate tumor xenograft model (McNamara et al., Citation2006). In another study 2´-F modified anti-PSMA aptamer (A-10) was covalently linked to sense strand of 21-mer siRNA hybridized to 21-mer antisense strand, the resulting chimeric was shown to selectively internalize into cells expressing PSMA and inhibit expression of targeted tumor survival genes (Plk1 and Bcl2) aptamer. A portion of PSMA-A10PLK1 chimera was truncated from 71 to 39 nucleotides, while still maintaining high binding affinity; structural changes were made to siRNA enabling efficient incorporation (Zhou & Rossi, Citation2010; Zhou et al., Citation2011).
In a similar work, a DNA-chimeric siRNA was generated against Plk1, which was more stable in human serum than nonchimeric siRNA and the chimeric Plk1-siRNA inhibited malignant mesothelioma (MM) cell proliferation through the induction of apoptosis in vitro. This DNA-modified siRNA chimera was constructed by substituting six ribonucleotides at the 5´ end of the guide strand with the cognate de-oxyribonuclotides; thus the siRNA could be protected from RNAse or nuclease degradation by partial substitution of ribonucleotides with de-oxyribonucleotides (Kawata et al., Citation2010). Another recent work describes an alternative approach in which expression of new and potent antigens is induced in tumor cells by inhibiting nonsense-mediated messenger RNA decay (NMD). In this study, PSMA aptamer-smg1/siRNA chimera was made to stimulate protective antitumor immunity and it was suggested that the dose treatment of aptamer–siRNA chimeras can be further optimized to inhibit tumor (Pastor et al., Citation2010). To down regulate gene expression through tryptophanase (TNA) interference in combination with aptamer therapy a 21-nucleotide double stranded (ds) siRNA and fluoro-modified aptamers were used. Substituting a borane phosphate for part of natural phosphodiester bond in RNA allows the use of siRNA to persist in cells for longer than unmodified siRNA and the boron neutron capture therapy (BNCT) to specifically kill prostate cancer cells (Shaw et al., Citation2008). Work done with aptamer–siRNA on other targets included use of anti-TAR aptamer (R06) as regulator of TAR-dependant gene expression to benefit affinity and resistance to RNAses (Duconge & Toulme, 1999). The binding between the aptamer and target occurs in form of kissing interactions as R06 has a hairpin RNA loop complementary to TAR and guanosine-adenosine (G-A) combination, stacking interactions and hydrogen bonds at stem/loop–loop junction aids in stability of complex (Duconge & Toulme, 1999; Lebars et al., Citation2007). Modification of R06 by N3´ to P5´ phosphoramidate, 2´-omethyl, hexitol nucleic acid, improved nuclease resistance; however, there was no increase in binding affinity (Lebars et al., Citation2007). A novel dual inhibitory functional anti-gp120 aptamer–siRNA chimera, in which both aptamer and siRNA have potent anti-HIV activities has also been developed (Zhou et al., Citation2008). In this approach, anti-gp120 aptamers (A-1 and B-68) were used which inhibited HIV-1 P24 production and provided more potent inhibition of HIV infectivity. The aptamer and siRNA were linked via sticky sequences consisting of 16 nucleotides at aptamer 3´ end, which were complementary to 16 bases on one of the two-siRNA strands (Zhou et al., Citation2008; Zhou et al., Citation2011). The sticky bridge base approach offered a major advantage in both the chemical synthesis and the opportunity to mix and match the aptamer with different siRNAs in a noncovalent fashion.
Micro RNAs (miRNAs) are short sequences of nucleotides occurring naturally in cells that are important in regulating gene function and expression of pathways by controlling the amount of miRNA that binds to a gene and is possible to upregulate or downregulate the genes (CitationVella & Slack, 2005). The miRNAs can be targeted to the stem cell signaling components or epithelial to mesenchymal transition (EMT) regulators and aptamers can be utilized for directing the application of miRNA as a potent therapeutic (Katoh & Katoh, Citation2008). Various miRNAs have tumor suppressing activity and may play a vital role in other disease prevention or development; hence, they are important in diagnosis and therapy of human diseases (Silahtaroglu A, Citation2010; Kanwar, Citation2010b). Isolation and characterization of RNA aptamers that specifically binds to primary transcript miRNA to the apical loop domain of the pri-miRNA has been done. The results showed that aptamers could be applied as agents for modulating pri-miRNA processing and also as tools for understanding the mechanism (Lunse, Citation2010).
Aptamer–drug conjugates
A work done with aptamer–siRNA chimera dealt with the development of a chimeric molecule using PSMA aptamer with small hairpin RNA (shRNA) against anti-apoptotic factor BCL-XL and drug doxorubicin (dox). Their conjugation into a polyplex of polyethyleneimine-polyethyleneglycol (PEI-PEG) was done for target-specific delivery, synergistic, and selective cancer cell death by aptamer mediated co-delivery of doxorubicin and shRNA was achieved by activation of an intrinsic apoptotic pathway in prostate cancer cells, both in vitro and in vivo (Kim et al., Citation2010). Nucleotide analog reverse transcriptase inhibitors (NRTI), for example, Azidothymidine triphosphate (AZTTP) and 2´, 3´-Dideoxy cytidine-5´-triphosphate (ddCTP), are chain terminators that prevent completion of viral genome replication following their incorporation in reverse transcriptase (RT) during DNA polymerization (Enomoto L, Citation2011). A recent work tested aptamers in combination with NRTI, which displayed significant synergy for inhibition of DNA polymerization by HIV1-RT, where the RNA and DNA aptamers were generated by SELEX, specific for HIV-1 RT (Held et al., Citation2006).
Aptamer–dye conjugates for nanodiagnostics
In presence of mercury, Hg(2+) and lead, Pb(2+) a thrombin-binding aptamer containing six thymine and nine guanine units changes into a hairpin like structure (Smirnov & Shaffer, Citation2000), thus thymine-containing thrombin-binding aptamers can be used to target mercury, Hg(2+) (Miyake et al., Citation2006). According to a recent chimeric approach, the domains of two aptamers can be linked, one that engages a fluorophore for signaling and other that binds a nonfluorescent target, into one sequence in such a way that the binding of the nonfluorescent target will strengthen or reduce the affinity of the chimeric aptamer for fluorophore. The signal is produced as a result of the aptamer–fluorophore association or separation that is accompanied by a change in fluorescence intensity. Similarly, multiple targets can be simultaneously reported by using two or more structure substituting aptamers labeled with different fluorophores, for example, the adenosine tri-phosphate (ATP) binding aptamer, ATP1.1, can form a signaling duplex with FDNA1 (carrying fluorescein) and QDNA2, while the guanosine tri-phosphate (GTP) binding aptamer, GTP1.2, can form a signaling duplex with FDNA2 (carrying cy3) and QDNA2. The resulting mixture can generate a real-time signal for both ATP and GTP when both reporters are mixed into a single solution (Nutiu & Li, Citation2005).
