Abstract
Eukaryogenesis, the origin of the eukaryotic cell, represents one of the fundamental evolutionary transitions in the history of life on earth. This event, which is estimated to have occurred over one billion years ago, remains rather poorly understood. While some well-validated examples of fossil microbial eukaryotes for this time frame have been described, these can provide only basic morphology and the molecular machinery present in these organisms has remained unknown. Complete and partial genomic information has begun to fill this gap, and is being used to trace proteins and cellular traits to their roots and to provide unprecedented levels of resolution of structures, metabolic pathways and capabilities of organisms at these earliest points within the eukaryotic lineage. This is essentially allowing a molecular paleontology. What has emerged from these studies is spectacular cellular complexity prior to expansion of the eukaryotic lineages. Multiple reconstructed cellular systems indicate a very sophisticated biology, which by implication arose following the initial eukaryogenesis event but prior to eukaryotic radiation and provides a challenge in terms of explaining how these early eukaryotes arose and in understanding how they lived. Here, we provide brief overviews of several cellular systems and the major emerging conclusions, together with predictions for subsequent directions in evolution leading to extant taxa. We also consider what these reconstructions suggest about the life styles and capabilities of these earliest eukaryotes and the period of evolution between the radiation of eukaryotes and the eukaryogenesis event itself.
Introduction
The origin of eukaryotes is considered, with justification, as one of the major evolutionary transitions for life on Earth (Maynard Smith & Szathmáry, Citation1995). It brought with it sophisticated intracellular compartmentalization, separation of translation and transcription (permitting increased complexity in gene expression (Martin & Koonin, Citation2006)), superior capabilities for genetic reassortment and, potentially, alterations to evolvability (Poole et al., Citation2003). Each advance individually is potential justification for the emergence of the eukaryotes, and so the coalescence of these mechanistic and cellular advances provides a compelling cohort of selective advantages along the pathway of prokaryote to eukaryote transition. However, an apparent burst of innovation is inevitably an oversimplification, due in large part to a paucity of data from the earliest periods of the eukaryotic period, suggested to have occurred up to two billion years ago (Butterfield et al., Citation1990; Chernikova et al., Citation2011; Parfrey et al., Citation2011; Peterson & Butterfield, Citation2005). Few unambiguous fossils (in the sense of having confidently assigned taxonomy) are documented from the sediments laid down in the Proterozoic era of the Precambrian (Cavalier-Smith, Citation2006), and the information content of many specimens is limited in terms of describing what cellular systems these organisms possessed and the molecules that facilitated construction of these systems. However, several specimens do suggest potentially complex life styles, with obvious molecular ramifications (Butterfield et al., Citation1988, Citation1990; Knoll et al., Citation2006). A fuller understanding of these earliest events requires a molecular paleontology, i.e. reconstructing ancient gene complements. With the improved availability of genome sequence data from diverse taxa, and the improved computational ability to analyses those data, the era of molecular paleontology is now upon us.
Eukaryotes are frequently considered a sister lineage to the archaea, on account of sharing multiple structures and features and as originally revealed from rRNA sequencing, a hypothesis known as the three primary domain model (Woese & Fox, Citation1977) (). The closer relationships between eukaryotic and archaeal transcription and translation systems, in particular, speaks strongly to this intimate relationship. However, what is unclear is precisely how the archaea and eukaryotes are related; while the concept of sister lineages (i.e. arising as separate but related branches) is supported by some molecular phylogenies, a second topology, the two primary domain model that suggests that eukaryotes emerged within the archaea and are hence monophyletic with them, is supported by other analyses. Confidently differentiating these models remains intractable (Forterre, Citation2011; Gribaldo et al., Citation2010). If eukaryotes arose after the archaea, as suggested by the two primary domain model, this predicts that phylogenetic reconstructions would reflect independent differentiation of multiple archaeal lineages, only one of which gave rise to eukaryotes. Most significantly, the two-domain model may imply that potentially more sophisticated and non-universal archaeal features were present in the ancestral lineage of the last eukaryotic common ancestor (LECA), as the eukaryotes represent only a single taxon within the archaea. Clearly, the order of events has important implications for the genetic repertoire that LECA would have inherited.
Figure 1. Unresolved questions in the early evolution of eukaryotes. (A) How many domains of life are there? The traditional view of the tree of life places all three of the major domains, i.e. bacteria, archaea and eukaryota as monophyletic (top tree). This implies that the eukaryotes branched from the Archaea as a separate and independent lineage, with a stepwise topology, i.e. bacteria emerged first, from which archaea arose and then finally the eukaryota. An alternate hypothesis, however, suggests that the eukaryotes are essentially a branch within the archaea, and that archaea and eukaryota are, therefore, monophyletic (lower tree), allowing for coevolution of an archaeal/eukaryote precursor prior to speciation. Attempts to reconstruct the topology of the achaeal/eukaryota differentiation have so far been inconclusive, with both models receiving support, although this support is far from unequivocal (Gribaldo et al., Citation2010). OoL; origin of life, FECA; first eukaryotic common ancestor. (B) When did the mitochondrial symbiont arrive? Many proposals for the origin of the first eukaryotic cell have been offered. Of the models that have garnered support, two simple common schema can be extracted, and a third more complex possibility. Left; A fusion event occurred between an archaea and an α-proteobacterium (the source of the mitochondrial genome and functions). Central; Significant development of endogenously-derived membranous and other structures by the Archaeal ancestor arose prior to endosymbiosis of the α-proteobacterium. The latter mechanism may have involved more complex fusion events, for example including methanogens or an endosymbiotic origin for the nucleus (discussed in Embley & Martin, Citation2006), while the metabolic capabilities of ancestral cells are essentially ignored. Right; A third possibility is that the mitochondrion arose comparatively late, after much of the complexity of the protoeukaryote had evolved, and following fusion between a bacterium (khaki lozenge) and an archaeon (right scheme). While multiple endosymbiont events are considered by many as highly unlikely, the point at which the mitochondrion came on board, as well as when a true nucleus arose remain controversial and unresolved. However, most models agree that the LECA possessed mitochondria, substantial internal differentiation and a well-defined nucleus. Probing beyond LECA is critical for understanding these earliest events. Gray lozenge; Archaeal ancestor, purple lozenge; α-proteobacteria (the mitochondrion is drawn in purple in the LECA), blue lozenge; protonuclear endosymbiont (the nucleus is drawn in blue in the LECA). (C) Eukaryotic tree of life with examples of sequenced organisms from currently recognized supergroups. The curved dotted-line indicates the separation of lineages included in the unikonts and the bikonts. SAR + CCTH: stramenopiles (heterokonts), alveolates, and Rhizaria plus cryptomonads, centrohelids, telonemids and haptophytes (see color version of this figure at www.informahealthcare.com/bmg).
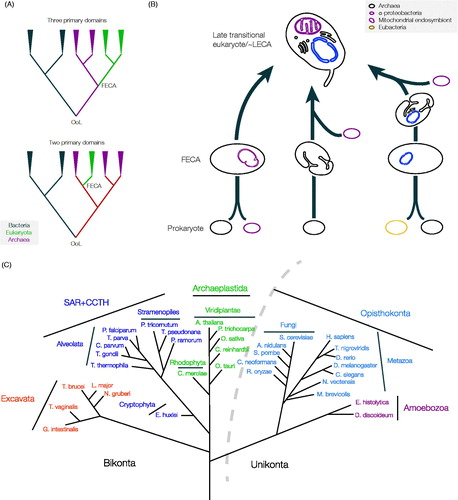
Further, there remains debate concerning the precise mechanisms behind eukaryogenesis, i.e. the events leading up to the first eukaryotic common ancestor (FECA) and the subsequent evolution of the LECA ( and ). For convenience here we are defining FECA as the first ancestor in a lineage that lead to LECA, and which is presumed to have acquired one or a few eukaryotic-specific cellular features, while LECA is the first eukaryote, minimally defined by having a mitochondrion and a nuclear envelope. However, as the discussion below will demonstrate, it is now clear that LECA most probably possessed most of the sophistication of modern eukaryotes, i.e. multiple intracellular compartments, a cytoskeleton and also complex metabolic and gene regulatory mechanisms.
Figure 2. Possible scenarios for the FECA to LECA transition. The top schema depicts the periods of prokaryotic (blue) and eukaryotic (red) evolution, separated by a transition period, which is expanded for clarity. Relative distances on the x-axis are arbitrary, and note that the earliest times shown are post origin of life. It is assumed that prokaryotic and eukaryotic evolution resulted in an increase in cellular complexity, denoted by the blue and red triangles, respectively. The possibility that eukaryotes evolved before prokaryotes is not discussed. It is unknown if FECA (red arrow head) and the origin of the nucleus, acquisition of the mitochondrion or internal compartments (green, purple and yellow arrow heads) are coincident, or near coincident events, despite the possibility that the nucleus evolved from simpler progenitor structures. It is also unclear if the origin of the nucleus is the earliest event in the transition period; for example it is possible to envisage other scenarios, i.e. where endosymbiosis of the mitochondrion ancestor came before acquiring the nucleus, and that this event, rather than formation of a nucleus (either by gradual steps or by fusion), was the initial event that produced FECA. During the transition period the LECA ancestor’s trajectory is shown as a solid line with a sharp increase in complexity, but other possibilities cannot be discounted (faint line; multiple transitions). Other trajectories that could be envisaged are not shown for purposes of clarity only. It is assumed that the LECA ancestor was just one of many lineages that arose from the single eukaryogenesis event, but that it came to dominate or integrate with other lineages. Examples of extant taxa and their approximate complexity given at right in the top panel, simply to illustrate that some extant eukaryotes are likely less complex than LECA, and that there is overlap in complexity between prokaryotic and eukaryotic organisms; note that complexity itself is a difficult term, and here is taken as a composite of genomic and cellular functional complexity/differentiation. The lower schemas illustrate two of the major hypotheses, the syntrophic and phagotrophic models (left and centre, respectively), that suggest that the mitochondrion (purple arrowhead, MT) was the first event or that evolution of the nucleus (green arrowhead, N) and more complex intracellular structures (yellow arrowhead, internal membranes, IM) occurred prior to phagocytosis of the mitochondrial ancestor. A third complex path, that incorporates additional evolvable systems like a sophisticated cytoskeleton (blue arrowhead, other), leading to a double transition after the mitochondrion/nucleus is also shown. Excellent arguments in favour of the first two models have been advanced, but due to the contingent nature of eukaryogenesis, a great many possibilities remain (see color version of this figure at www.informahealthcare.com/bmg).
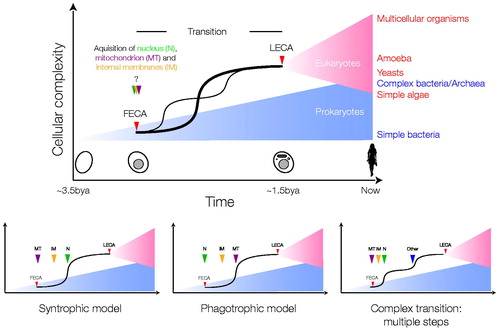
Taking a very simplistic view, and ignoring many excellent but less well-supported models, the proposed trajectories for eukaryotic cell origins can be grouped into two major categories; a fusion first model where an endosymbiosis event delivering the mitochondrion came extremely early, or a fusion later model where endosymbiosis occurred after development of several intracellular structures (discussed in Embley & Martin, Citation2006; O’Malley, Citation2010). Arguments for the mitochondrion first model are based primarily on energetic considerations (e.g. Lane & Martin, Citation2010), while the second model places emphasis on a requirement for phagocytosis-like mechanisms to be present to facilitate endosymbiont acquisition. There is also some speculation about a prior fusion event between archaea and bacteria, to produce a FECA which then took on the mitochondrial endosymbiont, reflected in a third model (, rightmost).
As strong as the connection with the archaea is, there is also evidence uniting eukaryotes and bacteria. This comes not only from the use of ester-linked phospholipids, but also from the eubacterial origins of many eukaryotic metabolic pathways. While this contribution could be dismissed as either derived from mitochondrial endosymbiosis or as a result of horizontal gene transfer, it may represent evidence for a fusion between archaea and bacteria to produce the FECA, which subsequently phagocytosed the primordial mitochondrion (Forterre, Citation2011), essentially a synthesis of the two previous models. Again, excellent arguments for all models have been made, but definitive discriminatory evidence remains lacking. There is also the very real need to appreciate that these are all singular events and hence stochastic influences are likely. Where all models agree is that, at a very early point in their evolution, eukaryotes possessed a fully functional mitochondrion, a nucleus and additional intracellular structures (Dacks & Doolittle, Citation2001; Embley & Martin, Citation2006; Roger, Citation1999). What remains is determining the origins of these features, which ones FECA possessed and how the rest arose post-FECA, and significantly, how long these processes took (Chernikova et al., Citation2011).
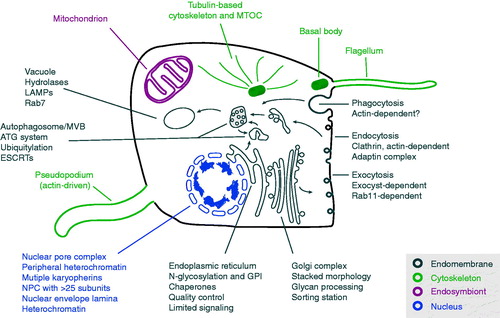
Much insight has been made possible recently through analysis of the genomes of extant organisms (). The greatly increased availability of molecular data, combined with improved phylogenetic tools and reconstructions of eukaryotic phylogeny, permits piecemeal reconstruction of likely LECA biology: the methodology has been discussed elsewhere (e.g. Koumandou & Field, Citation2011). A major surprise is that when molecular-level reconstructions of major cellular systems or protein families have been attempted, frequently these predict that LECA possessed a remarkably modern configuration, extending from cytoskeletal systems, through endomembrane and protein processing, metabolic capabilities and on to encompass meiosis, organization of the genome into linear chromosomes with telomeric ends and RNA processing (). Perhaps even more remarkable is the realisation that LECA may have been, in multiple aspects, more sophisticated than a significant number of extant eukaryotes. Here we aim, in overview, to assemble some of the complexity inferred for the LECA and to consider what type of organism or organisms LECA would have been.
Figure 3. A generalized model for LECA with emphasis on the major systems proposed as present and discussed here. It is now clear that the LECA was both a flagellate and capable of movement by actin-based pseudopodia and possessed a sophisticated cytoskeleton, including large families of kinesin and dynein motors (not shown for clarity). It possessed a complex and likely very flexible, metabolism and a fully functional mitochondrion. Endomembrane compartments would have been essentially indistinguishable from modern cells, and included the endoplasmic reticulum, the Golgi complex, endosomes, autophagosomes and others (many not shown for clarity). The LECA was also capable of both conventional endocytosis and phagocytosis. The nucleus was fully differentiated with nuclear pore complexes and a sophisticated system for organization and regulation of chromatin. A high energy burden is clearly implied by this architecture and required to construct and maintain these compartments and systems plus a differentiated cytoskeleton to coordinate location and function. Heterochromatin in some form could also support life-cycle and/or environmental cue-dependent coordinate gene expression. LECA also supported meiosis. Systems are exploded with examples of the complex aspects associated with a reconstructed LECA (see color version of this figure at www.informahealthcare.com/bmg).
Systems
The nucleus and the nuclear envelope
The defining feature of eukaryotic cells, the nucleus, is responsible for packaging the genetic material and coordinating gene expression, amongst other roles (Martin & Koonin, Citation2006). An expansion in the physical size of eukaryotic genomes, with a greater frequency of noncoding DNA also likely necessitates more sophisticated organization and structural support. This is provided in large part by histones, which are universal amongst eukaryotes, and most of which are also present in archaea and bacteria, and therefore, likely arose pre-FECA (Kasinsky et al., Citation2001; Sandman & Reeve, Citation2005). Importantly, separation of transcription and translation also facilitated evolution of complex gene expression regulatory mechanisms, which may in part result from evolutionary exploitation of histone packaging systems, and allowed splicing and other RNA processing events to emerge (Martin & Koonin, Citation2006). We have little de novo information on mechanisms controlling histone assembly and function beyond basic chromatin regulatory processes and most are shared between animals, fungi and other lineages; where data exist these aspects appear near universal (e.g. Figueiredo et al., Citation2009). At this level the system is probably very highly conserved. Furthermore, while RNA and DNA polymerases, despite eukaryotic elaborations, are clear prokaryotic descendants, transcriptional mechanisms are remarkably variable, with polycistronic transcription versus one-gene one-promoter systems providing an example of a process where even a well-conserved gene cohort operates in a distinct manner between lineages, albeit still retaining significant mechanistic similarities (Daniels et al., Citation2010; Moore & Russell, Citation2012; Morton & Blumenthal, Citation2011).
Much understanding of the structural organization of the nucleus derives from metazoan lamin proteins. These 60 kDa coiled-coil proteins have multiple functions, interacting with the nuclear pore complex (NPC), organizing heterochromatin and positioning of chromosomes and also subtending interactions with the cytoskeleton via interactions with the LINC complexes that span the nuclear envelope (Starr & Fridolfsson, Citation2010). Further, lamins, coiled coil intermediate filament proteins, are essential for the structural integrity of the nucleus as well as other higher order organizational functions (Simon & Wilson, Citation2011). Until recently, lamins appeared specific to metazoa, with no evidence for a presence in any other lineage, including the related fungi. The recent discovery of a lamin-like protein in the amoeba Dictyostelium discoideum (Krüger et al., Citation2012) suggests lamins originated as early as the unikont root (Cavalier-Smith, Citation2003; ). Lamins remain unikont specific at this time, despite evidence for heterochromatin and other lamina-requiring functions in the bikonts. Putative lamin analogs have been identified in Arabidopsis thaliana while a small family of coiled-coil LINC proteins are associated with the A. thaliana nucleus and nuclear periphery and appear to control nuclear size and chromosomal segregation (Dittmer et al., Citation2007), but these proteins also seem to be land plant specific. Recently NUP-1, a large coiled-coil protein was described in trypanosomes, which performs many of the roles ascribed to lamins, including maintaining nuclear structure and defining heterochromatin (DuBois et al., Citation2012). While similarities between NUP-1 and lamin functions are striking, in silico analysis has not identified NUP-1 orthologs outside of the kinetoplastida, or common ancestry with unikont lamins or plant LINC proteins. This may reflect both evolutionary distance and the low complexity amino acid composition of coiled-coil domains, but at present data are nondiscriminatory concerning the ultimate origins of lamins, LINC proteins and NUP-1 and their presence in the LECA. However, what is clear is that many taxa possess lamin functional equivalents, so it is rather likely that such functions were part of the LECA cellular physiology.
Integration of nuclear and other cellular functions requires bidirectional transport across the nuclear envelope. As translation is cytoplasmic this requires that all tRNA, rRNA and mRNAs be exported, while proteins required for DNA replication, transcription, transcriptional regulation, RNA processing and overall nuclear organization are imported. Import and export across the nuclear envelope is the function of the NPC, a huge structure that in Saccharomyces cerevisiae comprises ∼30 different proteins at multiple copy numbers, with a total subunit tally exceeding 430 (Alber et al., Citation2007). Transport is mediated by several mechanisms, mainly via recognition of a cis-acting signal within the transported protein by a karyopherin (KAP). Transport is powered by a gradient of GDP- versus GTP-bound Ran, a small Ras-like GTPase (Grossman et al., Citation2012). Export of mRNA, in some systems at least, utilises non-KAP factors, including Mex67, and is Ran-independent (Oeffinger & Zenklusen, Citation2012). There are differences in molecular mechanisms of nucleocytoplasmic transport between yeast, plants and mammals, and specifically in how the Ran gradient is controlled; in metazoa, RanGAP activity (GAPs stimulate GTPase activity converting the GTPase from a GTP to GDP-bound form) is associated with the nuclear pore complex, but in A. thaliana the RanGAP is targeted to the nuclear envelope by nuclear envelope-embedded trans-membrane domain proteins (Meier et al., Citation2007; Xu et al., Citation2007), while in S. cerevisiae RanGAP is not targeted to the nuclear envelope at all.
There are fourteen β-KAP subclasses, together with a single α-KAP that has undergone paralogous expansions in metazoa (Mason et al., Citation2009; O’Reilly et al., Citation2011). Some KAPs are highly specific, while others remain less well understood. Moreover, there is functional promiscuity, with distinct β-KAP families able to assume the roles of others when the primary β-KAP is deleted; the evolutionary basis for retention of such secondary functions is unclear, but may contribute to KAP cohort evolvability (O’Reilly et al., Citation2011). The fourteen basal β-KAP clades are represented in all eukaryotic supergroups indicating that the basic repertoire was present early in eukaryotic evolution, i.e. pre-LECA. KAP paralogs expanded in some lineages, while secondary loss is common. Most remarkably, with the exception of a plant-specific KAP clade, there has been little evolution of lineage-specific KAPs, and little evidence for evolution of new subfamilies post-LECA (O’Reilly et al., Citation2011). Mex67, important for mRNA export, is mainly restricted to animals and fungi (Serpeloni et al., Citation2011), but its presence in many excavates suggests it is in fact more wide-spread and hence more ancient than previously believed (Kramer et al., Citation2010). The flexible specificity of KAPs to recognise cargo may permit evolution of new specificity, however, without necessitating the emergence of a new KAP clade.
In silico identification of nuclear pore subunits has proved less straightforward than for KAPs, with indications that many are not conserved (Mans et al., Citation2004). Nucleoporins fall into two major groups: scaffold and FG repeat. Proteomics demonstrates that, in fact, the nucleoporins, and hence NPCs, are well conserved and that in silico failures in the identification of nucleoporins are due to poorly conserved sequence, although the proteins retain recognisable secondary structures (deGrasse et al., Citation2009; Tamura et al., Citation2010). A minor proportion of nucleoporins are probably lineage specific and provide evidence for the evolution of functional diversification, however, as the precise roles of most nucleoporins remain unclear the consequences of these changes are not known (Cronshaw et al., Citation2002; deGrasse et al., Citation2009; Rout et al., Citation2000; Tamura et al., Citation2010). While secondary structure is significantly more conserved than sequence, nucleoporin divergence does suggest significantly relaxed selective pressure for retention of specific sequences in this particular complex.
Both the mode of evolution and the structure of the NPC and its interactions with transport cargoes may reflect fundamental functional requirements. NPC interactions with transport substrates are based more on physicochemical properties than primary structure per se, and very distinct from KAP recognition of specific cargo via short amino acid sequences (Tetenbaum-Novatt & Rout, Citation2010). This may be a consequence of a need to transport thousands of different substrates through the NPC. Significantly, a specific sequence-based recognition system would likely have generated evolutionary inflexibility, essentially locking the NPC/KAP interaction system, and providing a barrier to further evolution. The flexibility of KAP recognition, despite its dependence more closely on amino acid sequence, also likely speaks to this requirement. Further, scaffold nucleoporins are members of the β-propeller/α-solenoid protocoatomer superfamily (Devos et al., Citation2004), and while conserved overall in architecture, both the β-propeller and α-solenoid are inherently flexible domains which can tolerate considerable sequence diversity (see Field et al., Citation2011). Such flexibility may underpin the wide exploitation of the β-propeller and α-solenoid domains by proteins involved in many trafficking complexes.
In summary, nucleocytoplasmic transport is ancient and the molecules, complexity, mechanisms and architectures of these systems were established pre-LECA, with comparatively minor post-LECA innovation. Organization of the lamina is apparently more divergent, with the possibility that proteins with distinct evolutionary histories assume the lamina role in different taxa. However, the basic lamina functions are conserved, suggesting that LECA possessed heterochromatin and the ability to strongly repress specific gene sets.
Endomembrane compartments and trafficking
Organelles of the endomembrane system include the entirety of the secretory/exocytic and endocytic pathways and the nuclear envelope, which is contiguous with the endoplasmic reticulum (). Proteins destined for the surface or to be secreted, enter the system by co- or post-translational translocation across the ER, are folded by numerous chaperones and monitored by quality control mechanisms (Alberts et al., Citation2002). Most protein export proceeds via the Golgi complex, where molecules enter the cis-face, traverse several cisternae and are exported from the trans-most cisternae. Plasma membrane delivery is achieved via secretory vesicles budding from the trans-Golgi compartment. Endocytic pathways originate by invagination of the plasma membrane, fusion or maturation of the resulting vesicles into endosomes and subsequent sorting to one of several destinations. The major destinations are recycling to the cell surface, a common pathway for nutrient receptors for example, and which intersects post-Golgi exocytic routes, or delivery to terminal degradative endosomes, variously termed lysosomes, vacuoles or reservosomes. This latter pathway, mediated by ubiquitination, is important for turnover of surface proteins, signaling receptors and destruction of immune factors for example, as well as degradation of material for nutritional purposes.
In a general sense all of these pathways are well understood and were present in the LECA. There is excellent evidence for universal conservation of the ER translocation machinery, with origins in the prokaryotic SecY system (Jungnickel et al., Citation1994). The major chaperone subclasses are also likely ancient, encompassing categories involved in folding, disulphide bond rearrangement and quality control, including sensing mechanisms based on N-glycan glycosylation and retro-translocation of terminally mal-folded proteins (Field et al., Citation2010). It is unclear if any of the proteins mediating these latter functions have direct prokaryotic ancestors, although several are part of the huge and universal HSP family. The Golgi complex was almost definitely present in the LECA (Klute et al., Citation2011), but interestingly has taken multiple evolutionary trajectories. At its most extreme the canonical Golgi stacked cisternae morphology has been lost, and in several cases there is no microscopic evidence for the organelle. This occurred in multiple lineages, indicating convergent evolution (Mowbrey & Dacks, Citation2009). However, the basic functions of protein targeting and N-glycan processing are retained in such lineages, which raises the issue of a potentially cryptic Golgi complex or repurposing of other compartments that assume these functions (Dacks et al., Citation2003). Further, the Golgi complex demonstrates quite extreme morphological variability. While many microbial eukaryotes possess a single Golgi complex, as seen in trypanosomes, others including prominent organisms such as apicomplexans (T. gondii aside) and ciliates have a reduced Golgi complex, i.e. a single cisterna (reviewed in Mowbrey & Dacks, Citation2009). Golgi organelles have also expanded in some lineages, such as the ribbon like, interconnected Golgi stacks in mammalian tissue culture cells, extensive and mobile stacks in A. thaliana (Staehelin & Kang, Citation2008) and beautifully expanded Golgi bodies of parabasalid taxa with large numbers of both stacks and cisternae per stack (e.g. Brugerolle, Citation2004). Even closely related yeast species display variance in Golgi complex morphology (Suda & Nakano, Citation2011). While it is near certain that LECA possessed a stacked Golgi apparatus, it is unclear what the precise LECA configuration would have been, and the molecular steps that facilitate extreme morphological plasticity within a central organelle of the eukaryotic cell are unknown.
Endocytosis, at least as far as we presently understand it, presents a more complex evolutionary story. The basic system is once more a feature of LECA, with the major endosomal coat protein, clathrin, being (so far) universal. In several lineages, including Trypanosomatids, Apicomplexa and plants, clathrin-mediated endocytosis may represent the sole mechanism for endocytosis (Field et al., Citation2007). By contrast, in metazoan cells multiple modes of endocytosis co-exist, with caveolin-, Cdc42-, RhoA- and flotillin-mediated pathways being frequently viewed as restricted to the Opisthokonta. Some of these pathways, for example Cdc42 and RhoA-dependent pathways, likely await more detailed functional dissection, as the presence of these multifunctional GTPases is, of itself, insufficient to define an endocytic route as mechanistically distinct from others (Sandvig et al., Citation2011). The evolutionary history of flotillin-mediated endocytosis, associated with both clathrin-dependent and -independent endocytic pathways, is less clear (Otto & Nichols, Citation2011). Flotillin orthologs are present in bacteria and archaea, and in Bacillus associate with detergent-resistant membranes (DRMs); the precise function is unknown or even if this represents true endocytosis (Lopez & Kolter, Citation2010). These findings are particularly significant as they suggest that bacteria, as well as eukaryotes, segregate their plasma membranes based on protein-lipid physicochemical properties, and these similar biochemical underpinnings, i.e. isoprenoid-derived metabolites and lipid-binding flotillins, operate (Lopez & Kolter, Citation2010). Additionally, signaling complex proteins are enriched in DRMs in both prokaryotes and eukaryotes, suggesting conserved function. The presence of GPI-anchored proteins in DRMs is apparently a result of association with pre-existing aspects of membrane physiology, and potentially involvement of flotillin could likewise represent recruitment to endocytic functions. Finally, while the distribution of flotillin in eukaryotes is broad, there is frequent secondary loss. The functions of these additional endocytic pathways are unclear, and while there may be other undiscovered taxon-specific pathways, it is firmly established that the LECA possessed a clathrin-based endocytic system, and possibly also a flotillin-mediated mode.
Later endocytic processes include the sorting of proteins by the ESCRT system, intimately involved in the generation of multi-vesicular bodies and pre-lysosomal compartments. ESCRTs were originally identified as class E vacuolar sorting mutants (vps) in S. cerevisiae, but their importance has become more extensive with subsequent analysis (Field & Dacks, Citation2009); their principal function in the endocytic system is the recognition of ubiquitylated endocytic cargo as well as the invagination of membrane in late endosomes to create multi-vesicular bodies (MVBs), a function which appears to be intrinsic to the snf7 and vps4 ATPase subunits (Hanson et al., Citation2008). The entire system is comprised of five subcomplexes, which together contain ∼25 distinct proteins. The core of this system is near universal, and indicates that the LECA possessed ESCRT machinery and hence likely the ability to sort ubiquitylated endocytic cargo and form MVBs, although the absence of one subcomplex (ESCRT 0) outside of animals and fungi, and responsible for cargo recognition suggests that some lineage-specific mechanisms must be present (Leung et al., Citation2008). Broad representation is unsurprising as several ESCRT components are present in the archaea, and significantly the snf7/vps4 orthologs play a role in the curving of membrane during cytokinesis, where they are recruited by CdvA to form helical fibers (Dobro et al., Citation2013; Samson et al., Citation2011). This more ancient role for a subset of ESCRT proteins seems to be have been maintained in eukaryotes, where ESCRT factors are recruited late during membrane scission and appear to participate in the final steps of cytokinesis, together with a number of eukaryotic-specific proteins of the endosomal system (reviewed in Chen et al., Citation2012).
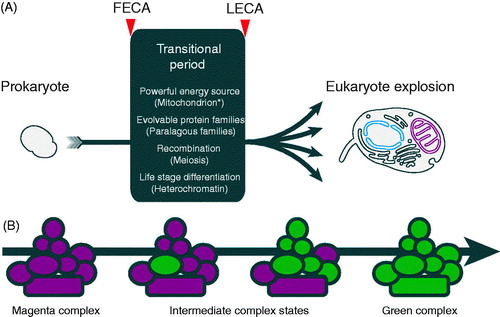
Control of vesicular transport and definition of compartments is highly dynamic and the result of collaborations between large cohorts of proteins. Much of this complexity is the result of expansions of several gene families (Dacks & Field, Citation2007). Major players include SM proteins, tethers, Rab and ARF family GTPases, SNAREs, adaptins and coat proteins, the evolution of which have now been investigated in considerable detail. The presence of paralogs at the core of these systems helps to explain two important features; how new compartments arise and why there is plasticity within the endomembrane system when considering the diverse configurations present in divergent taxa. We suggested a model for the evolution of new endomembrane compartments, which we term organellar paralogy, and which suggests a simple mechanism for the integration of new paralogs into pre-existing complexes and subsequent neofunctionalisation (Dacks & Field, Citation2007; Elias et al., Citation2012; ).
Figure 4. FECA to LECA transitions and flexible evolution of paralog complexes. (A) Transition of prokaryotes to eukaryotes during the period between FECA and LECA, and which incorporated a number of highly significant features. It is unresolved as to which of these occurred first, and only in the case of the acquisition of the mitochondrion, is it well agreed that this a singular event. It was only once all of these features were in place that the LECA was poised for the explosive differentiation of the eukaryotic lineage. (B) Flexibility in protein complex evolution. Rapid success for the LECA ancestors may have required evolvability within protein complexes, resulting in the large number of paralogs in modern eukaryotes. Complexes built from paralogs have an intrinsic evolutionary advantage in allowing new paralogs rapid access to functionality; if a substantial proportion of a complex is built using paralogs this potential is increased. For example, if a single subunit of the magenta complex is replaced by a paralog (green), but which initially is identical to the original paralog, this provides the opportunity for one of the paralogs to drift by acquisition of mutations. This process can then either relax sequence restraints on other subunits or even select for changes that facilitate neofunctionalisation. These other subunits can also be replaced by new paralogs, which is made more probable by the original paralogous expansion. The process is completed by the achievement of a fully green complex, but there are many examples of subunits being shared between complexes with bona fide distinct functions; this may reflect either the achievement of some maximal functionality or reflect an incomplete evolutionary change. See Dacks & Field (Citation2007) for a more detailed discussion of this concept as applied to the trafficking system (see color version of this figure at www.informahealthcare.com/bmg).