To facilitate the diagnosis, identification, and study of mechanism of various diseases and pathways, highly efficient aptamer-based detection systems have also been validated. To detect the presence of cocaine in samples, the functionalized quantum dots (QDs) consisting of two subunits of aptamers were used, where one was linked to QD and the other was specific to cocaine and formed the cocaine–aptamer–aptamer–QD complex in presence of cocaine, where cocaine induced a conformational change in the complex leading to change in QD behavior (Freeman, Citation2009; Zhang & Johnson, Citation2009). Detection of ATP has been achieved by adsorption and covalent coupling of ATP-binding DNA aptamers onto cellulose, where the properties of cellulose enhance the DNA aptamer activity (Su et al., Citation2007). Another study introduced a new fluorescent method for detection of ATP using the following three different oligonucleotides in a complex; (i) A 3´-biotin modified DNA specific to streptavidin bound to streptavidin coated on QDs; (ii) 3´-cy5 labeled DNA; and (iii) an aptamer specific to ATP. In the absence of ATP, all three oligonucleotides formed a stable complex and thus normal fluorescence was observed, whereas, in the presence of ATP, the 3´cy5 labeled DNA got dissociated from the complex and the fluorescence intensity changed (Chen et al., Citation2008). Angiogenin is a potent angiogenic protein which has a role in tumor angiogenesis and RNA transcription (Xua et al., Citation2002). Cellular internalization of angiogenin was investigated by a highly specific fluorophore-labeled aptamer for angiogenin by real-time protein imaging in living cells using confocal laser scanning microscopy (CitationLi, 2008). Other examples include detection of C-reactive protein (CRP) which is highly expressed in inflammation; it also serves as a biomarker (Tsimikas et al., Citation2006). The detection was based on a sandwich format where a biotinylated RNA aptamer specific to CRP was immobilized on streptavidin-coated magnetic beads; the modified beads were incubated with CRP solution and recoupled with same biotinylated aptamer. After the binding, the extent of affinity was evaluated by addition of enzymatic substrate, which was transformed into an electroactive product and the detection of the enzymatic reaction was done by differential pulse voltametry (DPV) (Centi et al., Citation2010). Super paramagnetic iron oxides (SPION) were used as nanoparticles; A10 RNA aptamer against PSMA was tagged with doxorubicin (dox) and covalently bound to the nanoparticles, which induced magnetization inside a magnetic field (Wang et al., Citation2008).
For imaging applications, the aptamers can be bound to imaging agents such as fluorophores, QDs, magnetic resonance imaging (MRI) agents, magnetic nanoparticles, etc. The method of visualization mainly depends on the type of nanoparticle used or the modifications made in the aptamer and aptamer chimerics which aid in their detection. In a recent work, the AS1411 aptamer specific to nucleolin protein was conjugated to cobalt-ferrite nanoparticles surrounded by fluorescent rhodamine within a silica shell and was incorporated into cancer cells for imaging. The imaging was based on magnetic fluorescence nanoparticles inside cancer cells by MRI (Hwang et al., Citation2010). Theophylline is a bronchodilator used in case of asthma, bronchitis, and emphysema; since its overdose can lead to toxic effects, it is important to detect its levels in body. The AuNP were conjugated to theophylline-specific aptamer. In the presence of theophylline, the aptamer dissociates from AuNP leading to an aggregation of AuNP and an increase in AuNP plasmonic peak. The observed intensity enables the detection of theophylline (Chavez et al., Citation2010). In another study, theophylline was studied and detected by ferrocene (Fc) redox-labeled RNA aptamer. The aptamer was able to quantify the theophylline in serum and thus proved to be an efficient biosensor (Ferapontova & Gothelf, Citation2009). Redox active probes are used in combination with aptamers, and by measuring the changes in their redox states, the electrochemical detection is carried out, such as in case of thrombin-bound aptamer (Smirnov & Shaffer, Citation2000). Lysozyme detection has been made possible by using an aptamer-functionalized silica nanoparticles where once the aptamer interacts with the protein, a fluorescence signal is emitted by anionic poly(fluorene-alt-vinylene) (PFVSO3) via electrostatic interaction. This detection can be done by naked eye and is a remarkable achievement in recent trend (Wang et al., Citation2010a) (). In another work, atomic force microscopy-based imaging of nucleosomal arrays was done by acetylation of histones at 16th lysine residue and recognized by a DNA aptamer specific for histones (Lin et al., Citation2009).
Table 2. Aptamer based detection systems.
Hence a lot of work has been done on various targets with different types of aptamer chimeras; however, the strategy and the parameters considered for their selection remains quite the same. The most successful and commonly used aptamer chimeras are the aptamer–siRNA chimeras, which have been used extensively to target various diseases.
Mechanisms and strategies of delivery of chimeras
It is known that aptamers cannot enter the cell membrane directly by simple diffusion due to their hydrophobic nature; hence, various studies have been conducted to study the mechanism of entry of aptamer into a cell.
Carrier peptides
Due to the poor permeability and selectivity of the cell membrane, there is a need of a strategy that can transport aptamers or drugs into large number of cells. The studies indicate that the mechanism of endocytosis best explains the intracellular trafficking of aptamers. Endocytosis and related pathways like classic clathrin-coated pit pathway, caveolar pathway, clathrin-independent noncaveolar pathway, macropinocytosis, and phagocytosis are the mechanisms that explain the entry of aptamers in cells (Kirkham & Parton, Citation2005; Perret et al., Citation2005). The complex flow of endomembrane traffic is regulated by Rab family of GTPases and by tethering complexes (Stenmark & Olkonnen, Citation2001), while vesicular fusion phenomenon is controlled by SNARE proteins; the sorting nexins (SNX) are also important in sorting and cargo retrieval from endosomes (Carlton et al., Citation2005; Van dergoof, Citation2006; Cai et al., Citation2007). Carrier peptides are small protein domains with the ability to cross biological membranes efficiently with or without specific receptors to promote the delivery of desired drug into target cells (Morris et al., Citation2001).There are examples where cell targeting ligands (CTLs) that enhance transmembrane permeation have been employed to deliver aptamers (Morris et al., Citation1997) as there is a wide variety of ligands available including antibodies, polypeptides derived from phage display libraries, and small organic molecules (Hollinger & Hudson, Citation2005). Cell penetrating peptides (CPPs) are small peptides of 30 amino acid residues with a net positive charge which can translocate across plasma or endomembranes, transport biomacromolecular cargos into cytoplasm and nucleus; few examples included are: R9 (arginine 9), MPG peptide, transportan, Tat, penetratin, etc. (Morris et al., Citation1997; Krissansen et al., Citation2006; Baratchi et al., Citation2010; Cheung et al., Citation2010) Several reports of noncovalent complexes between anionic siRNA chain aptamer and cationic CPPs confirm that they provided effective delivery to cells in culture (Morris et al., Citation1997; Lundberg et al., Citation2007).
Nanoparticle systems
Aptamer technology has evolved in a multidirectional fashion finding importance and playing essential roles in fields of biosensing, imaging, drug delivery, and modern molecular medicine for therapy for various diseases and disorders. Taking the advantages of rapidly expanding nanobiotechnology-based developments; aptamer–nanoparticle conjugation forms the basis of a new chemical and biological strategy with wide application starting from assembly to detection. Because of their small size, nanoparticles can interact readily with biomolecules both on surface and within the cells and thus they are considered as a revolutionary approach for detection and therapy of various diseases. Various conjugation principles have been described leading to the application of aptamers in cancer and other inflammatory diseases as a nanodelivery system (Kanwar et al., Citation2010c). Since the aptamers are negatively charged moieties, the charge of the nanoparticle surface is very important for the conjugation between the nanoparticle and the aptamers (Farokhzad et al., Citation2004). Covalent conjugation of aptamers to nanoparticles is mainly induced by succinimidyl-ester-amine conjugations, whereas, the noncovalent conjugations include affinity interactions, metal co-ordinations, surface adsorption, etc. (Farokhzad et al., Citation2005) The conjugation can be quantitatively confirmed by various methods, such as flow cytometry, X-ray photoemission spectroscopy, Fourier transform infrared spectroscopy (FTIR), fluorescent microscopy, etc. (Hicke & Stephens, Citation2000). Interestingly, a chimeric phage RNA (pRNA)-aptamer (specific to CD4 antigen on T-helper cells) conjugate was synthesized and labeled with fluorescein isothyocyanate (FITC) for detection of CD4 and with this a trimeric complex of pNA-aptamer (CD4), pRNA-FITC, and pRNA-rhodamine was prepared. Using this as a nanocarrier for therapy and diagnosis (Khaled et al., Citation2005), the cellular internalization was studied with confocal microscopy. When siRNA was attached to pRNA trimer complex for silencing the CD4 surface receptor’s gene expression, a decrease in CD4 expression in FITC positive cells was observed (Khaled et al., Citation2005). Another work comprised of synthesis of polylactic acid (PLA)-polyethyleneglycol (PEG) copolymers incorporated with paclitaxel (ptxl). The ptxl was incorporated into the PLA-PEG copolymer which was then attached to A10 PSMA aptamer with 5´-NH2 modifications and was tested for successful uptake in human prostate cancer cells (LNCAP and PC3) (Tong et al., Citation2010). The A10 RNA aptamer with 2´-F (fluoropyrimidine) modification was tagged to poly(lactide-co-glycolide) (PLGA) as a model-controlled release polymer system and PEG (a model hydrophilic polymer with anti-biofouling properties), to develop a triblock copolymer comprising of PLGA-PEG-A10 aptamer, which is a targeted nanodelivery system (GU et al., Citation2008).
Nanocapsule-based targeted multidrug delivery
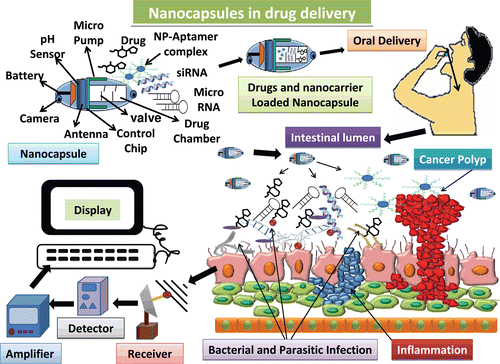
Most of these methods are well characterized and ensure appropriate detection of the desired molecule. Yet, modern developments are able to show light on the drawbacks of current methods. Nanocarriers are vehicles mainly used for drug delivery to enhance the stability and to prolong the circulation of the drug in the target tissue by promoting their permeability and retention. There are several drawbacks of nanocarriers, such as their limited drug loading capacity, difficulty in tracking the movement of the drug, imaging and diagnosis, no real-time diagnostics, lack of pH, and thermal stability. All these factors lead to lack of knowledge of mechanism of drug uptake by the cells. The mechanism of internalization of nanocarrier into target cells mainly depends on physiochemical property of the nanocarrier itself. Thermal stability and pH of nanocarrier are keys and essential factors for appropriate drug delivery. Over the time, it is evident that nanocarriers have a great potential in drug delivery. The need is to exploit, optimize, and enhance the intracellular and subcellular delivery of drugs, which are unstable or stable in physiological conditions to the target site. There is also a need to develop a multidrug nanocarrier for delivery of specific drugs for diverse functions in a particular or in various target sites. Recently multifunctional nanocapsules have emerged that are capable of providing therapy and diagnosis in real time (). These nanocapsules are portable and thus can be used on and off the field; they possess thermal stability, pH sensibility and are capable of carrying several different drugs at the same time. They are equipped with a camera for real-time imaging. Such efficiency will help to study the intracellular fate of the various drugs as well as real-time imaging and monitoring them. These drugs can be monitored distantly as well by means of satellite by more than one personnel in different places at the same time. The release of drug can be monitored well through the computed systems and once the nanocapsules reach the target the valves can be opened in order to release the drugs. Use of biodegradable nanocapsules will minimize the chance of drug toxicity and size of nanocapsules can be regulated considering the amount of drug to be loaded. The aptamer will bind to the target due to its high specificity, whereas the other drugs will bind to their respective target due to expression of various markers and receptors. This development will increase the possibility of targeting the nanocarriers to the desired cells and to study the nanocarrier cell interaction (Hillaireau & Couvreur, Citation2009; Huertas et al., Citation2010; Shen et al., Citation2010; Wang & Ho, Citation2010b).
Figure 4. Representation of future generation drug delivery by means of nanocapsules that are equipped with real-time imaging and wireless monitoring chips. These nanocapsules can be loaded with several drugs at the same time, and can be taken orally due to lack of any toxic effects. Once present in intestine the nanocapsules can be monitored by the controlling system to release the drugs by opening the valves. The drugs can act specifically on the respective target sites due to presence of various receptors and thus multiple diseases can be targeted at the same time and can be monitored as well in real time. These drugs can be monitored by means of satellite at different places simultaneously hence providing better diagnosis by the experts (Huertas et al., Citation2010; Shen et al., Citation2010).