GTPases play an important role in vesicle trafficking, in both the formation and fusion of transport intermediates. The Rab subfamily is the prime mediator of compartmental identity and controls the fusion events between transport intermediates and organelles. This family is perhaps the premier example of a large paralogous family with well-detailed evolutionary histories. Recent data indicate a substantial family of over twenty Rab proteins in the LECA (Diekmann et al., Citation2011; Elias et al., Citation2012). Importantly, reconstruction of Rab evolution indicates emergence of the major organelle-specific subfamily members in the LECA, and also pre-LECA emergence of primordial endocytic and exocytic Rabs (Elias et al., Citation2012), suggesting a stepwise, ongoing increase in complexity. This view is further supported by the continual emergence and elimination of Rab5 isoforms, which contribute to early endocytosis (Dacks et al., Citation2008; Pereira-Leal, Citation2008), indicating that evolution of intracellular compartments is continuing in modern lineages. However, there appears to be a limit to the level of specialization possible, and which may be due to energetic constraints or more simply no need for more than about three distinct routes. Further, there is increased Rab GTPase family complexity within animals and fungi, but also evidence for the emergence of lineage-specific Rab proteins in all supergroups (Diekmann et al., Citation2011; Elias et al., Citation2012). Finally, the LECA Rab complement is greater than some well-studied extant organisms, many of which, however, are highly derived taxa, suggesting secondary loss as a major driver for sculpting the endomembrane system. Recent analysis also suggests a large complement of Rab GTP-activating proteins and GTP-effector proteins in LECA (Gabernet-Castello et al., Citation2013).
The ARF GTPases provide a contrast to Rab evolution and offer one of a restricted number of examples of primordial simplicity in the LECA (Li et al., Citation2004). While there is good evidence for ARF participation in membrane transport, the contribution to the LECA was likely modest. ARF expansions in multiple taxa suggest a single ARF LECA ancestor, and that elaboration of the family is supergroup specific (Berriman et al., Citation2005). The basis for this curious evolutionary history is unclear at present, and the drivers that propelled expansion of the ARF families are less obvious than for Rabs, despite the clear widespread influence and requirement for a substantial ARF family. Interestingly, a recent analysis of Arf-GAPs shows much greater complexity in the LECA, with at least six ancient subfamilies (Schlacht et al., Citation2013), suggesting that perhaps GAPs provided an early source for functional Arf diversity.
The other prominent example of simplicity in the LECA is that the multiple Qa-SNAREs involved in anterograde transport in the early and late endosomes respectively in animals, fungi and plants appear to have evolved from a single primordial endosomal SNARE (Dacks et al., Citation2008). Nonetheless, while analysis is less extensive than for Rabs, a large SNARE cohort was also present in the LECA. Both comparative genomics (Yoshizawa et al., Citation2006) and phylogenetics (Dacks & Doolittle, Citation2002, Citation2004; Vedovato et al., Citation2009) have reconstructed not only the Qa, Qb, Qc and R families of SNAREs present in the LECA, but several major organelle-specific subfamilies as well.
Heterotetrameric adaptin (AP) complexes act as cargo receptors, and only four AP complexes were known until recently (Dacks et al., Citation2008; Robinson, Citation2004). Recently a fifth, highly divergent AP-5, operating in late endosomal transport, was identified and was probably present in the LECA (Hirst et al., Citation2011). While the AP family is less extensive than the Rab/SNARE families, it appears more stable over time. Most taxa possess at least AP-1, 2, 3 and 4, with a significant level of secondary loss of AP-5 from many lineages, for example, the trypanosomatids. However, examples of losses of other AP complexes have also been described, including AP-2, 3 and 4 (Berriman et al., Citation2005; Field et al., Citation2007; Manna et al., Citation2013; Nevin & Dacks, Citation2009), suggesting sculpting of this feature of the endomembrane system as well. Furthermore, detection of a divergent AP complex provides a salient lesson on the ability of search methods to call the limits of protein families: it is essential to maintain an open mind as to where such limits may lie.
SM proteins and tether complexes are involved in control of vesicle fusion and interact intimately with the Rab and SNARE proteins. While the total number of gene products involved is quite large, their evolution appears to be comparatively stable. As far as we are aware, there are four broadly distributed SM proteins, likely present in the LECA. The tether system is more complex, as these factors comprise several complexes of varying size, and in many taxa several subunits are lost, but most lineages have at least a representative of all examined tethering complexes, consistent with a complement of at least seven complexes in the LECA (Koumandou et al., Citation2007). Limited sequence similarity is apparent between the tether complex subunits, but structural biology has identified a similar “CATCHR” fold in subunits from multiple tether complexes, suggesting the presence of paralogous subunits (Bröcker et al., Citation2010; Spang, Citation2012). Extreme examples of specific expansions of tether subunits are known and include Exo70, an exocyst subunit, in A. thaliana, where there are over 20 paralogs (Chong et al., Citation2010). At least some of the expansion appears to be due to tissue-specific expression (Li et al., 2010), but the absence of such numbers of Exo70 paralogs from other complex multicellular organism lineages, e.g. metazoa, suggests that there is a more sophisticated driver at work.
A unification between the large cohorts of proteins involved in cytoplasmic membrane transport and nucleocytoplasmic transport hinges on the presence of a highly conserved architecture within these proteins (Devos et al., Citation2004). Specifically, clathrin, β-COP, adaptins, Sec13/31 of COPII and several subunits of the intraflagellar transport system share the same β-propeller and α-solenoid secondary structure present in many NUPs (deGrasse et al., Citation2009; Devos et al., Citation2004; van Dam et al., Citation2013). Further there are suggestions that similar architectures are also present in the HOPS/Corvet and SEA complexes which have roles in endocytosis, although formal solution of the structures themselves remains to be achieved (Dokudovskaya et al., Citation2011). As all of these protein families were fully established before the LECA, this also provides a model by which a primitive membrane deforming complex could have given rise to the numerous systems present in the LECA cell, essentially through simple paralogous expansion during the transition period between the FECA and LECA.
Prokaryotic origins of eukaryotic trafficking systems
Although intracellular vesicle trafficking is a hallmark of eukaryotic cells, many of the components have ancestral prokaryotic orthologs. Most were identified by comparing 3D structures as sequence identity between prokaryotic and eukaryotic orthologs is frequently insignificant, and many examples have only been identified recently due to increased genomic data from a variety of bacteria and archaea. For example, prokaryotic V4R proteins, which currently have no clear function, have low sequence, but significant structural, similarity to the Bet3 subunit of TRAPPI (Podar et al., Citation2008), a tethering complex component involved in attachment of TRAPPI to Golgi membranes (Kim et al., Citation2005). Similarly, prokaryotic members of a family of protein cargo receptors possibly involved in vesicle formation and protein trafficking in eukaryotes have recently been identified through PSI-BLAST, HHMer, and secondary structure predictions (Saudek, Citation2012), and the secreted MPT63 protein of Mycobacterium tuberculosis has structural similarity to adaptins (Goulding et al., Citation2002). However, at present these links are tentative. Several prokaryotic trafficking-related factors have been studied in considerable detail, and here the evidence for common descent is compelling. For example, bacterial dynamin-like proteins have been studied in the cyanobacterium Nostoc punctiforme (BDLP) (Low & Lowe, Citation2006; Low et al., Citation2009) and the gram-positive bacterium Bacillus subtilis (DynA) (Burmann et al., Citation2011), and homologs are found in many bacterial lineages as well as in certain archaea (Methanomicrobia) (Bramkamp, Citation2012). Structural and functional studies suggest a role in cytokinesis and/or membrane fission, similar to eukaryotic dynamin. Rab and Arf GTPases are central to vesicle transport, and Ras homologs are present in several bacteria and archaea, suggesting a possible prokaryotic origin for the Rab and Arf GTPases central to vesicle transport (Dong et al., Citation2007). The bacterial Ras-like GTPase MglA, along with its cognate GAP, MglB, are required for sporulation and motility in the gram-negative soil bacterium Myxococcus xanthus (Hartzell, Citation1997), and also in the regulation of cell polarity, i.e. the localization of proteins to the leading or lagging cell pole of motile cells (Bulyha et al., Citation2011; Leonardy et al., Citation2010; Zhang et al., Citation2010). The polar localization of motility proteins by MglA may also involve the actin-like protein MreB (Mauriello et al., Citation2010). MglB does have homologs in eukaryotes but not to known eukaryotic GAPs; rather, it contains a dynein light chain domain and may define a novel GAP family (Wanschers et al., Citation2008). MglA and MglB orthologs are present in many phylogenetically distant bacteria and archaea (Koonin & Aravind, Citation2000), suggesting that regulation of polarity by a Ras-like G-protein and a GAP is possibly a general prokaryotic feature.
Ubiquitination is a key sorting signal for protein sorting and degradation in eukaryotes. A functionally similar system, PUPylation, targets PUPylated proteins to the proteasome for degradation in Mycobacterium. Similar to ubiquitin, Pup is post-translationally transferred to proteins on lysine residues, but the enzymes involved are fewer than in eukaryotes and do not exhibit significant sequence or structural similarity to the eukaryotic ubiquitin-ligase system (Burns & Darwin, Citation2010; Pearce et al., Citation2008). A ubiquitin-like proteasome system has also been described in the halophilic archaeon Haloferax volcanii, and is shared with various archaeal species (Humbard et al., Citation2010). In addition, the genome of the archaea Candidatus “Caldiarchaeum subterraneum” harbors an operon-like gene cluster encoding homologs of eukaryote-type E1 and E2 ubiquitin ligases, as well as a small Zn-finger protein containing a RING finger motif that, in eukaryotes, mediates the ubiquitin ligase activity of RING-type E3s. This suggests the presence of an unprecedented eukaryote-type ubiquitin ligase system in archaea. HGT from eukaryotes is considered unlikely, given that the individual components acquired by HGT would need to have reorganized to form a gene cluster; however, such an arrangement may facilitate coordinate expression with significant selective advantage (Nunoura et al., Citation2011).
In the Crenarchaea, where FtsZ and MreB are absent, cell division is mediated by the Cdv complex, with several components orthologous with eukaryotic genes, including the Vps2/Snf7 subunits of the ESCRT-III system and Vps4 (Bernander & Ettema, Citation2010; Lindas et al., Citation2008; Makarova et al., Citation2010; Samson et al., Citation2008). Interestingly, an archaeal-specific factor recruits Cdv to the membrane (Samson et al., Citation2011). Cdv forms ring structures between segregating nucleoids, which constrict during cell division. In eukaryotes, ESCRT-III-derived curved filaments are involved in vesicle formation during endosomal protein sorting; Vps2/Snf7 mediates membrane bending itself while Vps4, an ATPase, is responsible for disassembly. The Cdv system may be a preferential cytokinesis mechanism in some thaumarchaeal species that possess both FtsZ and Cdv, and which may be an archaeal lineage most closely related to eukaryotes (Busiek & Margolin, Citation2011; Pelve et al., Citation2011). The bacterial proteins PspA and Vipp1 also have homology to Vps2, and Vipp1 is thought to function in membrane stabilisation or vesicle traffic for thylakoid biogenesis within chloroplasts of cyanobacteria (Vothknecht et al., Citation2012).
The Cdv system is also involved in outer membrane vesicle (OMV) formation in archaea (Ellen et al., Citation2010). Various archaeal and bacterial species lacking known ESCRT-like factors can release proteins packaged into small (10–300 nm in diameter) membrane vesicles that emerge from the cell surface. OMVs are implicated in a variety of processes, including release of bacterial toxins and quorum-sensing factors (Ellen et al., Citation2010; Ellis & Kuehn, Citation2010; Lee et al., Citation2009). In fact, the quorum-sensing hydrophobic molecules packaged into OMVs directly affect OMV biogenesis (Mashburn & Whiteley, Citation2005; Mashburn-Warren et al., Citation2008). Furthermore, OMVs have a distinct lipopolysaccharide composition from the outer membrane, which could influence sorting of specific proteins into these structures (Haurat et al., Citation2011). While no prokaryotic vesicle coats have been formally demonstrated yet, some evidence suggests that vesicle budding may simply be a physicochemical process in cells synthesizing excess membrane for their surface and/or that lack the factors to support a certain cell shape (Bendezu & de Boer, Citation2008; Erickson & Osawa, Citation2010; Leaver et al., Citation2009). Imbalances in protein associations between outer/inner membrane and the peptidoglycan wall (Deatherage et al., Citation2009; Moon et al., Citation2012), as well as hydrophobic molecules preferentially intercalating into the outer leaflet of the membrane bilayer (Schertzer & Whiteley, Citation2012) also induce membrane curvature and OMV budding. Furthermore, two bacterial proteins, SpoVM and DivIVA, preferentially associate with positively or negatively curved membranes, respectively, and apparently without the need for adaptors or other sorting signals (Shapiro et al., Citation2009). Lipid microdomains also affect protein localization in bacteria. For example, cardiolipin preferentially associates with negatively curved membranes and mediates polar positioning of the proteins MinD and ProP (Renner & Weibel, Citation2011; Romantsov et al., Citation2007), while sterol rich flotillin-containing microdomains in B. subtilis and other bacteria have already been mentioned (Lopez & Kolter, Citation2010).
Internal membranes are far from unique to eukaryotes and present in multiple bacterial and archaea lineages (reviewed in Fuerst & Sagulenko, Citation2012). Considerable interest in the Planctomycete bacteria and Gemmata obscuriglobus, in particular, has led to detailed examination of these bacteria, and how their internal membrane systems relate to eukaryotes (Fuerst, Citation2005; Santarella-Mellwig et al., 2013). The observation that G. obscuriglobus DNA is contained within a membrane-bounded structure has been used as evidence for a nucleus-like precursor in these organisms (Fuerst, Citation2005; Fuerst & Sagulenko, Citation2011), and evidence for an energy-dependent, endocytosis-like mechanism by a Planctomycete, has also been presented (Lonhienne et al., Citation2010). However, most recently it has been shown that the putative nucleoid is open and that the endomembrane system within G. obscuriglobus, while being highly complex, is similar to those found more widely in bacterial cells (Santarella-Mellwig et al., 2013). Structure prediction suggests the presence in Planctomycetes of proteins with the β/α architecture, a hallmark of the protocoatomer superfamily (Devos et al., Citation2004; Santarella-Mellwig et al., Citation2010). These findings have caused considerable controversy, with suggestions for HGT being put forward as an explanation for the presence of β/α proteins in Planctomycetes (Devos, Citation2012; McInerney et al., Citation2011). However, the β and α architectures are incredibly common, and while the type of α-solenoid that features in protocoatomers is a specific subfamily (Field et al., Citation2011), it is difficult to imagine that the β/α topology can be used alone as evidence for common ancestry between bacterial and eukaryotic proteins, rather than simple convergence. Forterre has argued for a model whereby the protoeukaryote arose through fusion of a Planctomycete, or close relative, with a thaumarchaeon, which is potentially consistent with direct descent (Forterre, Citation2011), although definitive functions for the Planctomycete β/α proteins remain to be reported. Regardless of their status as eukaryotic precursors or not, these β/α proteins are of considerable interest to prokaryotic biology and the general understanding of membrane trafficking in a broader context.