Multitargeted LNA-aptamer complexes with anticancer-loaded iron-saturated bovine lactofeerin dopamine surface modified Fe3O4 nanoparticles for specific killing of tumor cells
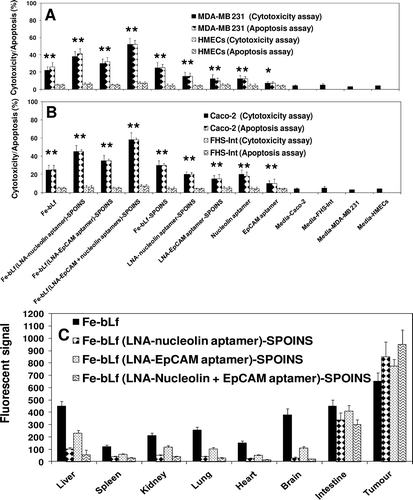
We showed that 100% iron-saturated bovine lactofeerin (Fe-bLf) acts as a potent natural adjuvant and fortifying agent for augmenting cancer chemotherapy with broad utility in the treatment of cancer (Kanwar et al., Citation2008). The key findings of the study revealed: (i) Fe-bLf bound to the intestinal epithelium and was preferentially taken up within Peyer’s patches(); (ii) Fe-bLf enhanced antitumor activity in combination with major anticancer drugs (paclitaxel, doxorubicin, epirubucin, or flurouracil), the combination being capable of completely eradicating large tumors of EL4 lymphoma, Lewis lung carcinoma, B16 melanoma (0.6 cm diameter) that were otherwise completely insensitive to chemotherapy; (iii) Natural bLf, 4% iron-saturated or 50% iron-saturated bLf were less effective at potentiating cancer chemotherapy, and did not cause tumor eradication; (iv) Fe-bLf increased the production of Th1 and Th2 cytokines within the intestine and tumor, including TNF-α, IFN-γ, as well as nitric oxide (NO) that have been reported to sensitize tumors to chemotherapy; (v) Fe-bLf almost completely reduced tumor vascularity (angiogenesis) and blood flow, and increased tumor apoptosis regulated by survivin, Bcl-2, caspases 9/3, and Fas molecules; (vi) Fe-bLf increased leukocyte infiltration (CD4+,CD8+, NK, and dendritic cells) to tumors, lamina propria, and spleen; and (vii) importantly, Fe-bLf restored both red (RBC) and white (WBC) blood cell numbers depleted by chemotherapy, potentially fortifying the mice against cancer (Kanwar et al., Citation2008; Gibbons et al., Citation2011; Kanwar RK et al., Citation2011). The Fe-bLf was prepared (Kanwar et al., Citation2008). Superparamagnetic iron oxide (Fe3O4) nanoparticles (SPIONs), which are nontoxic, biodegradable, inexpensive and candidate platforms for the build-up of theranostic nanostructures were prepared as described (Xie et al., Citation2010, Soundararajan et al., Citation2008). Here, we used dopamine to modify the surface of SPIONs for yielding nanoconjugates that can be easily encapsulated into anticancer Fe-bLf protein. These nanocarrier systems were well-suited for dual encapsulation of SPIONs for imaging or monitoring therapy and drug molecules, because the encapsulation or conjugation is achieved in a way that is similar to common drug loading. The LNA-nucleolin DNA aptamer (molecule present on cancer cell surfaces and translocated to nucleus) (Soundararajan et al., Citation2008) and LNA-EpCAM RNA aptamer (Shigdar et al., Citation2011), known to be over-expressed on the apical side of the cancer cell surfaces were prepared (Hertoghs et al., Citation2003) and loaded individually or in combination on these Fe-bLf-SPOINS nanocarriers. The average size of the nanocarriers in various steps of their preparations were found to be (170 ± 35 nm) as determined by dynamic light scattering (DLS), scanning electron microscopy (SEM), and transmission electron microscopy (TEM), Fourier transform infrared spectroscopy (FTIR), differential scanning calorimeter (DSC), thermogravimetric analysis (TGA), and X-ray diffraction (XRD). These LNA-aptamers-loaded Fe-bLf nanocarriers were treated with human cancer cell line Caco-2 (human colon cancer), MDA-MB 231 (human breast cancer), normal HMECs (human mammary epithelial cells), and normal FHs 74 Int (an adherent human primary fetal small intestinal) cell lines obtained from the American Type Culture Collection (ATCC, USA). These LNA-aptamer complexes of Fe-bLf nanocarriers induce massive cell death in breast and colon cancer cells in vitro and spare normal HMECs and FHs 74 Int cells in an in vitro co-culture model ( and ). A rotational magnetic field frequency of 1Hz has shown a maximum amount of LDH release or cytotoxicity and cell death through apoptosis measured by Tunnel and Annexin-V positive cells compared to normal cells (Kanwar et al., Citation2001; Kanwar et al., Citation2008). After applying magnetic field with 1Hz, we found that the cancer cells massively increased in breast and colon as compared to normal HMECs and normal FHs 74 Int cell lines (data not shown). This indicates that SPIONs-loaded LNA-aptamers have the potential to kill cancer cells more specifically, effectively, and spare normal cells. We evaluated the tissue biodistribution of these nanocarriers using a fluorescent marker, 6-coumarin and the fluorescent quantum yield characters enabled the selective tracking of the in vivo distribution of nanocapsule (Woodrow, Citation2009) (. These signals were captured by magnetic sensors and release of the Fe-bLf -loaded LNA-aptamers from nanocarriers was monitored on computer from outside (unpublished information). The release of nanocarriers can be controlled in in vivo situations not only in human gut associated microbial (parasitic or viral or bacterial) infections, inflammations, and cancers but also anywhere in the body by LNA-aptamers targeting nanocarriers (). Thus, we developed LNA-modified nucleolin aptamers and LNA-modified EpCAM aptamers conjugated “Fe-bLf natural anticancer protein-loaded nanobullet nanocarriers,” which specifically target cancer cells and spare normal cells. Hence, we developed natural nanomedicinal-based war against cancer cells with targeted nanobullet nanocarriers that specifically induce their traumatic death and spare normal cells. Superparamagnetic iron oxide (Fe3O4) nanoparticles (SPIONs) nature of these nanocarriers was used to monitor the size of tumors in the treated mice through different imaging systems such as in vivo positron emission tomography (PET)/near-infrared fluorescence (NIRF)/magnetic resonance imaging (MRI) for future potential contrast material for cancer diagnosis as described earlier (Xie et al., Citation2010).
Figure 5. Cell cytotoxicity determined by LDH release assay and cell death (apoptosis) by TUNEL assay of (A) breast cancer cells and (B) colon cancer cells and compared with normal cells following treatment with 1600 μg/mL of iron saturated lactoferrin (Fe-bLf) and other control nanoformulations and control aptamers (nucleolin and EpCAM). Cells were treated for 24 h with different nanoformulations and stained by TUNEL analysis for apoptotic cells. Cell death is shown here in terms of apoptotic (% apoptosis) and LDH release (% cytotoxicity). All treatments were performed in triplicate and assay was repeated three times independently with similar results. The mean for representative experiment was calculated and presented as a mean ± SD values; ** indicates a highly significant p < 0.001 value from the normal control cell lines and with media only; * indicates a significant p < 0.05 value from the normal control cell lines and control with media only, (C) Biodistribution study of nanocarriers compared with Fe-bLf in oral administrations. Fluorescent signal of tissue extracts after 36 h of oral administration. Nanocarriers were labeled with coumarin (10mg/mL).
Aptamers in clinical trials
With new developments in nanotechnology and chemistry, few aptamers have found their way into modern medicine. The pharmaceutical industries are ready to invest more on recent nano-aptamer technology which has added benefits when compared to the traditional drugs. In a very short span of time, aptamers have attained critical importance in the field of modern molecular medicine and nanomedicine. Several aptamers are in clinical trial such as, EYE001 targeting vascular endothelial growth factor receptor (VEGFR); phase II/III (Carrasquillo et al., Citation2003). Edifoligide aptamer which is used to treat vein graft failure of heart and leg; phase III (C. Inc, 2004). The E100300 to target platelet derived growth factor (PDGF) (phase II) (JO et al., Citation2006). The Nu172 to target thrombin; phase II (Wagner-Whyte et al., Citation2007), REG1, which targets factor IXa; phase IIb (Becker, Citation2009), ARC1779, which inhibits the acute coronary syndromes by preventing the binding of platelet receptor glycoprotein Ib to its receptor; phase I/II (ClinicalTrials.gov, 2009), AS1411, which is being used to treat cancer by inhibiting their DNA replication; phase III (Stuart et al., Citation2009), ARC1905, which prevents age-related macular degeneration (AMD); phase I (ClinicalTrials.gov, 2010), LY2181308, which is used in treatment of nonsmall cell lung cancer (NSLC); phase III (Cancerhelp.org.uk, 2010), and many others. However, macugen was the first aptamer to get the FDA approval in 2004 for treatment of wet AMD (NG et al., Citation2006).