In summary, while no unequivocal prokaryotic vesicle coat or SNARE-like proteins have been reported, there are candidate prokaryotic homologs for Rab-like GTPases, cytoskeletal components (see below), putative adaptors, tethering complex precursors, a subset of the ESCRT system and a ubiquitin-like sorting system, as well as membrane microdomains characteristic of lipid rafts, as well as possible β/α architecture proteins. Together, this suggests a probable prokaryotic origin for at least some portions of the eukaryotic trafficking machinery, and it is tempting to hypothesize that the eukaryotes did not innovate all trafficking factors de novo, but instead repurposed pre-existing prokaryotic systems as vesicle budding and trafficking machinery within a cell with multiple membrane-bounded compartments. The prokaryotic ESCRT system and the actin/tubulin/dynamin homologs are likely involved in cell division (Field & Dacks, Citation2009), whereas prokaryotic lipid-rafts and GTPases mediate cell polarity. However, many of these connections must be treated with caution as the ability to detect common ancestry in silico falls at the edge of statistical significance, and we must be aware that precise mechanistic details are lacking for many of these examples, and which are an important step in confirming putative relationships. It may also be that various combinations of prokaryotic trafficking factors were exploited in lineages pre-LECA, with HGT and novel innovations such as coatomer proteins also involved, but that the LECA was so successful that the vast majority of these other configurations failed to survive into the extant eukaryotic lineage.
Cytoskeletal filaments
The cytoskeletons of all extant eukaryotes are dominated by two filament-forming protein families; tubulin and actin. Both protein families were already present as multiple paralogs in LECA. Tubulin had already diversified into the two major microtubule paralogs (α and β), a nucleating paralog (γ), and two paralogs associated with the axoneme/basal body (δ and ε) (Vaughan et al., Citation2000), while actin had expanded to produce several actin-related protein (ARP) families, including paralogs associated with dynactin (ARP1), nucleation (ARP2 and ARP3), and four nuclear families (ARP4, 5, 6 and 8) (Sehring et al., Citation2007; Schafer & Schroer, Citation1999). Three tubulin paralogs, α, β and γ, and actin, appear to be ubiquitous to all eukaryotes studied to date, while the other paralogs have been lost from at least some lineages. As expected from their primary functions, δ and ε-tubulin are absent from species lacking cilia/flagella, but curiously are also absent from Thalassiosira pseudonana. Caenorhabditis elegans, dipterans and lepidopterans (Hodges et al., Citation2010). Both ARP2 and ARP3 are lost from several protistan/algal lineages including some diatoms, the red alga Cyanidioschyzon merolae and the Apicomplexa (Wickstead & Gull, Citation2011a). This is rather surprising, since together ARP2 and ARP3 are part of an otherwise highly-conserved actin nucleator, which is essential in yeast and nematodes (Lees-Miller et al., Citation1992; Sawa et al., Citation2003; Schwob & Martin, Citation1992). ARP1 has also been lost several times: in ciliates, Theileria annulata, metamonads (e.g. Giardia lamblia) and trypanosomes (Wickstead & Gull, Citation2011a). In Trypanosoma and Leishmania, the absence of ARP1 is part of a general loss of all dynactin complex components, except cytoplasmic dynein 1 itself (Berriman et al., Citation2005). These findings indicate plasticity in the eukaryotic cytoskeleton that is not apparent from a consideration of any one lineage, and similarly to some Rab GTPases, genes that are essential in some taxa can be lost from others. Further, organisms in particular groups have reduced their dependence on specific aspects of the cytoskeleton while elaborating others; the excavate Giardia lamblia reduced its actin cytoskeleton to the extent that the actin gene is highly divergent and no families of ARPs or actin-based motors have been identified in the genome (Morrison et al., Citation2007).
Both tubulin and actin have prokaryotic ancestors with low sequence similarity but clear tertiary structural conservation (Bork et al., Citation1992; de Boer et al., Citation1992; Mukherjee et al., Citation1993; RayChaudhuri & Park, Citation1992). Tubulin is homologous to the prokaryotic proteins FtsZ, TubZ and RepX, the latter two encoded by bacterial plasmids. Due to divergence it is difficult to robustly determine which prokaryotic gene is the true eukaryotic tubulin ortholog. However, given wide-spread occurrence of FtsZ in both bacteria and archaea (although, notably not the Crenarchaeota), it is reasonable to assume that this is the nearest extant relative. Heterodimeric BtubA/B, found in some Prosthecobacter species, is much more similar to eukaryotic tubulins than other bacterial homologs (Jenkins et al., Citation2002; Vaughan et al., Citation2004). Lack of strong phylogenetic affinity for any extant tubulin families, coupled with an ability to fold in the absence of chaperones, has been argued to point to these proteins being representatives of an ancient tubulin ancestor (Pilhofer et al., Citation2011). However, BtubA/B are extremely limited in their distribution and show strong evidence of horizontal gene transfer (Pilhofer et al., Citation2007), making divergence of sequence following transfer of tubulin from a eukaryote the more likely scenario.
FtsZ is also found in some eukaryotes alongside tubulin. This eukaryotic FtsZ is plastid-derived and serves a similar role in the division of the chloroplast and/or mitochondrion that it once did in their free-living ancestors. FtsZ mediates prokaryotic cell division, and mitochondrial and plastid division in eukaryotes, by forming a dynamic ring between prospective daughter cells (or daughter organelles) before cytokinesis (see Wickstead & Gull, Citation2011a).
Actin is a member of a large superfamily of ATPases that includes prokaryotic MreB, FtsA, AlfA and ParM, but also Hsp70 chaperones and several classes of sugar/sugar alcohol kinases (Derman et al., Citation2009; Flaherty et al., Citation1991; Jockusch & Graumann, Citation2012). Actin and MreB have similar fold structures (Kabsch & Holmes, Citation1995), but until recently it was unclear which of the many prokaryotic ATPases was most closely related to eukaryotic actin/ARPs. This was resolved by the discovery of “crenactin” – an archaeal actin-like protein with a localization similar to bacterial MreB in bacteria, but which is monophyletic with eukaryotic actin (Ettema et al., Citation2011; Yutin et al., Citation2009). Interestingly, this actin ortholog is only present in Crenarchaeota and some basal archaeal lines. This, together with the distribution of FtsZ, suggests the prokaryotic ancestor of the FECA probably arose near the base of the archaea (see Wickstead & Gull, Citation2011a).
MreB filaments are involved in maintenance of cell shape, forming a helix below the cell membrane and influencing cell wall synthesis. Another prokaryotic actin homolog, ParM, along with the tubulin homolog TubZ, have roles in plasmid segregation. In contrast, eukaryotic DNA segregation is dependent on the tubulin-based cytoskeleton in all systems studied, whereas cytokinesis involves actin–myosin (Wickstead & Gull, Citation2011a). Surprisingly, recent data show that in yeast a form of rudimentary nuclear division can proceed in the absence of a microtubule-based spindle and that this auxillary system is potentially actin-based (Castagnetti et al., Citation2010). This possibly reflects a persistence of an ancestral mechanism, although given the ubiquity of tubulin-based mitosis in eukaryotes and the absolute requirement of other systems on the presence of the spindle, this interpretation is rather unlikely.
Intermediate filaments (IF) are a distinct class of cyloskeletal elements and, unlike tubulin and actin, lack directionality and cytomotivity in their own right, and have no known dedicated motor proteins (see Fuchs & Weber, Citation1994). Vertebrates possess many IF protein classes, including keratins, vimentin and desmin, α-internexin and lamins. It is likely that lamins were the first IF proteins to evolve within the unikonts and from these all other IF proteins evolved (Erber et al., Citation1998; Weber et al., Citation1989). Interestingly, in the cnidarian Hydra spp., a lamin-related protein, lacking both a nuclear-localization signal and farnesylation site, is present in the mechanosensory cilia of nematocysts (Hwang et al., Citation2008). This gene arose through duplication of the nuclear lamin gene and provides an example of the evolution of cytoplasmic IFs from lamins in a manner independent of that which occurred in animals more generally. No good candidates for prokaryotic ancestors of eukaryotic IF proteins have been identified. The “IF-like” protein crescentin in the bacterium Caulobacter crescentus is more likely to be an example of convergence than true homology (see Wickstead & Gull, Citation2011a), and the absence of detectable lamin homologs outside of the unikonts may indicate the absence of the IF class of cytoskeletal proteins from other arms of the eukaryotic lineage ().
Cytoskeletal motors
Eukaryotic cytoskeletal function is hugely augmented by the recruitment of motors, kinesins and dyneins, to the tubulin-based cytoskeleton, and myosins, to F-actin. None of the eukaryotic cytoskeletal motors have characterized prokaryotic homologs with a similar motor function. However, systems such as the AglQRS system for gliding in the bacterium Myxococcus xanthus, suggest that trafficking on the bacterial cytoskeleton analogous to that seen extensively in eukaryotes has evolved (Sun et al., Citation2011). Kinesin and myosin motors share a common structure and are distant relatives (Kull et al., Citation1998, Citation1996). It is likely that they do not share a single common motor ancestor that walked on both actin- and tubulin-based filaments, but evolved independently from the same superfamily of proteins (Leipe et al., Citation2002). The ancestral superfamily of P-loop NTPases also gave rise to many other eukaryotic families, including the Ras-superfamily GTPases and additional G protein families (Leipe et al., Citation2002).
In contrast to kinesin/myosin, dyneins belong to the large, diverse AAA+ superfamily. Each dynein heavy chain contains six AAA+ domains of ∼220 residues which form a hexameric ring (Carter et al., Citation2011; Samsó et al., Citation1998). Most prokaryotic AAA+ proteins contain a single AAA+ domain, but many assemble into homomeric rings (Lupas & Martin, Citation2002). Dynein most likely evolved by duplication and subsequent divergence of a single AAA+ domain. Much of this divergence occurred before the LECA and prior to emergence of the major dynein classes. Due to their small size and degree of divergence, the evolutionary history of AAA+ domains is also extremely challenging, but there is some evidence that the closest prokaryotic family to dynein may be MoxR and its relatives (Iyer et al., Citation2004; Snider & Houry, Citation2006). MoxR family members have diverse roles in prokaryotes and no known motor function, but have instead chaperone-like properties, suggestive of a similar role for the original ancestor of dynein in the pre-eukaryotic cell.
The cytoskeletal motors comprise superfamilies containing several paralogous classes, many of which are ancient. At least eleven ancient kinesin paralogs were present in the LECA under any likely model of early eukaryotic branching – namely, Kinesin-1, 2, 3, 4/10, 5, 8, 9 A, 9B, 13, 14 and 17 (Wickstead et al., Citation2010). This provides molecular evidence for several key aspects of LECA biology. It can be inferred that the LECA built a bidirectional spindle with antagonistic plus-end directed (Kinesin-5) and minus-end directed (Kinesin-14) motors, which was modulated by microtubule depolymerizing motors (Kinesin-8 and -13). Also, LECA trafficked membrane-bounded organelles within the cytoplasm using Kinesin-1 and -3 and built a cilium/flagellum containing a 9 + 2 axoneme (Kinesin-9 A) by intraflagellar transport (Kinesin-2). However, post LECA all of these families have experienced multiple losses, so that no family is now ubiquitous. In spite of this, no eukaryote entirely lacking kinesins has been discovered, although the apicomplexan T. annulata completes its life-cycle with only two kinesins, both depolymerizing motors (Kinesin-8 and -13) (Wickstead et al., Citation2010).
Myosin diversity may be even higher than that of kinesins, with descriptions of up to 35 classes (Foth et al., Citation2006; Odronitz & Kollmar, Citation2007). However, much of this diversity may result from difficulties in phylogenetic reconstruction, as seen in the large number of apparently lineage-specific myosin families, and more conservative estimates are similar to kinesins (Richards & Cavalier-Smith, Citation2005). Even with these considerations, at least three myosin families can be traced back to the LECA. Myosins are entirely absent from excavates (G. lamblia and Trichomonas vaginalis) and the red alga Cyanidioschyzon merolae (Richards & Cavalier-Smith, Citation2005).
Recent phylogenetic analyses of dynein heavy chains suggests nine major classes (Morris et al., Citation2006; Wickstead & Gull, Citation2007; Wilkes et al., Citation2008), which encompass seven classes built into the axoneme of motile cilia/flagella and two cytoplasmic classes. All nine dynein classes were present in the LECA, but multiple losses during eukaryote diversification are clear. Cytoplasmic dynein 2 is the retrograde motor for IFT and required for construction of the axoneme in almost all lineages (Briggs et al., Citation2004; Rosenbaum & Witman, Citation2002). Unsurprisingly, loss of cilia/flagella is associated with loss of flagellar dyneins or cytoplasmic dynein 2 (Wickstead & Gull, Citation2007). Cytoplasmic dynein 1 has also been lost independently at least three times, and the amoeba E. histolytica, red alga C. merolae and all angiosperms lack dyneins entirely (Lawrence et al., Citation2001; Wickstead & Gull, Citation2011b).
These analyses provide both molecular evidence for the existence of key motor-related functions in LECA and show that a large proportion of motor family diversity had already arisen in this ancient lineage. The advent of motor proteins was likely critical for eukaryotic cellular compartmentalization and facilitated the increased cellular complexity between the FECA, with a prokaryote-like cytoskeleton, and the more sophisticated LECA.
The axoneme
In many lineages, the cytoskeleton is used to form flagella and/or cilia, constructed from a microtubule axoneme extending from the basal body. In spite of notable absences, in angiosperms and most fungi, flagella/cilia are widely distributed (Carvalho-Santos et al., Citation2011; Hodges et al., Citation2010). In nearly all organisms the axoneme and basal body retain their iconic nine-fold symmetry. The basal body of flagella/cilia is an identical structure to the centriole, which is embedded in the primary microtubule organising centre of metazoan cells, the centrosome (see Azimzadeh & Marshall, Citation2010). Given their distribution in extant eukaryotes, cilia/flagella arose pre-LECA (Cavalier-Smith, Citation1978) and molecular data suggest that the LECA flagellum possessed both sensory and motility functions (Mitchell, Citation2007).
There are obvious cytoplasmic analogs for much of the axonemal machinery, including core microtubules, dynein motors and several IFT components, presenting a plausible route for evolution of the axoneme from cytoskeletal factors; an autogenous origin for the axoneme is now accepted (Mitchell, Citation2007; Pickett-Heaps, Citation1974). However, the precise pathway by which the flagellum formed is still unclear and three alternative hypotheses have been proposed. The “sensation-first” model suggests axoneme-like structures evolved from microtubule-based protrusions with an exclusively sensory function (Cavalier-Smith, Citation1978), “beat-first” models place motility as the most ancestral function, while “gliding-first” models propose that the original function was motility, but driven by gliding resulting from an IFT-like motor, rather than microtubule sliding (Mitchell, Citation2004, Citation2007). Since dynein and axonemal evolution are intimately linked, dynein phylogenies can distinguish between these alternate hypotheses if the tree can be accurately rooted, such that the order of dynein family emergence can be inferred. If the ancestral dynein is assumed as a cytoplasmic dynein 1 (Hartman & Smith, Citation2009; Wilkes et al., Citation2008), then analyses support the sequential appearance of IFT and then axonemal beating, consistent with sensation-first and gliding-first hypotheses. Gibbons suggested that the homodimeric nature of cytoplasmic dynein 1 represents a more “primitive” arrangement than some of the axonemal dyneins (Gibbons, Citation1995), but the subsequent discovery of simple axonemal single-headed dyneins refutes this. Moreover, proposing that the axoneme evolved from cytoplasmic components does not necessitate cytoplasmic dynein 1 as the progenitor dynein. In contrast, rooting of dynein phylogenies using the closest eukaryotic relative of dynein, midasin (Garbarino & Gibbons, Citation2002; Iyer et al., Citation2004), suggests that microtubule sliding was much closer to the origin of proto-cilia and evolved before the specialized IFT machinery (Wickstead & Gull, Citation2011b). This implies that the cilium may have evolved not from an immotile protrusion, but from a motile cytoplasmic microtubule bundle analogous to the axostyles of oxymonads (McIntosh, 1973; McIntosh et al., Citation1973). This bundle would have been assembled in an IFT-independent manner, as are the axonemes of several extant organisms (Briggs et al., Citation2004; Witman, Citation2003).
Cell division
There are two major modes of cell division in eukaryotes, mitosis and meiosis. The former is the mechanism that underpins somatic or non-reductive division, resulting in two daughter cells with similar DNA content, while meiosis is a sexual process resulting in production of gametes. Depending on the configuration of the genome, this proceeds either by reductive cell division from a diploid state prior to gamete production and fusion or mating between two haploid cells prior to the reductive divisions, although other modes have been described. The primary mechanisms of eukaryotic cell division require participation of the cytoskeleton, for construction of the spindle, nuclear division and cytokinesis itself. Prokaryotic origins for the cytokinesis machinery and the role of the cytoskeleton in nuclear events have been described above. Meiosis, which for most organisms is a facultative mode of division, requires the participation of a specialised set of gene products that are frequently only expressed during the meiotic process, and which include Spo11, Hop1, Dmc1 and others (Peacock et al., Citation2011; Ramesh et al., Citation2005, Schurko & Logsdon Citation2008). Meiosis is clearly very distinct from bacterial conjugation. The ability to reassort genomes during meiosis is a major step in evolution and can facilitate the sweep of new traits more rapidly through a population.
Early phylogenetic analysis suggested that sex was likely widespread and that the LECA was a faculative sexual organism, despite evidence for loss of such activity in some lineages (Dacks & Roger, Citation1999), and the inherent difficulty in observing such behaviour in most taxa (e.g. Peacock et al., Citation2011). Importantly, a facultative sex mode ensures that the major cost to meiotic division, i.e. disruption of potentially successful genotypes, is only paid when the environmental conditions change. Comparative genomics further demonstrated the presence of meiotic genes in most lineages (Malik et al., Citation2007a; Ramesh et al., Citation2005; Schurko & Logsdon Citation2008), although significantly, even in organisms where meiosis clearly occurs, the entire meiotic gene cohort may not be present, or in taxa where a substantial cohort are present, the precise mechanism may be distinct. For example, Giardia, considered by some to be descended from an early branching eukaryotic lineage, is able to exchange chromosomes during specific stages in the life cycle without a true meiosis (Carpenter et al., Citation2012; Ramesh et al., Citation2005).
There is some evidence for the evolution of meiosis genes prior to the LECA. Spo11, a critical topoisomerase essential for meiosis is derived from archael topoisomerase VI (Bin3), and which is also retained by eukaryotes (Malik et al., Citation2007b). Based on the conservation of Spo11 paralogs in extant taxa, there were likely three Spo11 genes present in the LECA. Only Spo11-1 and Spo11-2 paralogs are meiosis specific, and significantly at least one of these is retained by all major lineages, but with evidence for frequent secondary losses. Only higher plants retain all three Spo11 paralogs, while Spo11-3 (Top6A) and Bin3 (Top6B) were lost from the unikont common ancestor. Hence the conclusion that the LECA possessed meiotic capability is well supported, with clear implications for its life style.
Mitochondrial origins and LECA
Unlike the majority of intracellular compartments, the mitochondrion has a non-endogenous origin, arising from once free-living bacteria, specifically an α-proteobacterium-like organism. The non-endogenous origin of mitochondria was first postulated by Portier & Wallin nearly a century ago (reviewed in Martin, Citation2007). The presence of DNA (Nass & Nass, Citation1963), semi-autonomous replication (Mitchell & Mitchell, Citation1952) and a supposed role in oxygen removal allowed others to postulate an endosymbiotic mitochondrial origin (Sagan, Citation1967). Subsequent accumulation of mainly molecular data provided clear evidence for a prokaryotic origin but the timing remains contentious with, in principle, two conflicting hypotheses: the phagotrophy and syntrophy models (O’Malley, Citation2010; and ). The phagotrophy model argues that the protoeukaryote required a cytoskeleton to facilitate endocytosis, after which an α-proteobacterium was engulfed (Sagan, Citation1967; Whatley et al., Citation1979). Based on metabolic and energetics arguments, the syntrophy model postulates that the origin of eukaryotes and establishment of the mitochondrion was one event (Lane & Martin, Citation2010; Martin & Müller, Citation1998).
Initial ultrastructural studies of various eukaryotes believed to be primitive, including Entamoeba. Giardia and Trichomonas, suggested the absence of typical cristate mitochondria, leading to proposal of the Archezoa as a primitively amitochondriate eukaryote group, whose extant representatives are these amitochondrial taxa (Cavalier-Smith, Citation1987). This implied that LECA could have lacked a mitochondrion. However, subsequent unambigous identification of diagnostic mitochondrial features in each of these organisms plus the discovery of highly derived mitochondrial organelles in all other studied Archezoan taxa, resulted in the hypothesis being rejected. Mitochondrial variants go by various names as mitosomes, hydrogenosomes, mitochondrial relicts or mitochondrial-like organelles (Müller et al., Citation2012; van der Giezen, Citation2009). Discovery and assignment of these organelles is strong evidence against any eukaryotic amitochondrial lineage, with the consequence that, if not one and the same event, eukaryogenesis and the origin of mitochondria were chronologically closely linked, so that while the status of FECA remains uncertain, LECA definitely possessed mitochondria (). More recently, thermodynamic and bioenergetic arguments have been used to support a syntrophic origin of eukaryotes and, simultaneously, mitochondria (Lane et al., Citation2010; Lane & Martin, Citation2010; Lane, Citation2011).
One early event during mitochondrial establishment was loss of much of the genome of the α-proteobacterial progenitor, and transfer of many of these genes to the nucleus. In comparison with the >1000 proteins imported into aerobic mitochondria, very few mitochondrial proteins are encoded by extant mitochondrial genomes (Barbrook et al., Citation2010; Bullerwell & Gray Citation2004; Gray et al., Citation2004). Based on modern bacteria, the endosymbiont likely possessed a ∼1–2 Mb genome encoding at least a thousand proteins and RNAs. Extant mitochondrial genomes can be over 2 Mb in size, but in terms of gene content currently range from three protein-coding genes on a genome of ∼6 kb to 97 protein coding genes on a ∼69 kb genome (Bullerwell & Gray Citation2004). Therefore, the vast majority of proto-mitochondrial genes that were originally endosymbiont-encoded, have been lost with substantial portions transferred to the genome of the host (Rivera et al., Citation1998). Analyses of extant nuclear genomes indicate that most operational genes, i.e. encoding proteins involved in DNA and RNA functions, have an archaeal origin, while genes encoding metabolic proteins processes are eubacterial (Pisani et al., Citation2007; Rivera et al., Citation1998). As the host must have possessed genes encoding metabolic proteins, irrespective of the mechanism of mitochondrial origin (O’Malley, Citation2010), this suggests that FECA had archaeabacterial metabolism and LECA replaced this system using endosymbiont genes (Doolittle, Citation1998; Ginger et al., Citation2010; Müller et al., Citation2012). An immediate consequence of transferring any essential genes from endosymbiont to host is to make the endosymbiont dependent, and potentially requiring the host to return essential proteins whose genes have been transferred to the nuclear genome. This has been viewed as enslavement, but as the process of endosymbiotic gene transfer seems inevitable, it seems the endosymbiont and host had no choice as to the outcome of their relationship (Doolittle, Citation1998). This is clearly a further massive revolution in function in the transitional period between FECA and LECA.
Extant eukaryotes use complex protein import mechanisms to target nuclear encoded proteins to various mitochondrial compartments (Neupert & Herrmann, Citation2007), although more recent studies indicate that many microbial eukaryotes seem able to survive with simpler import mechanisms (Basu et al., Citation2013; Burri et al., Citation2006; Dagley et al., Citation2009; Dolezal et al., Citation2010; Eckers et al., Citation2013). Significantly, evolution of mitochondrial targeting signals is not that difficult in terms of sequence evolution and synthetic evolution experiments suggest that such signals can arise readily, while selective pressure is likely greater for prevention of import of inappropriate polypeptides than for failure to translocate a bona fide mitochondrial protein (Allison & Schatz, Citation1986; Lemire et al., Citation1989). Further, the mitochondrial import machinery did not arise de novo in the early eukaryotes but via existing bacterial membrane proteins, such as OmpA (Clements et al., Citation2009; Hewitt, et al., Citation2011; Selkrig et al., Citation2012), further lowering the selective barrier.
A major mitochondrial function is to support the electron transport chain (ETC). The ETC transfers electrons from donors (NADH) to an acceptor (O2 in aerobes), via several enzyme complexes residing in the mitochondrial inner membrane, while also pumping protons across that membrane. The proton gradient drives ATP synthesis using a specialised mitochondrial ATPase (F0F1 ATPase). The ETC is an ATP generation mechanism common to prokaryotes and eukaryotes, suggesting that LECA possessed a mitochondrial ETC. Studies focusing on aerobes suggested an ETC containing five complexes (Complex I; NADH:quinone oxidoreductase, Complex II; succinate dehydrogenase, Complex III; the cytochrome bc1 complex, Complex IV; cytochrome c oxidase Complex V; F0F1 ATPase). On the one hand, taxonomically broader sampling suggests that the five complex ETC is lineage-restricted and that a variety of shorter ETCs exist across the eukaryotes, although these are likely derived states (Müller et al., Citation2012). On the other hand, many eukaryotes (some animals, plants and unicellular eukaryotes) possess an alternative oxidase, which sits in the inner mitochondrial membrane and passes electrons directly from a reduced quinone to oxygen. Here electron transfer to/from a quinol is not coupled to proton-pumping. The distribution of alternative oxidase is broad, suggesting that alternative oxidase was present in LECA (Müller et al., Citation2012), but was subsequently lost from many lineages, rather than being an enzyme introduced by repeated lateral gene transfer. However, alternative oxidase is seldom present in extant α-proteobacteria or archaea for which complete genome sequences are available. This leaves uncertainty as to whether alternative oxidase was present in the endosymbiont that became the proto-mitochondrion; rather, the alternative oxidase likely provides an example of a feature of the core metabolism introduced early into the eukaryotic lineage, but coming neither from the α-proteobacterial endosymbiont nor necessarily from the archaeal host. This is a theme that is repeated in other core aspects of eukaryotic metabolism.
Recently, it has become evident that mitochondria play a crucial role in the production of iron-sulphur clusters, cofactors with essential roles for many different enzymes (Lill & Kispal, Citation2000). In fact, FeS clusters are so widespread that it is probable that iron and sulphur played an important role in the origin of life (Hall et al., Citation1971). There are three principal distinct FeS cluster synthesizing mechanisms: a nitrogen fixation system (NIF), the iron-sulphur cluster system (ISC) and the mobilization of sulphur system (SUF) (Bandyopadhyay et al., Citation2008; Xu & Møller, Citation2011). Until recently, the ISC system was viewed as the sole essential mitochondrial pathway, as all FeS proteins in the cell depended on this biosynthetic mechanism (Lill & Kispal, Citation2000), but more recently evidence for a eukaryotic SUF system has appeared (Tsaousis et al., Citation2012). As archaeabacteria generally use SUF and eubacteria use ISC, it is likely that the mitochondrial endosymbiont brought ISC to the eukaryotes. An important component of SUF is the cysteine desulphurase, which contains the essential Isd11 protein in eukaryotes (Richards & van der Giezen, Citation2006). Isd11 is absent from eubacteria; instead bacterial cysteine desulphurase requires a distinct protein, YfhJ to function (Pastore et al., Citation2006; Shimomura et al., Citation2005). It seems YfhJ was lost early in eukaryotic evolution and was rapidly replaced by the eukaryotic Isd11.
General cellular metabolism
As mentioned above, a eubacterial origin for many eukaryotic central metabolic enzymes was evident from early genomic comparisons (e.g. Esser et al., Citation2004), and is supported by recent analyses (e.g. Thiergart et al., Citation2012). However, several major pathways, such as glycolysis, have probably experienced significant HGT and gene displacement post-LECA, most notably during the evolution of anaerobic/microaerophilic lineages (Liapounova et al., Citation2006; Stechmann et al., Citation2006). Thus catabolism of glucose, the carbon source for ATP production preferred by the majority of extant eukaryotes, is catalyzed by orthologs of classic Embden-Meyerhof glycolytic enzymes, and not the variants present in some archaea (Sato & Atomi, Citation2011; Siebers & Schönheit, Citation2005). If endosymbiosis with the α-proteobacterial mitochondrial progenitor was the key event in eukaryogenesis, facilitating enhanced energy production via oxidative phosphorylation (Lane & Martin, Citation2010), the preponderance of eubacterial homologs within eukaryotic metabolism suggests that balancing metabolic regulation from the eubacterial endosymbiont might have been easier than integrating and co-regulating metabolism from both the endosymbiont and archaeal host during the FECA to LECA transition.
The presence of mitochondrial sirtuins in animals and trypanosomes (Alsford et al., Citation2007; Katada et al., Citation2012) may indicate the possible extent of integration between mitochondrial and nuclear-encoded metabolism in LECA. Sirtuins catalyse NAD+-dependent deacetylation of a variety of substrates, including histones. Together with other chromatin re-modelling enzymes, sirtuins regulate nuclear gene expression in response to metabolite changes in mammalian cells and yeast (discussed in Katada et al., Citation2012). Thus far in mitochondria, the identified targets of sirtuins are metabolic enzymes. However, the compact packaging of mitochondrial DNA from taxonomically diverse sources, including histone-like proteins in trypanosomatids (Avliyakulov et al., Citation2004), possibly points towards the presence of a sophisticated mechanism for dual regulation of both mitochondrial and nuclear genomes, and that epigenetic regulation of metabolism was established in the LECA.
Potential similarities between metabolic regulation in LECA and extant eukaryotes are further illustrated by conservation of AMP-activated kinase (AMPK) in all eukaryotes except the microsporidian Encephalitozoon cuniculi (Hardie, Citation2011). In animals, some plants and yeasts, AMPK is responsible for maintaining cellular energy homeostasis, acting on key catabolic and anabolic targets and regulating mitochondrial biogenesis and turnover, suggesting that in LECA the ancestral AMPK functioned similarly (Hardie, Citation2011; Hardie et al., Citation2012; Thelander et al., Citation2004). In E. cuniculi the absence of AMPK is an example of reductive evolution, and likely a result of reliance on the host for ATP (Tsaousis et al., Citation2008). Even in Giardia, an excellent example of genomic minimalism, orthologs of AMPK subunits are retained despite a simplified metabolism where mitochondria do not contribute directly to ATP production (Hardie et al., Citation2003; Morrison et al., Citation2007). However, determining if an ancestral role for AMPK was to trigger a switch to oxidative metabolism in response to nutrient deprivation requires experimental analysis of taxonomically diverse protists (Hardie, Citation2011).
For anabolic biochemistry, the biosynthesis of heme, nucleotides and multiple coenzymes is undertaken by many unicellular and multicellular eukaryotes. In contrast, biosynthetic pathways leading to the “essential” amino acids have been lost from animals and many taxonomically diverse protists (Guedes et al., Citation2011; Payne & Loomis, Citation2006). The latter organisms tend to be either phagotrophs or parasites. However, retention of highly conserved pathways for essential amino acid biosynthesis, as well as for and sulphate assimilation in fungi, strongly suggests that LECA was capable of extensive macromolecular biosynthesis and similar to many bacteria, for example Escherichia coli, which can grow in minimal media. Although LECA was almost certainly a heterotroph, capable of utilising glucose, fatty acids or amino acids as carbon sources, retention of a broad repertoire of amino acid biosynthetic pathways may have been required to support the evolution of the cellular complexity underpinning the FECA-LECA transition. Since the transition was unlikely to have occurred in ecological isolation, FECA and its immediate descendants would have likely been competitively disadvantaged in comparison to prokaryotes if their anabolic metabolism was more limited.
Beyond a conserved core there are clear eukaryote-specific metabolic innovations. A hallmark of eukaryotic biology is sterol biosynthesis, and the few bacteria capable of sterol biosynthesis are often presumed to do so via HGT from eukaryotes (e.g. Desmond & Gribaldo, Citation2009). In contrast, sterols are either synthesized or acquired by all eukaryotes, testifying to an ancient origin and function of a biosynthetic pathway, with over 25 enzyme-catalyzed reactions required for de novo synthesis. This also explains the potential of persistent sterol breakdown products as eukaryotic biomarkers within the geological record (Love et al., Citation2009; Summons et al., Citation2006). Sterols function most obviously as membrane constituents, regulating membrane fluidity and microdomain partitioning, but also encompass other activities, including key roles in many aspects of development and survival in mammals (Entchev & Kurzchalia, Citation2005; Kurzchalia & Ward, Citation2003). A so-called “sparking” role for trace amounts of sterol or sterol-derived metabolites in cell cycle regulation of yeasts and some protists has also been proposed (Nes et al., Citation2012; Parks et al., Citation1995), and this raises the possibility of a role for sterols in cell cycle “sparking” in either LECA or an older eukaryotic ancestor.