Conclusion
Various chimeras such as aptamer–aptamer, aptamer–siRNA, aptamer–miRNA, aptamer–enzyme, aptamer–antibody, and aptamer–NRTI have been established. All of these chimeras show better stability and functional capability than the parent molecules used to form the chimera. Several methodologies have been established to produce and select the chimeras based on their specificity such as chimeric SELEX. Specific delivery of chimeric aptamers has been achieved by their conjugation with various nanoparticles and according to the nature of nanoparticles or the modifications in the aptamer chimeras and can be used for imaging and diagnosis. Aptamer chimerism offers a wide range of activity due to the presence of two molecules playing two or more different roles simultaneously. They have an additional advantage over the existing methodology and are highly stable, more flexible with various types of modifications, and cost effective. Since aptamers are nonimmunogenic, most of the aptamer chimerics can be expected to be less or nonimmunogenic. Aptamer chimerics have various applications, such as tumoricidal activity, detection of small analytes, HIV inhibitory properties and detection of pathogens in food samples, personalized drug and vaccine development against parasites, bacterial, and viral agents, targeting cancer cells at a subcellular level, such as the cell organelles, and targeting different cellular cascade pathways. Well-characterized chimerics can be brought into therapeutic use for various inflammatory disorders as well. Because aptamer chimeras can be generated for several biomarkers and receptors, they offer a better specificity, target recognition, and improved therapeutics than any antibodies or synthetic peptides. Recently, various drug delivery and imaging systems have been developed which can be further improved with the help of aptamer chimerics. Nanocapsules with modern technologies for real-time imaging can be used to monitor the interaction between the aptamer chimeras and target cells. Multivalent nanoparticles, such as dendrimers with high potential for performing several functions simultaneously can be developed by the use of chimeric aptamers. Thus, aptamer chimeras may prove to be useful in future for a wide range of diseases.
Declaration of interest
The work was supported by the grants from Institute of Biotechnology, Institute for Technology and Research Innovation and the Australia-India Strategic Research Fund (AISRF BF030016).
Michael M. Cox
References
- Alaoui AT et al. 2002. High affinity nucleic acid aptamers for streptavidin incorporated into bi-specific capture ligands. Nucl Acids Res 30:45–49.
- Alonso MJ, Gupta RK, Min C, Siber GR, Langer R. 1994. Biodegradable microspheres as controlled-release tetanus toxoid delivery systems. Vaccine 12:299–306.
- Altieri DC. 2003. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 22:8581–8589.
- Baratchi S, Kanwar RK, Kanwar JR. 2010. Survivin: A target from brain cancer to neurodegenerative disease. Crit Rev Biochem Mol Biol 45:535–554.
- Barciszewski J, Medgaard M, Koch T, Kurreck J, Erdmann VA. 2009. Locked nucleic acid aptamers. Methods Mol Biol 535:165–186.
- Barry P, Robert JD, Gold L. 1996. High affinity nucleic acid ligands that discriminate between theophylline and caffeine. US Patent No 5, 580,737.
- Bassett SE, Fennewald SM, King DJ, Li X, Herzog NK, Shope R, Aronson JF, Luxon BA, Gorenstein DG. 2004. Combinatorial selection and edited combinatorial selection of phosphorothioate aptamers targeting human nuclear factor-kappaB RelA/p50 and RelA/RelA. Biochemistry 43:9105–9115.
- Becker RC, Chan MY. 2009. REG-1, a regimen comprising RB-006, a Factor IXa antagonist, and its oligonucleotide active control agent RB-007 for the potential treatment of arterial thrombosis. Curr Opin Mol Ther 11:707–715.
- Berezovski M, Krylov SN. 2004. Using nonequilibrium capillary electrophoresis of equilibrium mixtures for the determination of temperature in capillary electrophoresis. Anal Chem 76:7114–7117.
- Bivalkar-Mehla Svj Mehla, R, Abreha M, Kanwar JR, Tikoo A, Chauhan A. 2010. Viral Rna silencing suppressors (Rss): Novel strategy of viruses to ablate the host Rna interference (Rnai) defense system. Virus Res 155:1–9.
- Bruno JG, Kiel JL. 2002. Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. BioTechniques 32:178–80–182.
- Burke DH, Gold L. 1997a. RNA aptamers to the adenosine moiety of S-adenosyl methionine: Structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Res 25:2020–2024.
- Burke DH, Hoffman DC, Brown A, Hansen M, Pardi A, Gold L. 1997b. RNA aptamers to the peptidyl transferase inhibitor chloramphenicol. Chem Biol 4:833–843.
- Burke DH, Willis JH. 1998. Recombination, RNA evolution, and bifunctional RNA molecules isolated through chimeric SELEX. RNA 4:1165–1175.
- Cheng, C, Li, LM. 2008. Inferring MicroRNA activities by combining gene expression with MicroRNA target prediction. Plos One 3:1–9.
- Corgentech Inc (2004, 07 Dec). From Edifoligide (E2F Decoy) Phase 3 Trial For Peripheral Bypass Graft. http://www.medicalnewstoday.com/articles/17439.php.
- Cai H, Reinisch K, Ferro-Novick S. 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12:671–682.
- Cancerhelp.,Org.UK (2010). A trial of LY2181308 alongside chemotherapy for non small cell lung cancer. http://www.cancerhelp.org.uk/trials/a-trial-of-ly2181308-alongside-chemotherapy-for-non-small-cell-lung-cancer
- Carlton J, Bujny M, Rutherford A, Cullen P. 2005. Sorting nexins–unifying trends and new perspectives. Traffic 6:75–82.
- Carrasquillo KG, Ricker JA, Rigas IK, Miller JW, Gragoudas ES, Adamis AP. 2003. Controlled delivery of the anti-VEGF aptamer EYE001 with poly(lactic-co-glycolic)acid microspheres. Invest Ophthalmol Vis Sci 44:290–299.
- Centi S, Tombelli S, Palchetti I. & Mascini M. 2010. Development of an aptamer-based electrochemical sandwich assay for the detection of a clinical biomarker. Sensors and Lecture Notes in Electrical Engineering 54:207–210.
- Chávez JL, Lyon W, Kelley-Loughnane N, Stone MO. 2010. Theophylline detection using an aptamer and DNA-gold nanoparticle conjugates. Biosens Bioelectron 26:23–28.
- Chen Z, Li G, Zhang L, Jiang J, Li Z, Peng Z, Deng L. 2008. A new method for the detection of ATP using a quantum-dot-tagged aptamer. Anal Bioanal Chem 392:1185–1188.
- Cheung CH, Sun X, Kanwar JR, Bai JZ, Cheng L, Krissansen GW. 2010. A cell-permeable dominant-negative survivin protein induces apoptosis and sensitizes prostate cancer cells to TNF-a therapy. Cancer Cell Int 10:36.
- Chi-Hong B, Chen KRD, Christopher PO, Cecilia DS, Meikana L, George AC, Michelle G, Francis EB, Audrey LF, Rachel M, Elizabeth FN. 2008. Aptamer-based endocytosis of a lysosomal enzyme. Proc Natl Acad Sci USA 105:15908–15913.
- Clinicaltrials.,Gov. 2010. A Study of Arc1905 (Anti-C5 Aptamer) in Subjects With Dry Age-related Macular Degeneration. http://clinicaltrials.gov/show/Nct00950638
- Clinicaltrials.,Gov. 2009. Arc1779 Injection in Patients With Von Willebrand Factor-Related Platelet Function Disorders. http://clinicaltrialsfeeds.org/clinical-trials/show/Nct00632242
- Collett JR, Cho EJ, Ellington AD. 2005. Production and processing of aptamer microarrays. Methods 37:4–15.