Further, peroxisomes provide an excellent example of metabolic organelles that are the products of eukaryotic innovation. The presence of peroxisome-bearing organisms in all major eukaryotic groups, albeit with considerable diversity in likely composition between distinct taxa, indicates that peroxisomes were present in the LECA and it is probable that these organelles functioned in diverse lipid metabolism (Gabaldón et al., Citation2006, Citation2010). The possibility that peroxisomes had endosymbiotic origins has been overturned by recent phylogenetic analysis of the peroxisomal protein import apparatus and proteomes (Bolte et al., Citation2011; Gabaldón et al., Citation2006; Gabaldón & Capella-Gutiérrez, Citation2010) together with cytological analysis of trafficking pathways of essential peroxisomal membrane proteins (reviewed in Tabak et al., Citation2008). Crucially, de novo peroxisome formation in S. cerevisiae is dependent upon routing of Pex3 and Pex19, also required for peroxisome formation in mammals, to newly forming peroxisomes via the endoplasmic reticulum, suggesting that peroxisomal membranes arise from the ER (Hoepfner et al., Citation2005). Lipid-associated pathways reconstructed for LECA peroxisomes include long-chain fatty acid β-oxidation, α-oxidation of branched-chain fatty acids, the glyoxylate cycle, ether lipid biosynthesis and several enzymes of the mevalonate pathway. In many instances the peroxisomal location of these pathways also dictates the presence within the organelles of enzymes associated with NADPH formation (e.g. isocitrate dehydrogenase, glucose-6-phosphate dehydrogenase) and the detoxification of reactive oxygen species (most notably catalase and superoxide dismutase) (Gabaldón et al., Citation2006; reviewed in Gabaldón, Citation2010), suggesting that similar metabolic functions plausibly featured in the peroxisomes of LECA.
Of the more extreme examples of metabolic compartmentalization is the essential and exclusive compartmentalisation to peroxisomes of either the first six or seven glycolytic enzymes in trypanosomatids (depending upon the species or life cycle stage examined) (see Gualdrón-López et al., Citation2012). A recent report of peroxisomal targeting of glycolytic enzyme isoforms in diverse fungi and possible peroxisomal targeting of glycolytic enzymes in mammals (Freitag et al., Citation2012), suggests either a fascinating example of convergent evolution or implies that the metabolic capabilities of peroxisomes in extant eukaryotes, LECA, or both are significantly under-appreciated.
As metabolic compartmentalisation clearly arose early during eukaryotic evolution, so would a need to regulate and co-ordinate Fe-S cluster assembly for incorporation into proteins across multiple cellular compartments, including mitochondria, cytosol, nucleus and ER (Balk & Pilon, Citation2011). As discussed earlier, mitochondria seemingly retain a universal and conserved role in assembly of Fe-S clusters (see also Lill et al., Citation2012), and mitochondrial involvement is at several points: in provision of the Fe-S clusters themselves, regulating overall cellular iron homeostasis and providing an activated sulphur compound for export to the cytosol and thence cytosolic Fe-S cluster assembly. The cytosolic Fe-S cluster machinery appears to be another eukaryotic-specific innovation (Lill, Citation2009; Stehling et al., Citation2012) that is conserved in all eukaryotes and required for the maturation of essential cytosolic and nuclear proteins, albeit with individual components whose origins can be readily traced to prokaryotes (Allen et al., Citation2008, Basu et al., Citation2013; Boyd et al., Citation2009; Horner et al., Citation2002). The eukaryotic-specific cytosolic Fe-S cluster assembly pathway is particularly relevant if one considers the enzymes and other proteins present in LECA and potentially FECA, but generally absent from extant α-proteobacteria and archaea. Cytosolic Fe-S cluster assembly requires a protein closely related to Fe-hydrogenase, the defining enzyme of hydrogenosomes and responsible for anaerobic H2 production in eukaryotes (Balk et al., Citation2004; Luo et al., Citation2012; Müller et al., Citation2012; Song & Lee, Citation2011). The Fe-hydrogenase-like protein (NAR1 in yeast or iron-only hydrogenase-like protein 1 (IOP1) in animals) probably evolved from a bona fide Fe-hydrogenase (Horner et al., Citation2002). However, Fe-hydrogenase is present widely in prokaryotes, but poorly represented in extant α-proteobacteria or archaea, suggesting that the eukaryotic Fe-hydrogenase was unlikely derived from the mitochondrial progenitor or archaeal host. A presumed early origin of eukaryotic Fe-hydrogenase may suggest either a more complex syntrophic model for eukaryotic origins (e.g. López-Garćia & Moreira, Citation1999) or HGT during early eukaryogenesis, with subsequent neofunctionalisation. As most Fe-hydrogenases are oxygen-labile, the finding that Fe-hydrogenase was present early in eukaryotic evolution is also significant in considering whether FECA evolved in an aerobic or microxic/anoxic environment (e.g. Hug et al., Citation2010; Müller et al., Citation2012).
A final example of a metabolic regulatory mechanism is autophagy, which encompasses various pathways for selective and non-selective remodelling of cellular architecture through lysosomal degradation. Macroautophagy in S. cerevisiae utilises over 30 ATG gene-products (Yang & Klionsky, Citation2010), and the majority are conserved across the breadth of eukaryotic evolution, including TOR kinases that act as master autophagy regulators. This indicates a major presence in the LECA (Brennand et al., Citation2011; Rigden et al., Citation2009). Macroautophagy also has several roles in cellular differentiation (Duszenko et al., Citation2011; Yang & Klionsky, Citation2010), while a function in turnover of whole organelles in diverse eukaryotes suggests the presence of similar pathways in versatile early eukaryotes (Herman et al., Citation2008; Manjithaya et al., Citation2010). Curiously, while understanding the mechanistic basis and regulation of autophagy in model systems has advanced during the last 15–20 years, the origin of much of the canonical, conserved autophagy machinery remains enigmatic, including the origin of the autophagosome membrane (Mari et al., Citation2011).
Despite the more limited metabolic repertoire in extant eukaryotes generally when compared with prokaryotic taxa, an essential Fe-hydrogenase-related protein in cytosolic Fe-S cluster assembly and glycolytic enzymes imported to peroxisomes from diverse taxa demonstrate that the full appreciation of the metabolic repertoires of FECA and LECA remains a way off. Regulation of metabolic fluxes in LECA was also likely complex; AMPK was present and data suggest potential epigenetic regulation of metabolism, while a clear role for autophagy in LECA is strongly indicated based on conservation of the ATG genes across the eukaryotes.
FECA to LECA: A severe bottleneck?
The events leading to eukaryogenesis, and the transitional period between FECA and LECA, may have become clearer, but the precise sequence of events, what drove them, how much diversity arose and how much was lost during this period remain less well defined. Moreover, it is unknown what the duration of the transition period was in terms of cell/organismal generations. We previously suggested that colonization of the eukaryotic endomembrane system by protocoatomer-based membrane deforming complexes may reflect a form of intracellular competition and natural selection (Field et al., Citation2011). While the presence of bona fide prokaryotic protocoatomer is in doubt (McInerney et al., Citation2011), the protein architectural elements are clearly present. With expansions of many paralogous families critical for eukaryotic cells, one can speculate that similar selections for protein families able to undergo neofunctionalization during the period prior to LECA occurred, so that specific gene families dominated. With these elements in place, massive and rapid expansion of eukaryotic diversity was perhaps inevitable. The organellar paralogy hypothesis (Dacks & Field, Citation2007) is an example of this mechanism, and posits evolution of new compartments based on ratchet-like replacement of paralogs within complexes, generating new functions, and could be applied to any modular system (). This also implies that alternate evolutionary strategies by post-FECA/pre-LECA organisms were unsuccessful and lost, and the complexity of transition period organisms remains unknown.
What was required to progress from FECA to LECA? Due to prokaryotic relatives of many gene families that mediate eukaryote-specific features (however distant), only a moderate level of protein structural invention may have been required, regardless of how critical such inventions were. What permitted elaboration and expansion of gene families and the consequent rise in cellular complexity? If most of the pieces were already in place in many prokaryotes, why did this transition not happen repeatedly? One attractive explanation is that the acquisition of the mitochondrion, generally agreed to have occurred only once, massively increased energy production, and may have facilitated elaboration of sophisticated membrane structures, the cytoskeletal systems to subtend them and the eukaryotic flagellum (Lane & Martin, Citation2010). Membrane transport and flagellum-mediated motility are extremely expensive activities in both biosynthetic demand and direct ATP requirements. An alternate model, that phagocytosis was required for acquisition of the α-proteobacterial mitochondrial endosymbiont, is also conceivable. Specifically, an endomembrane system may have evolved early, and could even have been sufficient for dominance within the transitional period. The ability to eat, or at least out-eat, the competition would also have been a powerful selective advantage.
Is this then simply an example of contingency; the first organism to acquire the mitochondrion rapidly dominated the local environment, eliminating all but a restricted lineage of eukaryotes and their descendants? Conceivably, mitochondrial acquisition facilitated even greater exploitation of primitive eukaryotic systems, delivering the coup de grâce to all amitochondrial eukaryotes. For example, it is now clear that the IFT system is related to protocoatomer, and recent data suggest evolution from COP-I (Satir et al., Citation2007; van Dam et al., Citation2013); an interesting hypothesis would be that evolution of the flagellum had to await an enabling set of conditions, but increased the selective advantage of transitional eukaryotes once this occurred. This may have been comparatively late as all IFT subunits were present in LECA. One other possible answer to the question of why the transition did not happen frequently is that perhaps it did. Comparative genomics suggests the presence of the meiotic system in the LECA (Ramesh et al., Citation2005). The ability to recombine and resort genes has the obvious ability to allow rapid innovation. Furthermore, comparative analysis of sexual cycles across eukaryotes implies that facultative sexual stages are common and likely ancestral, providing benefits of meiosis without the disadvantages and dangers of obligately linking meiosis with cell replication (Dacks & Roger, Citation1999). The other facet to the process is syngamy, or fusion of gametes, which facilitates the sweep of advantageous alleles through a population and allows for both genetically encoded traits (e.g. complex Golgi) and cytoplasmic traits such as mitochondria to arise in independent lineages and then spread through a reticulating population. This even raises the question of whether the observed complexity arose in a single FECA lineage or through multiple transitional lineages that, via interbreeding, “multiplexed” their innovations. This idea, conceptually similar to a fusion origin of eukaryotes, emphasizes that what we reconstruct through comparative genomics is a LECA. All points prior to this are still very much Terra incognita, and determining the order of emergence of the various cellular innovations remains an important goal for resolution by future work.
Reconstructing the LECA
What has emerged from comparative analysis of cellular systems is the great complexity in the reconstructed LECA, implying an ancestral which organism possessed capabilities exceeding those of many extant eukaryotes. LECA had a mitochondrion, meiotic machinery, a sophisticated and flexible metabolic capability, a fully differentiated endomembrane system, phagocytosis, actinomyosin and tubulin-based cytoskeletal systems together with a large complement of motor proteins, and finally a nucleus subcompartmentalised into hetero- and euchromatin, together with sophisticated nucleocytoplasmic transport. Subsequently, many lineages simplified various cellular aspects, losing genes that played important roles in their forebears. Due to the limitations of phylogenetic reconstruction, the LECA may have been even more complex than these studies suggest; for example, the presence of multiple subfamily paralogs cannot be reconstructed for LECA, with the consequence that estimates of a LECA Rab complement of twenty could easily be an underestimate (Elias et al., Citation2012). This caveat may be relevant for many of the gene families considered here, especially those where paralogous families are drivers of new function, including G proteins, SNAREs, protocoatomers, kinesins, dyneins and karyopherins, as well as expanded metabolic enzyme isoforms. Finally, as our understanding of eukaryotic biology is biased towards functions described in animals and fungi, it remains unknown how many gene families present in the LECA may emerge from broader sampling, but which have been specifically lost from animals and fungi. Several such examples have emerged within the membrane-trafficking system recently (Elias et al., Citation2012; Gabernet-Castello et al., Citation2013; Schlacht et al., Citation2013).
Why was the LECA so complex? The answer may simply be that a high level of complexity was required to dominate the early eukaryotic landscape and to occupy a successful position within the ecosystem. Having on board phagocytic capabilities, possibly amoeboid locomotion and a flagellum permits multiple modes of motility, together with the ability to feed on other organisms. As the LECA was a heterotroph and lacked a plastid, it was probably dependent on such abilities. The presence of the mitochondrion would at least partly offset energetic costs of increased complexity, while a highly sophisticated metabolic network with little dependence on a specific nutrient source allowed exploitation of a wide range of carbon sources. The LECA was an aerobe and possessed a TCA cycle, as evidence suggests that anaerobic eukaryotes have arisen from secondary losses of mitochondrial metabolic capacity. The presence of meiosis further implies that the LECA was capable of the generation of gametes, while the probable differentiation of LECA chromatin into hetero- and euchromatin, indicates potential developmental progression via repression of selected gene cohorts and the presence of distinct life stages. This latter aspect could, for example, have facilitated the emergence of quiescent forms allowing survival during unfavorable conditions, as present in testate amoebae, and which may have been present quite early on in post-LECA evolution (Butterfield, Citation2007). This feature both protects against transient environmental changes and aids in dispersal via traversing hostile environments between favorable locales. In essence, what has emerged is a sophisticated and potentially predatory organism, with great flexibility allowing survival in varied ecological niches.
Two modes of post-LECA evolution have now emerged. Firstly, many taxa acquired significant complexity, for example, vascular plants, metazoa and multiple protist lineages. Here, examples of paralogous expansions and the evolution of novel gene families abound. By contrast, there are many lineages where complexity decreased, encompassing many fungi, some algae, most kinetoplastids, apicomplexans and diplomonads. Many reductions are almost certainly a result of parasitism, frequently associated with reduced metabolic potential, as well as more limited trafficking and cytoskeletal arrangements. In other cases, streamlining may be a result of adaptation to specific environments, where reductive pressure includes energetic reasons to facilitate shorter cell cycle times, and increase competitive advantage (yeasts), or for protection (C. merolae has a reduced endocytic system, likely to protect against a low pH environment). Critically, a LECA with a broad metabolic and cellular functional repertoire would have been best placed for subsequent exploitation of novel niches, a well-equipped explorer, with ample capacity from which to build greater complexity, plus access to a smörgåsbord of functionality from which more limited activities could be selected, providing opportunity for adaptation to a broad range of conditions. Hence a complex LECA may explain the enormous range of life styles and cellular forms that are exhibited by living taxa.
Conclusions, perspectives and many unanswered questions
Progress in the last decade on describing the earliest events in eukaryote evolution has been spectacular, and has advanced from a rather skewed view of ever increasing complexity based predominantly on assumptions, to the appreciation and description of a LECA cellular sophistication that is based on substantial molecular data. The overriding conclusion from all of these studies is of great functional differentiation, and that the LECA was, in many ways, a surprisingly modern organism. While we may never fully understand the life cycle and life style of the LECA, we now have a far more sophisticated view of its likely capabilities, functions and even modes of gene regulation. The importance of secondary reduction to subsequent evolution is also highlighted by the evidence of simpler extant eukaryotes.
The transitional period remains poorly reconstructed, and while there are now several clear biological principles running between FECA and LECA, the ordering of events such as acquisition of the mitochondrion and evolution of the endomembrane system and flagellum remain to be resolved ( and ); not least concerning is the understanding of the state of the FECA/LECA transitional form that acquired the mitochondrion, and how the nuclear membrane itself arose. Resolution of some of these steps from molecular data may become possible in the future, but both this issue and the more accurate analysis of extant eukaryotes require several technical advances.
First, there remains a sensitivity issue: many homologs fail to be detected, or detected with sufficient confidence in diverse genomes for robust conclusions to be drawn. Detailed analysis is time consuming, and even with improved algorithms, models of sequence evolution or phylogenetic approaches there is a clear need for new and more robust methods to eliminate both much of this burden as well as, if possible, erroneous calls. Coupled to this is the continual demand for more sequence data, and specifically data from taxa residing at critical positions within the eukaryotic phylogeny. Second, it is critical that such taxa, or at least a well-chosen subset, can be analyzed functionally; such approaches are frequently the only way in which sequence-based studies can be thoroughly validated, or even understood at the cellular level. The impact of neofunctionalisation, for example, amongst paralog families may be a major evolutionary driver, and more detailed insights into these processes can only be gleaned through experiment. Third is the issue of asymmetry, the biasing of analysis due to the significantly greater understanding that we have for Opisthokont taxa, and relatively poor details in most other supergroups. Taken together, tackling each of these issues will provide a greater appreciation of eukaryotic diversity and the role this plays in the context of ecology and disease mechanisms, potentially opening up the transitional period and finally break the “asymmetry” problem (Dacks & Field, 2007). An era of molecular paleontology may have arrived.