- DI Primo C et al. 2007. Systematic screening of Lna/20-O-methyl chimeric derivatives of a Tar Rna aptamer. Febs Letters 581:771–774.
- Ducongé F, Toulmé JJ. 1999. In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA 5:1605–1614.
- Eckstein F, Gish G. 1989. Phosphorothioates in molecular biology. Trends Biochem Sci 14:97–100.
- Eli G. 2010. Aptamer-targeted sirna to prevent attenuation or suppression of a T cell function. United States patent application, 12/752,802.
- Ellington, AD, Szostak, JW. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346,818–822.
- Ellington ADS, Szostak W. 1992. Selection in vitro of single-stranded Dna molecules that fold into specific ligand-binding structures. Nature 355:850–852.
- Enomoto, L, Anderson, PL, Li, S, Edelstein, CL, Weinber, GA. 2011. Effect of Nucleoside and Nucleotide Analog Reverse Transcriptase Inhibitors on Cell-Mediated Immune Functions. AIDS Research and Human Retroviruses 27:47–55.
- Eulberg D, Klussmann S. 2003. Spiegelmers: Biostable aptamers. Chembiochem 4:979–983.
- Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. 2004. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res 64:7668–7672.
- Farokhzad OC, Khademhosseini A, Jon S, Hermmann A, Cheng J, Chin C, Kiselyuk A, Teply B, Eng G, Langer R. 2005. Microfluidic system for studying the interaction of nanoparticles and microparticles with cells. Anal Chem 77:5453–5459.
- Ferapontova EE, Gothelf KV. 2009. Effect of serum on an RNA aptamer-based electrochemical sensor for theophylline. Langmuir 25:4279–4283.
- Freeman Rea. 2009. Self assembly of superamolecular aptamer structures for optical or electrochemical sensing. Analyst 134:653–656.
- Gibbons JA, Kanwar RK, Kanwar JR. 2011. Lactoferrin and cancer in different cancer models. Front Biosci (Schol Ed) 3:1080–1088.
- Goldstein NI, Prewett M, Zuklys K, Rockwell P, Mendelsohn J. 1995. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res 1:1311–1318.
- Green LS, Jellinek D, Bell C, Beebe LA, Feistner BD, Gill SC, Jucker FM, Janjic N. 1995. Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem Biol 2:683–695.
- GU F et al. 2008. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. PNAS 105:2586–2591.
- Haller AA, Sarnow P. 1997. In vitro selection of a 7-methyl-guanosine binding RNA that inhibits translation of capped mRNA molecules. Proc Natl Acad Sci USA 94:8521–8526.
- Heckel A, Buff MC, Raddatz MS, Müller J, Pötzsch B, Mayer G. 2006. An anticoagulant with light-triggered antidote activity. Angew Chem Int Ed Engl 45:6748–6750.
- Held DM, Kissel JD, Saran D, Michalowski D, Burke DH. 2006. Differential susceptibility of HIV-1 reverse transcriptase to inhibition by RNA aptamers in enzymatic reactions monitoring specific steps during genome replication. J Biol Chem 281:25712–25722.
- Hernandez FJ, Kalra N, Wengel J, Vester B. 2009. Aptamers as a model for functional evaluation of LNA and 2’-amino LNA. Bioorg Med Chem Lett 19:6585–6587.
- Hertoghs KM, Ellis JH, Catchpole IR. 2003. Use of locked nucleic acid oligonucleotides to add functionality to plasmid DNA. Nucleic Acids Res 31:5817–5830.
- Hicke BJ, Stephens AW. 2000. Escort aptamers: A delivery service for diagnosis and therapy. J Clin Invest 106:923–928.
- Hillaireau H, Couvreur P. 2009. Nanocarriers’ entry into the cell: Relevance to drug delivery. Cell Mol Life Sci 66:2873–2896.
- Holliger P, Hudson PJ. 2005. Engineered antibody fragments and the rise of single domains. Nat Biotechnol 23:1126–1136.
- Homann M, Göringer HU. 1999. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res 27:2006–2014.
- Huertas CEM, Fessi H, Elaissari A. 2010. Polymer-based nanocapsules for drug delivery. International Journal of Pharmaceutics 383:113–142.
- Hwang DW et al. 2010. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. The Journal of Nuclear Medicine 51:98–105.
- Jenison SGRD, Pardi A, Polisky B. 1994. High-resolution molecular discrimination by RNA. Science 263:1425–1429.
- Jensen KB, Atkinson BL, Willis MC, Koch TH, Gold L. 1995. Using in vitro selection to direct the covalent attachment of human immunodeficiency virus type 1 Rev protein to high-affinity RNA ligands. Proc Natl Acad Sci USA 92:12220–12224.
- JO MCN, JU M, Cheung E, Nishijima K, Robinson GS, Adamis AP, Shima DT. 2006. Inhibition of platelet-derived growth factor B signaling enhances the efficacy of anti-vascular endothelial growth factor therapy in multiple models of ocular neovascularization. Am J Pathol 168:2036–2053.
- Justin PD et al. 2009. Systemic administration of optimized aptamer-sirna chimeras promotes regression of Psma-expressing tumours. Nat Biotechnol 27:839–849.
- Kanwar JR, Kamalapuram SK, Kanwar RK. 2010a. Targeting survivin in cancer: Patent review. Expert Opin Ther Pat 20:1723–1737.
- Kanwar JR, Mahidhara G, Kanwar RK. 2010b. MicroRNA in human cancer and chronic inflammatory diseases. Front Biosci (Schol Ed) 2:1113–1126.
- Kanwar JR, Mohan RR, Kanwar RK, Roy K, Bawa R. 2010c. Applications of aptamers in nanodelivery systems in cancer, eye and inflammatory diseases. Nanomedicine (Lond) 5:1435–1445.
- Kanwar JR, Palmano KP, Sun X, Kanwar RK, Gupta R, Haggarty N, Rowan A, Ram S, Krissansen GW. 2008. ‘Iron-saturated’ lactoferrin is a potent natural adjuvant for augmenting cancer chemotherapy. Immunol Cell Biol 86:277–288.
- Kanwar JR, Shen WP, Kanwar RK, Berg RW, Krissansen GW. 2001. Effects of survivin antagonists on growth of established tumors and B7-1 immunogene therapy. J Natl Cancer Inst 93:1541–1552.
- Kanwar JR, Singh N, Kanwar RK. 2011. Role of nanomedicine in reversing drug resistance mediated by ATP binding cassette transporters and P-glycoprotein in melanoma. Nanomedicine (Lond) 6:701–714.
- Kanwar RK, Cheung CH, Chang JY, Kanwar JR. 2010. Recent advances in anti-survivin treatments for cancer. Curr Med Chem 17:1509–1515.
- Kanwar RK, Kanwar JR. 2011. Immunomodulatory lactoferrin in the regulation of apoptosis modulatory proteins in cancer. Protein & Peptide Letters. Accepted on 05-08-2011.
- Kasahara Y, Kitadume S, Morihiro K, Kuwahara M, Ozaki H, Sawai H, Imanishi T, Obika S. 2010. Effect of 3’-end capping of aptamer with various 2’,4’-bridged nucleotides: Enzymatic post-modification toward a practical use of polyclonal aptamers. Bioorg Med Chem Lett 20:1626–1629.
- Katoh Y, Katoh M. 2008. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int J Mol Med 22:271–275.
- Kawata E, Ashihara E, Nakagawa Y, Kiuchi T, Ogura M, Yao H, Sakai K, Tanaka R, Nagao R, Yokota A, Takeuchi M, Kimura S, Hirai H, Maekawa T. 2010. A combination of a DNA-chimera siRNA against PLK-1 and zoledronic acid suppresses the growth of malignant mesothelioma cells in vitro. Cancer Lett 294:245–253.
- Keefe AD, Cload ST. 2008. SELEX with modified nucleotides. Curr Opin Chem Biol 12:448–456.
- Khaled A, Guo S, Li F, Guo P. 2005. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett 5:1797–1808.
- Kim E, Jung Y, Choi H, Yang J, Suh JS, Huh YM, Kim K, Haam S. 2010. Prostate cancer cell death produced by the co-delivery of Bcl-xL shRNA and doxorubicin using an aptamer-conjugated polyplex. Biomaterials 31:4592–4599.
- Kirkham M, Parton RG. 2005. Clathrin-independent endocytosis: New insights into caveolae and non-caveolar lipid raft carriers. Biochim Biophys Acta 1745:273–286.
- Krissansen GW, Singh J, Kanwar RK, Chan YC, Leung E, Lehnert KB, Kanwar JR, Yang Y. 2006. A pseudosymmetric cell adhesion regulatory domain in the beta7 tail of the integrin alpha4beta7 that interacts with focal adhesion kinase and src. Eur J Immunol 36:2203–2214.
- Kusser W. 2000. Chemically modified nucleic acid aptamers for in vitro selections: Evolving evolution. J Biotechnol 74:27–38.
- Laity JH, Lee BM, Wright PE. 2001. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol 11:39–46.
- Lebars I, Richard T, Di Primo C, Toulmé JJ. 2007. LNA derivatives of a kissing aptamer targeted to the trans-activating responsive RNA element of HIV-1. Blood Cells Mol Dis 38:204–209.
- Li, W, Yang, X, Wang, K, Tan, W, He, Y, Guo, Q, Tang, H, Liu, J. 2008. Real-time imaging of protein internalization using aptamer conjugates. Anal Chem 80:5002–8.
- Lin L, Fu Q, Williams BA, Azzaz AM, Shogren-Knaak MA, Chaput JC, Lindsay S. 2009. Recognition imaging of acetylated chromatin using a DNA aptamer. Biophys J 97:1804–1807.
- Lin Y, Nieuwlandt D, Magallanez A, Feistner B, Jayasena SD. 1996. High-affinity and specific recognition of human thyroid stimulating hormone (hTSH) by in vitro-selected 2’-amino-modified RNA. Nucleic Acids Res 24:3407–3414.
- Lu D, Searles MA, Klug A. 2003. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 426:96–100.
- Lundberg P, El-Andaloussi S, Sütlü T, Johansson H, Langel U. 2007. Delivery of short interfering RNA using endosomolytic cell-penetrating peptides. FASEB J 21:2664–2671.
- Lünse CE, Michlewski G, Hopp CS, Rentmeister A, Cáceres JF, Famulok M, Mayer G. 2010. An aptamer targeting the apical-loop domain modulates pri-miRNA processing. Angew Chem Int Ed Engl 49:4674–4677.
- Majumder, P, Gomes, KN, Ulrich, H. 2009. Aptamers: from bench side research towards patented molecules with therapeutic applications. Expert Opin Ther Pat 19:1603–13.
- Mathews L, Kanwar RK, Zhou S, Punj V, Kanwar JR. 2010. Applications of nanomedicine in antibacterial medical therapeutics and diagnostics. The Open Tropical Medicine Journal 3:1–9.
- Mayer G, Kröck L, Mikat V, Engeser M, Heckel A. 2005. Light-induced formation of G-quadruplex DNA secondary structures. Chembiochem 6:1966–1970.
- McNamara JO 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. 2006. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 24:1005–1015.
- Mendonsa SD, Bowser MT. 2004. In vitro evolution of functional DNA using capillary electrophoresis. J Am Chem Soc 126:20–21.
- Miyachi Y, Shimizu N, Ogino C, Kondo A. 2010. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res 38:e21.
- Miyake Y, Togashi H, Tashiro M, Yamaguchi H, Oda S, Kudo M, Tanaka Y, Kondo Y, Sawa R, Fujimoto T, Machinami T, Ono A. 2006. MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J Am Chem Soc 128:2172–2173.
- Molpeceres J, Chacón M, Guzmán M, Berges L, del Rosario Aberturas M. 1999. A polycaprolactone nanoparticle formulation of cyclosporin-A improves the prediction of area under the curve using a limited sampling strategy. Int J Pharm 187:101–113.
- Morris MC, Vidal P, Chaloin L, Heitz F, Divita G. 1997. A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res 25:2730–2736.
- Morris MC, Depollier J, Mery J, Heitz F, Divita G. 2001. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol 19:1173–1176.
- NG EWM, Calias P, Cunningham ETJR, Guyer DR, Adamis AP. 2006. Pegaptanib, a targeted anti-Vegf aptamer for ocular vascular disease. Nature Reviews 5:123–132.
- Nutiu R, Li Y. 2005. Aptamers with fluorescence-signaling properties. Methods 37:16–25.
- Ohk SH, Koo OK, Sen T, Yamamoto CM, Bhunia AK. 2010. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J Appl Microbiol 109:808–817.
- Orum H, Wengel J. 2001. Locked nucleic acids: A promising molecular family for gene-function analysis and antisense drug development. Curr Opin Mol Ther 3:239–243.
- Pastor F, Kolonias D, Giangrande PH, Gilboa E. 2010. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465:227–230.
- Perret E, Lakkaraju A, Deborde S, Schreiner R, Rodriguez-Boulan E. 2005. Evolving endosomes: How many varieties and why? Curr Opin Cell Biol 17:423–434.
- Petersen M, Wengel J. 2003. LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol 21:74–81.
- Qian J, Lou X, Zhang Y, Xiao Y, Soh HT. 2009. Generation of highly specific aptamers via micromagnetic selection. Anal Chem 81:5490–5495.
- Riechmann L, Clark M, Waldmann H, Winter G. 1988. Reshaping human antibodies for therapy. Nature 332:323–327.
- Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 1998. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 273:20556–20567.
- Russell SJ, Llewelyn MB, Hawkins RE. 1992. The human antibody library. BMJ 304:585–586.
- Sampson T. 2003. Aptamers and Selex: The technology. World Patent Information 25:123–129.
- Seferos DS, Giljohann DA, Rosi NL, Mirkin CA. 2007. Locked nucleic acid-nanoparticle conjugates. Chembiochem 8:1230–1232.
- Sengle G, Jenne A, Arora PS, Seelig B, Nowick JS, Jäschke A, Famulok M. 2000. Synthesis, incorporation efficiency, and stability of disulfide bridged functional groups at RNA 5’-ends. Bioorg Med Chem 8:1317–1329.
- Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. 2006. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA 103:11838–11843.