Declaration of interest
The authors acknowledge various funding agencies that have supported many aspects of our work in this area: the Wellcome Trust (program grant 082813 to MCF), the Medical Research Council (project grants to MCF), the Marie Curie fund (FP7-PEOPLE-2011-IEF to VLK), the Johannes Veldkamp Foundation (MvdG), the Biotechnology & Biological Sciences Research Council (BW and MLG), The Royal Society (MLG), the Leverhulme Trust (MLG) and the Natural Sciences and Engineering Research Council (of Canada) (JBD). BW is a recipient of University of Nottingham New Lecturer support. JBD is the Canada Research Chair in Evolutionary Cell Biology. None of the funding agencies were involved in the decision to publish or in the selection of content. The authors have no conflict of interest to declare.
Acknowledgements
MCF thanks Michael Rout for many insightful discussions concerning evolution of gene expression and trafficking systems and suggestions with figures.
References
- Alber F, Dokudovskaya S, Veenhoff LM, et al. (2007). The molecular architecture of the nuclear pore complex. Nature 450:695–701
- Alberts B, Johnson A, Lewis J, et al. (2002). Molecular biology of the cell. 4th ed. New York: Garland Science
- Allen JW, Ferguson SJ, Ginger ML. (2008). Distinctive biochemistry in the trypanosome mitochondrial intermembrane space suggests a model for stepwise evolution of the MIA pathway for import of cysteine-rich proteins. FEBS Lett 582:2817–25
- Allison DS, Schatz G. (1986). Artificial mitochondrial presequences. Proc Natl Acad Sci U S A 83:9011–15
- Alsford S, Kawahara T, Isamah C, Horn D. (2007). A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol 63:724–7
- Avliyakulov NK, Lukes J, Ray DS. (2004). Mitochondrial histone-like DNA-binding proteins are essential for normal cell growth and mitochondrial function in Crithidia fasciculata. Eukaryot Cell 3:518–26
- Azimzadeh J, Marshall WF. (2010). Building the centriole. Curr Biol 20:R816–25
- Balk J, Pilon M. (2011). Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 16:218–26
- Balk J, Pierik AJ, Netz DJ, et al. (2004). The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J 23:2105–15
- Bandyopadhyay S, Chandramouli K, Johnson MK. (2008). Iron-sulfur cluster biosynthesis. Biochem Soc Trans 36:1112–9
- Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. (2010). Organization and expression of organellar genomes. Philos Trans R Soc Lond B Biol Sci 365:785–99
- Basu S, Leonard JC, desai N, et al. (2013). Divergence of Erv1-associated mitochondrial import and export pathways in trypanosomes and anaerobic protists. Eukaryot Cell 12:343–55
- Bendezu FO, de Boer PA. (2008). Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190:1792–811
- Bernander R, Ettema TJ. (2010). FtsZ-less cell division in archaea and bacteria. Curr Opin Microbiol 13:747–52
- Berriman M, Ghedin E, Hertz-Fowler C, et al. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309:416–22
- Bolte K, Gruenheit N, Felsner G, et al. (2011). Making new out of old: recycling and modification of an ancient protein translocation system during eukaryotic evolution. Mechanistic comparison and phylogenetic analysis of ERAD, SELMA and the peroxisomal importomer. Bioessays 33:368–76
- Bork P, Sander C, Valencia A. (1992). An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A 89:7290–4
- Boyd JM, Drevland RM, Downs DM, Graham DE. (2009). Archaeal ApbC/Nbp35 homologs function as iron-sulfur cluster carrier proteins. J Bacteriol 191:1490–7
- Bramkamp M. (2012). Structure and function of bacterial dynamin-like proteins. Biol Chem 393:1203–14
- Brennand A, Gualdrón-López M, Coppens I, et al. (2011). Autophagy in parasitic protists: unique features and drug targets. Mol Biochem Parasitol 177:83–99
- Briggs LJ, Davidge JA, Wickstead B, et al. (2004). More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr Biol 14:R611–12
- Bröcker C, Engelbrecht-Vandré S, Ungermann C. (2010). Multisubunit tethering complexes and their role in membrane fusion. Curr Biol 20:R943–52
- Brugerolle G. (2004). Devescovinid features, a remarkable surface cytoskeleton, and epibiotic bacteria revisited in Mixotricha paradoxa, a parabasalid flagellate. Protoplasma 224:49–59
- Bullerwell CE, Gray MW. (2004). Evolution of the mitochondrial genome: protist connections to animals, fungi and plants. Curr Opin Microbiol 7:528–34
- Bulyha I, Hot E, Huntley S, Sogaard-Andersen L. (2011). GTPases in bacterial cell polarity and signalling. Curr Opin Microbiol 14:726–33
- Burmann F, Ebert N, van Baarle S, Bramkamp M. (2011). A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion. Mol Microbiol 79:1294–304
- Burns KE, Darwin KH. (2010). Pupylation versus ubiquitylation: tagging for proteasome-dependent degradation. Cell Microbiol 12:424–31
- Burri L, Williams BA, Bursac D, et al. (2006). Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc Natl Acad Sci U S A 103:15916–20
- Busiek KK, Margolin W. (2011). Split decision: a thaumarchaeon encoding both FtsZ and Cdv cell division proteins chooses Cdv for cytokinesis. Mol Microbiol 82:535–8
- Butterfield NJ. (2007). Macroevolution and macroecology through deep time. Palaeontology 50:41–55
- Butterfield NJ, Knoll AH, Swett K. (1988). Exceptional preservation of fossils in an Upper Proterozoic shale. Nature 334:424–7
- Butterfield NJ, Knoll AH, Swett K. (1990). A bangiophyte red alga from the Proterozoic of arctic Canada. Science 250:104–7
- Carpenter ML, Assaf ZJ, Gourguechon S, Cande WZ. (2012). Nuclear inheritance and genetic exchange without meiosis in the binucleate parasite Giardia intestinalis. J Cell Sci 125:2523–32
- Carter AP, Cho C, Jin L, Vale RD. (2011). Crystal structure of the dynein motor domain. Science 331:1159–65
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. (2011). Evolution: tracing the origins of centrioles, cilia, and flagella. J Cell Biol 194:165–75
- Castagnetti S, Oliferenko S, Nurse P. (2010). Fission yeast cells undergo nuclear division in the absence of spindle microtubules. PLoS Biol 8:e1000512
- Cavalier-Smith T. (1978). The evolutionary origin and phylogeny of microtubules, mitotic spindles and eukaryote flagella. Biosystems 10:93–114
- Cavalier-Smith T. (1987). Eukaryotes with no mitochondria. Nature 326:332–3
- Cavalier-Smith T. (2003). Protist phylogeny and the high-level classification of Protozoa. Eur J Protistology 39:338–48
- Cavalier-Smith T. (2006). Cell evolution and Earth history: stasis and revolution. Philos Trans R Soc Lond B Biol Sci 361:969–1006
- Chen CT, Hehnly H, Doxsey SJ. (2012). Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat Rev Mol Cell Biol 13:483–8
- Chernikova D, Motamedi S, Csürös M, et al. (2011). A late origin of the extant eukaryotic diversity: divergence time estimates using rare genomic changes. Biol Direct 19:26
- Chong YT, Gidda SK, Sanford C, et al. (2010). Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic., expression profiling, and subcellular localization studies. New Phytol 185:401–19
- Clements A, Bursac D, Gatsos X, et al. (2009). The reducible complexity of a mitochondrial molecular machine. Proc Nat Acad Sci 106:15791–5
- Cronshaw JM, Krutchinsky AN, Zhang W, et al. (2002). Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158:915–27
- Dacks J, Roger AJ. (1999). The first sexual lineage and the relevance of facultative sex. J Mol Evol 48:779–83
- Dacks JB, Doolittle WF. (2001). Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell 107:419–25
- Dacks JB, Doolittle WF. (2002). Novel syntaxin gene sequences from Giardia. Trypanosoma and algae: implications for the ancient evolution of the eukaryotic endomembrane system. J Cell Sci 115:1635–42
- Dacks JB, Doolittle WF. (2004). Molecular and phylogenetic characterization of syntaxin genes from parasitic protozoa. Mol Biochem Parasitol 136:123–36
- Dacks JB, Field MC. (2007). Evolution of the eukaryotic membrane-trafficking system: origin., tempo and mode. J Cell Sci 120:2977–85
- Dacks JB, Davis LA, Sjögren AM, et al. (2003). Evidence for Golgi bodies in proposed ‘Golgi-lacking' lineages. Proc Biol Sci 7:270
- Dacks JB, Poon PP, Field MC. (2008). Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci U S A 105:588–93
- Dagley MJ, Dolezal P, Likic VA, et al. (2009). The protein import channel in the outer mitosomal membrane of Giardia intestinalis. Mol Biol Evol 26:1941–7
- Daniels JP, Gull K, Wickstead B. (2010). Cell biology of the trypanosome genome. Microbiol Mol Biol Rev 74:552–69
- de Boer P, Crossley R, Rothfield L. (1992). The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254–6
- Deatherage BL, Lara JC, Bergsbaken T, et al. (2009). Biogenesis of bacterial membrane vesicles. Mol Microbiol 72:1395–407
- deGrasse JA, DuBois KN, Devos D, et al. (2009). Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics 8:2119–30
- Derman AI, Becker EC, Truong BD, et al. (2009). Phylogenetic analysis identifies many uncharacterized actin-like proteins Alps in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol 73:534–52
- Desmond E, Gribaldo S. (2009). Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol Evol 1:364–81
- Devos DP. (2012). Regarding the presence of membrane coat proteins in bacteria: confusion? What confusion? Bioessays 34:38–9
- Devos D, Dokudovskaya S, Alber F, et al. (2004). Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2:e380
- Diekmann Y, Seixas E, Gouw M, et al. (2011). Thousands of rab GTPases for the cell biologist. PLoS Comput Biol 7:e1002217
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. (2007). LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19:2793–803
- Dobro MJ, Samson RY, Yu Z, et al. (2013). Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggest spiraling filaments are involved in membrane scission. Mol Biol Cell in press
- Dokudovskaya S, Waharte F, Schlessinger A, et al. (2011). A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 10:M110.006478
- Dolezal P, Dagley MJ, Kono M, et al. (2010). The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog 6:e1000812
- Dong JH, Wen JF, Tian HF. (2007). Homologs of eukaryotic Ras superfamily proteins in prokaryotes and their novel phylogenetic correlation with their eukaryotic analogs. Gene 396:116–24
- Doolittle WF. (1998). You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet 14:307–11
- DuBois KN, Alsford S, Holden JM, et al. (2012). NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol 10:e1001287
- Duszenko M, Ginger ML, Brennand A, et al. (2011). Autophagy in protists. Autophagy 7:127–58
- Eckers E, Petrungaro C, Gross D, et al. (2013). Divergent molecular evolution of the mitochondrial sulfhydryl:cytochrome C oxidoreductase Erv in opisthokonts and parasitic protists. J Biol Chem 288:2676–88
- Elias M, Brighouse A, Gabernet-Castello C, et al. (2012). Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J Cell Sci 125:2500–8
- Ellen AF, Zolghadr B, Driessen AM, Albers SV. (2010). Shaping the archaeal cell envelope. Archaea 2010:608243
- Ellis TN, Kuehn MJ. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94
- Embley TM, Martin W. (2006). Eukaryotic evolution, changes and challenges. Nature 440:623–30
- Entchev EV, Kurzchalia TV. (2005). Requirements of sterols in the life cycle of the nematode Caenorhabditis elegans. Semin. Cell Dev Biol 16:175–82
- Erber A, Riemer D, Bovenschulte M, Weber K. (1998). Molecular phylogeny of metazoan intermediate filament proteins. J Mol Evol 47:751–62
- Erickson HP, Osawa M. (2010). Cell division without FtsZ – a variety of redundant mechanisms. Mol Microbiol 78:267–70
- Esser C, Ahmadinejad N, Wiegand C, et al. (2004). A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21:1643–60
- Ettema TJG, Lindås A, Bernander R. (2011). An actin-based cytoskeleton in archaea. Mol Microbiol 80:1052–61
- Field MC, Dacks JB. (2009). First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs., vesicle coat proteins., and nuclear pore complexes. Curr Opin Cell Biol 21:4–13
- Field MC, Gabernet-Castello C, Dacks JB. (2007). Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology. Adv Exp Med Biol 607:84–96
- Field MC, Sali A, Rout MP. (2011). Evolution: On a bender-BARs, ESCRTs, COPs, and finally getting your coat. J Cell Biol 193:963–72
- Field MC, Sergeenko T, Wang YN, et al. (2010). Chaperone requirements for biosynthesis of the trypanosome variant surface glycoprotein. PLoS One 5:e8468
- Figueiredo LM, Cross GA, Janzen CJ. (2009). Epigenetic regulation in African trypanosomes: a new kid on the block. Nat Rev Microbiol 7:504–13
- Flaherty KM, McKay DB, Kabsch W, Holmes KC. (1991). Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci USA 88:5041–5
- Forterre P. (2011). A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Res Microbiol 162:77–91
- Foth BJ, Goedecke MC, Soldati D. (2006). New insights into myosin evolution and classification. Proc Natl Acad Sci U S A 103:3681–6
- Freitag J, Ast J, Bölker M. (2012). Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485:522–5
- Fuchs E, Weber K. (1994). Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem 63:345–82
- Fuerst JA. (2005). Intracellular compartmentation in planctomycetes. Annu Rev Microbiol 59:299–328
- Fuerst JA, Sagulenko E. (2011). Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–13
- Fuerst JA, Sagulenko E. (2012). Keys to eukaryality: planctomycetes and ancestral evolution of cellular complexity. Front Microbiol 3:167
- Gabaldón T. (2010). Peroxisome diversity and evolution. Philos Trans R Soc Lond B Biol Sci 365:765–73
- Gabaldón T, Capella-Gutiérrez S. (2010). Lack of phylogenetic support for a supposed actinobacterial origin of peroxisomes. Gene 461:61–5
- Gabaldón T, Snel B, van Zimmeren F, et al. (2006). Origin and evolution of the peroxisome proteome. Biol Direct 1:8
- Gabernet-Castello C, O'Reilly AJ, Dacks JB, Field MC. (2013). Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol Biol Cell 24:1574–83
- Garbarino JE, Gibbons IR. (2002). Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics 3:18
- Gibbons IR. (1995). Dynein family of motor proteins: present status and future questions. Cell Motil Cytoskeleton 32:136–44
- Ginger ML, Fritz-Laylin LK, Fulton C, et al. (2010). Intermediary metabolism in protists: a sequence-based view of facultative anaerobic metabolism in evolutionarily diverse eukaryotes. Protist 161:642–71
- Goulding CW, Parseghian A, Sawaya MR, et al. (2002). Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at 1.5-A resolution. Protein Sci 11:2887–93
- Gray MW, Lang BF, Burger GE. (2004). Mitochondria of protists. Annu Rev Genet 38:477–524
- Gribaldo S, Poole AM, Daubin V, et al. (2010). The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat Rev Microbiol 8:743–52
- Grossman E, Medalia O, Zwerger M. (2012). Functional architecture of the nuclear pore complex. Annu Rev Biophys 41:557–84
- Gualdrón-López M, Brennand A, Hannaert V, et al. (2012). When, how and why glycolysis became compartmentalised in the Kinetoplastea. A new look at an ancient organelle. Int J Parasitol 42:1–20
- Guedes RL, Prosdocimi F, Fernandes GR, et al. (2011). Amino acids biosynthesis and nitrogen assimilation pathways: a great genomic deletion during eukaryotes evolution. BMC Genomics 12:S2
- Hall DO, Cammack R, Rao KK. (1971). Role for ferredoxins in the origin of life and biological evolution. Nature 233:136–8
- Hanson PI, Roth R, Lin Y, Heuser JE. (2008). Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180:389–402
- Hardie DG. (2011). AMP-activated protein kinase – an energy sensor that regulates all aspects of cell function. Genes Dev 25:1895–908
- Hardie DG, Ross FA, Hawley SA. (2012). AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Rev Mol Cell Biol 13:251–62
- Hardie DG, Scott JW, Pan DA, Hudson ER. (2003). Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett 546:113–20
- Hartman H, Smith TF. (2009). The evolution of the cilium and the eukaryotic cell. Cell Motil Cytoskeleton 66:215–19