- Shaw BR, Moussa L, Sharaf M, Cheek M, Dobrikov M. 2008. Boranophosphate siRNA-aptamer chimeras for tumor-specific downregulation of cancer receptors and modulators. Nucleic Acids Symp Ser (Oxf) 53:655–656.
- Shen Y, Jin E, Zhang B, Murphy CJ, Sui M, Zhao J, Wang J, Tang J, Fan M, Van Kirk E, Murdoch WJ. 2010. Prodrugs forming high drug loading multifunctional nanocapsules for intracellular cancer drug delivery. J Am Chem Soc 132:4259–4265.
- Su S, Nutiu R, Filipe CD, Li Y, Pelton R. 2007. Adsorption and covalent coupling of ATP-binding DNA aptamers onto cellulose. Langmuir 23:1300–1302.
- Shigdar S, Lin J, Yu Y, Pastuovic M, Wei M, Duan W. 2011. RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci 102:991–998.
- Silahtaroglu A, Stenvang J. 2010. MicroRNAs, epigenetics and disease. Essays Biochem 48:165–185.
- Smirnov I, Shafer RH. 2000. Lead is unusually effective in sequence-specific folding of DNA. J Mol Biol 296:1–5.
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. 2005. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol 23:709–717.
- Sooter LJ, Riedel T, Davidson EA, Levy M, Cox JC, Ellington AD. 2001. Toward automated nucleic acid enzyme selection. Biol Chem 382:1327–1334.
- Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. 2008. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res 68:2358–2365.
- Stenmark H, Olkkonen VM. 2001. The Rab GTPase family. Genome Biol 2:REVIEWS3007.
- Stuart RK, Stockerl-Goldstein K, Cooper M, Devetten M, Herzig R, Medeiros B, Schiller G, Wei A, Acton G, Rizzieri D. 2009. Randomized phase II trial of the nucleolin targeting aptamer AS1411 combined with high-dose cytarabine in relapsed/refractory acute myeloid leukemia (Aml). J Clin Oncol 27:7019.
- Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. 1990. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell 63:601–608.
- Tong R, Yala L, Fan TM, Cheng J. 2010. The formulation of aptamer-coated paclitaxel-polylactide nanoconjugates and their targeting to cancer cells. Biomaterials 31:3043–3053.
- Tsimikas S, Willerson JT, Ridker PM. 2006. C-reactive protein and other emerging blood biomarkers to optimize risk stratification of vulnerable patients. J Am Coll Cardiol 47:C19–C31.
- Tuerk C, Gold L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510.
- Ulrich H, Magdesian MH, Alves MJ, Colli W. 2002. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J Biol Chem 277:20756–20762.
- Van der Goot FG, Gruenberg J. 2006. Intra-endosomal membrane traffic. Trends Cell Biol 16:514–521.
- Vater A, Jarosch F, Buchner K, Klussmann S. 2003. Short bioactive Spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: Tailored-SELEX. Nucleic Acids Res 31:1–7.
- Wagner-Whyte JKS, Preiss J, Kurz JC, Olson K, Hatala P, Boomer RM, Fraone JM, Brosnan N, Makim A et al. 2007. Discovery of a potent, direct thrombin inhibiting aptamer. J Thromb Haemost (Isth Congress abstracts).
- Wang AZ, Bagalkot V, Vasilliou CC, Gu F, Alexis F, Zhang L, Shaikh M, Yuet K, Cima MJ, Langer R, Kantoff PW, Bander NH, Jon S, Farokhzad OC. 2008. Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy. ChemMedChem 3:1311–1315.
- Wang C, Zhang M, Yang G, Zhang D, Ding H, Wang H, Fan M, Shen B, Shao N. 2003. Single-stranded DNA aptamers that bind differentiated but not parental cells: Subtractive systematic evolution of ligands by exponential enrichment. J Biotechnol 102:15–22.
- Wang, W, ling-yun, J. 2009. Progress in aptamer screening methods. Chin J Anal Chem 37:454–460.
- Wang Y, Pu KY, Liu B. 2010a. Anionic conjugated polymer with aptamer-functionalized silica nanoparticle for label-free naked-eye detection of lysozyme in protein mixtures. Langmuir 26:10025–10030.
- Wang Z, Ho PC. 2010b. A nanocapsular combinatorial sequential drug delivery system for antiangiogenesis and anticancer activities. Biomaterials 31:7115–7123.
- Weiss TC, Zhai GG, Bhatia SS, Romaniuk PJ. 2010. An RNA aptamer with high affinity and broad specificity for zinc finger proteins. Biochemistry 49:2732–2740.
- White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M, Sullenger B. 2001. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol Ther 4:567–573.
- Wlotzka B, Leva S, Eschgfäller B, Burmeister J, Kleinjung F, Kaduk C, Muhn P, Hess-Stumpp H, Klussmann S. 2002. In vivo properties of an anti-GnRH Spiegelmer: An example of an oligonucleotide-based therapeutic substance class. Proc Natl Acad Sci USA 99:8898–8902.
- Wolfgang, P, Tasset, D, Janic, N, Gold, L, Kirschenheuter, GP. 1997. High affinity nucleic acid ligand containing modified nucleotides. US Patent No 5:660–985.
- Woodrow, KA, Cu, Y, Booth, CJ, Saucier-Sawyer, JK, Wood, MJ, Saltzman, WM. 2009. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater 8:526–33.
- Wu L, Curran JF. 1999. An allosteric synthetic DNA. Nucleic Acids Res 27:1512–1516.
- Xie J, Chen K, Huang J, Lee S, Wang J, Gao J, Li X, Chen X. 2010. PET/NIRF/MRI triple functional iron oxide nanoparticles. Biomaterials 31:3016–3022.
- XU W, LU Y. 2010. Label-Free Fluorescent Aptamer Sensor Based on Regulation of Malachite Green Fluorescence. Anal Chem 82:574–578.
- Xua Z, Takanori T, Riordana JF., and HU GF. 2002. The nuclear function of angiogenin in endothelial cells is related to rrna production. Biochemical and Biophysical Research Communications 294:287–292.
- Yang X, Bassett SE, Li X, Luxon BA, Herzog NK, Shope RE, Aronson J, Prow TW, Leary JF, Kirby R, Ellington AD, Gorenstein DG. 2002a. Construction and selection of bead-bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res 30:e132.
- Yang X, Bassette SE, LI X, Luxon BA, Herzog NK, Shope RE, Aronson J, Prow TW, Leary JF, Kirby R, Ellington AD, Gorenstein DG. 2002b. Construction and selection of bead-bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid Pcr-based sequencing. Nucleic Acids Res 30:1–8.
- Yang, X, Li, N, Gorenstein, DG. 2011. Strategies for the discovery of therapeutic aptamers. Expert Opinion on Drug Discovery 6:75–87.
- Zhang CY, Johnson LW. 2009. Single quantum-dot-based aptameric nanosensor for cocaine. Anal Chem 81:3051–3055.
- Zhou J, LI H, Zhang J, Piotr S, Rossi J. 2011. Development of cell-type specific anti-Hiv gp120 aptamers for sirna delivery. J Vis Exp 23:pii: 2954.
- Zhou J, Li H, Li S, Zaia J, Rossi JJ. 2008. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther 16:1481–1489.
- Zhou J, Rossi JJ. 2010. Aptamer-targeted cell-specific RNA interference. Silence 1:4.