- Hartzell PL. (1997). Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc Natl Acad Sci USA 94:9881–6
- Haurat MF, Aduse-Opoku J, Rangarajan M, et al. (2011). Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem 286:1269–76
- Herman M, Pérez-Morga D, Schtickzelle N, Michels PAM. (2008). Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy 4:294–308
- Hewitt V, Alcock F, Lithgow T. (2011). Minor modifications and major adaptations: The evolution of molecular machines driving mitochondrial protein import. Biochim Biophys Acta (BBA) – Biomembranes 1808:947–54
- Hirst J, Barlow LD, Francisco GC, et al. (2011). The fifth adaptor protein complex. PLoS Biol 9:e1001170
- Hodges ME, Scheumann N, Wickstead B, et al. (2010). Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci 123:1407–13
- Hoepfner D, Schildknegt D, Braakman I, et al. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122:85–95
- Horner DS, Heil B, Happe T, Embley TM. (2002). Iron hydrogenases–ancient enzymes in modern eukaryotes. Trends Biochem Sci 27:148–53
- Hug LA, Stechmann A, Roger AJ. (2010). Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol Biol Evol 27:311–24
- Humbard MA, Miranda HV, Lim JM, et al. (2010). Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463:54–60
- Hwang JS, Takaku Y, Chapman J, et al. (2008). Cilium evolution: identification of a novel protein, nematocilin, in the mechanosensory cilium of Hydra nematocytes. Mol Biol Evol 25:2009–17
- Iyer LM, Leipe DD, Koonin EV, Aravind L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 146:11–31
- Jenkins C, Samudrala R, Anderson I, et al. (2002). Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci U S A 99:17049–54
- Jockusch BM, Graumann PL. (2012). The long journey: actin on the road to pro- and eukaryotic cells. Rev Physiol Biochem Pharmacol 161:67–85
- Jungnickel B, Rapoport TA, Hartmann E. (1994). Protein translocation: common themes from bacteria to man. FEBS Lett 346:73–7
- Kabsch W, Holmes KC. (1995). The actin fold. FASEB J 9:167–74
- Kasinsky HE, Lewis JD, Dacks JB, Ausio J. (2001). Origin of H1 linker histones. FASEB J 15: 34–42
- Katada S, Imhof A, Sassone-Corsi P. (2012). Connecting Threads: epigenetics and metabolism. Cell 148:24–8
- Kim YG, Sohn EJ, Seo J, et al. (2005). Crystal structure of bet3 reveals a novel mechanism for Golgi localization of tethering factor TRAPP. Nat Struct Mol Biol 12:38–45
- Klute MJ, Melançon P, Dacks JB. (2011). Evolution and diversity of the Golgi. Cold Spring Harb Perspect Biol 3:a007849
- Knoll AH, Javaux EJ, Hewitt D, Cohen P. (2006). Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond B Biol Sci 361:1023–38
- Koonin EV, Aravind L. (2000). Dynein light chains of the Roadblock/LC7 group belong to an ancient protein superfamily implicated in NTPase regulation. Curr Biol 10:R774–6
- Koumandou VL, Dacks JB, Coulson RM, Field MC. (2007). Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol 7:29
- Koumandou VL, Field MC. (2011). The emergence of cellular complexity at the dawn of the eukaryotes: reconstructing the endomembrane system with in silico and functional analyses. In: Pontarotti P, ed. Evolutionary biology concepts, biodiversity, macroevolution and genome evolution. Berlin and Heidelberg: Springer Verlag, Chapter 10, 153–67
- Kramer S, Kimblin NC, Carrington M. (2010). Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei. Trypanosoma cruzi and Leishmania major. BMC Genomics 11:283
- Krüger A, Batsios P, Baumann O, et al. (2012). Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol Biol Cell 23:360–70
- Kull FJ, Vale RD, Fletterick RJ. (1998). The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil 19:877–86
- Kull FJ, Sablin EP, Lau R, et al. (1996). Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380:550–5
- Kurzchalia TV, Ward S. (2003). Why do worms need cholesterol? Nature Cell Biol 5:684–8
- Lane N. (2011). Energetics and genetics across the prokaryote-eukaryote divide. Biology Direct 6:35
- Lane N, Allen JF, Martin W. (2010). How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays 32:271–80
- Lane N, Martin W. (2010). The energetics of genome complexity. Nature 467:929–34
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK. (2001). Dyneins have run their course in plant lineage. Traffic 2:362–3
- Leaver M, Dominguez-Cuevas P, Coxhead JM, et al. (2009). Life without a wall or division machine in Bacillus subtilis. Nature 457:849–53
- Lee EY, Choi DY, Kim DK, et al. (2009). Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–36
- Lees-Miller JP, Henry G, Helfman DM. (1992). Identification of act2, an essential gene in the fission yeast Schizosaccharomyces pombe that encodes a protein related to actin. Proc Natl Acad Sci U S A 89:80–3
- Leipe DD, Wolf YI, Koonin EV, Aravind L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317:41–72
- Lemire BD, Fankhauser C, Baker A, Schatz G. (1989). The mitochondrial targeting function of randomly generated peptide sequences correlates with predicted helical amphiphilicity. J Biol Chem 264:20206–15
- Leonardy S, Miertzschke M, Bulyha I, et al. (2010). Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J 29:2276–89
- Leung KF, Dacks JB, Field MC. (2008). Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–716
- Li Y, Kelly WG, Logsdon JM, et al. (2004). Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J 18:1834–50
- Liapounova NA, Hampl V, Gordon PMK, et al. (2006). Reconstructing the mosaic glycolytic pathway of the anaerobic eukaryote Monocercomonoides. Eukaryot Cell 5:2138–46
- Lill R. (2009). Function and biogenesis of iron-sulphur proteins. Nature 460:831–8
- Lill R, Kispal G. (2000). Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem. Sci 25:352–6
- Lill R, Hoffmann B, Molik S, et al. (2012). The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta 1823:1491–508
- Lindas AC, Karlsson EA, Lindgren MT, et al. (2008). A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A 105:18942–6
- Lonhienne TG, Sagulenko E, Webb RI, et al. (2010). Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A 107:12883–8
- López-Garćia P, Moreira D. (1999). Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci 24:88–93
- Lopez D, Kolter R. (2010). Functional microdomains in bacterial membranes. Genes Dev 24:1893–902
- Love GD, Grosjean E, Stalvies C, et al. (2009). Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 457:718–21
- Low HH, Lowe J. (2006). A bacterial dynamin-like protein. Nature 444:766–9
- Low HH, Sachse C, Amos LA, Lowe J. (2009). Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139:1342–52
- Luo D, Bernard DG, Balk J, et al. (2012). The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 24:4135–48
- Lupas AN, Martin J. (2002). AAA proteins. Curr Opin Struct Biol 12:746-53
- Makarova KS, Yutin N, Bell SD, Koonin EV. (2010). Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol 8:731–41
- Malik SB, Pightling AW, Stefaniak LM, et al. (2007a). An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One 3:e2879
- Malik SB, Ramesh MA, Hulstrand AM, Logsdon JM Jr. (2007b). Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol Biol Evol 24:2827–41
- Manjithaya R, Nazarko TY, Farré JC, Subramani S. (2010). Molecular mechanism and physiological role of pexophagy. FEBS Lett 584:1367–73
- Manna PT, Kelly S, Field MC. (2013). Adaptin evolution in kinetoplastids and emergence of the variant surface glycoprotein coat in African trypanosomatids. Mol Phylogenet Evol 67:123–8
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. (2004). Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3:1612–37
- Mari M, Tooze SA, Reggiori F. (2011). The puzzling origin of the autophagosomal membrane. F1000 Biol Rep 3:25
- Martin W. (2007). Eukaryote and mitochondrial origins: two sides of the same coin and too much ado about oxygen. In: Falkowski P, Knoll AH, eds. Primary Producers of the Sea. New York: Academic Press
- Martin W, Koonin EV. (2006). Introns and the origin of nucleus-cytosol compartmentalization. Nature 440:41–5
- MartinW, Müller M. (1998). The hydrogen hypothesis for the first eukaryote. Nature 392:37–41
- Mashburn-Warren L, Howe J, Garidel P, et al. (2008). Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 69:491–502
- Mashburn LM, Whiteley M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–5
- Mason DA, Stage DE, Goldfarb DS. (2009). Evolution of the metazoan-specific importin alpha gene family. J Mol Evol 68:351–65
- Mauriello EM, Mouhamar F, Nan B, et al. (2010). Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J 29:315–26
- Maynard Smith J, Szathmáry E. (1995). The major transitions in evolution. Oxford, England: Oxford University Press
- McInerney JO, Martin WF, Koonin EV, et al. (2011). Planctomycetes and eukaryotes: a case of analogy not homology. Bioessays 33:810–7
- McIntosh JR. (1973). The axostyle of Saccinobaculus. II. Motion of the microtubule bundle and a structural comparison of straight and bent axostyles. J Cell Biol 56:324–39
- McIntosh JR, Ogata ES, Landis SC. (1973). The axostyle of Saccinobaculus. I. Structure of the organism and its microtubule bundle. J Cell Biol 56:304–23
- Meier I. (2007). Composition of the plant nuclear envelope: theme and variations. J Exp Bot 58:27–34
- Mitchell DR. (2004). Speculations on the evolution of 9+2 organelles and the role of central pair microtubules. Biol Cell 96:691–6
- Mitchell DR. (2007). The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv Exp Med Biol 607:130–40
- Mitchell M, Mitchell H. (1952). A case of maternal inheritance in Neurospora crassa. Proc Natl Acad Sci USA 38:442
- Moon DC, Choi CH, Lee JH, et al. (2012). Acinetobacter baumannii outer membrane protein a modulates the biogenesis of outer membrane vesicles. J Microbiol 50:155–60
- Moore AN, Russell AG. (2012). Clustered organization, polycistronic transcription, and evolution of modification-guide snoRNA genes in Euglena gracilis. Mol Genet Genomics 287:55–66
- Morris RL, Hoffman MP, Obar RA, et al. (2006). Analysis of cytoskeletal and motility proteins in the sea urchin genome assembly. Dev Biol 300:219–37
- Morrison HG, McArthur AG, Gillin FD, et al. (2007). Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–6
- Morton JJ, Blumenthal T. (2011). RNA processing in C. elegans. Methods Cell Biol 106:187–217
- Mowbrey K, Dacks JB. (2009). Evolution and diversity of the Golgi body. FEBS Lett 583:3738–45
- Mukherjee A, Dai K, Lutkenhaus J. (1993). Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci U S A 90:1053–7
- Müller M, Mentel M, Van Hellemond JJ, et al. (2012). Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76:444–95
- Nass MM, Nass S. (1963). Intramitochondrial fibers with DNA characteristics. I. Fixation and electron staining reactions. J Cell Biol 19:593–611
- Nes CR, Singha UK, Liu J, et al. (2012). Novel sterol metabolic network of Trypanosoma brucei procyclic and bloodstream forms. Biochem. J 443:267–77
- Neupert W, Herrmann JM. (2007). Translocation of proteins into mitochondria. Annu Rev Biochem 76:723–49
- Nevin WD, Dacks JB. (2009). Repeated secondary loss of adaptin complex genes in the Apicomplexa. Parasitol Int 58:86–94
- Nunoura T, Takaki Y, Kakuta J, et al. (2011). Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39:3204–23
- O'Malley MA. (2010). The first eukaryote cell: an unfinished history of contestation. Stud Hist Philos Biol Biomed Sci 41:212–24
- O'Reilly AJ, Dacks JB, Field MC. (2011). Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS One 6:e19308
- Odronitz F, Kollmar M. (2007). Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol 8:R196
- Oeffinger M, Zenklusen D. (2012). To the pore and through the pore: a story of mRNA export kinetics. Biochim Biophys Acta 1819:494–506
- Otto GP, Nichols BJ. (2011). The roles of flotillin microdomains–endocytosis and beyond. J Cell Sci 124:3933–40
- Parks LW, Smith SJ, Crowley JH. (1995). Biochemical and physiological effects of sterol alterations in yeast – a review. Lipids 30:227–30
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. (2011). Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A 108:13624–9
- Pastore C, Adinolfi S, Huynen MA, et al. (2006). YfhJ, a molecular adaptor in iron-sulfur cluster formation or a Frataxin-like protein? Structure 14:857–67
- Payne SH, Loomis WF. (2006). Retention and loss of amino acid biosynthetic pathways based on the analysis of whole-genome sequences. Eukaryot Cell 5:272–6
- Peacock L, Ferris V, Sharma R, et al. (2011). Identification of the meiotic life cycle stage of Trypanosoma brucei in the tsetse fly. Proc Natl Acad Sci U S A 108:3671–6
- Pearce MJ, Mintseris J, Ferreyra J, et al. (2008). Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322:1104–7
- Pelve EA, Lindas AC, Martens-Habbena W, et al. (2011). Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol 82:555–66
- Pereira-Leal JB. (2008). The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 9:27–38
- Peterson KJ, Butterfield NJ. (2005). Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc Natl Acad Sci U S A 102:9547–52
- Pickett-Heaps J. (1974). The evolution of mitosis and the eukaryotic condition. Biosystems 6:37–48
- Pilhofer M, Bauer AP, Schrallhammer M, et al. (2007). Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel Two-Step Gene Walking method. Nucleic Acids Res 35:e135
- Pilhofer M, Ladinsky MS, McDowall AW, et al. (2011). Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol 9:e1001213
- Pisani D, Cotton JA, Mcinerney JO. (2007). Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol 24:1752–60
- Podar M, Wall MA, Makarova KS, Koonin EV. (2008). The prokaryotic V4R domain is the likely ancestor of a key component of the eukaryotic vesicle transport system. Biol Direct 3:2
- Poole AM, Phillips MJ, Penny D. (2003). Prokaryote and eukaryote evolvability. Biosystems 69:163–85
- Ramesh MA, Malik SB, Logsdon JM. (2005). A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol 15:185–91
- RayChaudhuri D, Park JT. (1992). Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251–4
- Renner LD, Weibel DB. (2011). Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci U S A 108:6264–9
- Richards TA, Van Der Giezen M. (2006). Evolution of the Isd11/IscS complex reveals a single α-proteobacterial endosymbiosis for all eukaryotes. Mol Biol Evol 23:1341–4
- Richards TA, Cavalier-Smith T. (2005). Myosin domain evolution and the primary divergence of eukaryotes. Nature 436:1113–18
- Rigden DJ, Michels PAM., Ginger ML. (2009). Autophagy in protists. Examples of secondary loss, lineage-specific innovations, and the conundrum of remodelling a single mitochondrion. Autophagy 5:784–94
- Rivera MC, Jain R, Moore JE, Lake JA. (1998). Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA 95:6239–44
- Robinson MS. (2004). Adaptable adaptors for coated vesicles. Trends Cell Biol 4:167–74
- Roger AJ. (1999). Reconstructing early events in eukaryotic evolution. Am Nat 154:S146–S163
- Romantsov T, Helbig S, Culham DE, et al. (2007). Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol 64:1455–65
- Rosenbaum JL, Witman GB. (2002). Intraflagellar transport. Nat Rev Mol Cell Biol 3:813–25
- Rout MP, Aitchison JD, Suprapto A, et al. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148:635–51
- Sagan L. (1967). On the origin of mitosing cells. J Theoret Biol 14:225–74
- Samsó M, Radermacher M, Frank J, Koonce MP. (1998). Structural characterization of a dynein motor domain. J Mol Biol 276:927–37
- Samson RY, Obita T, Freund SM, et al. (2008). A role for the ESCRT system in cell division in archaea. Science 322:1710–13
- Samson RY, Obita T, Hodgson B, et al. (2011). Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell 41:186–96
- Sandman K, Reeve JN. (2005). Archaeal chromatin proteins: different structures but common function? Curr Opin Microbiol 8:656–61
- Sandvig K, Pust S, Skotland T, van Deurs B. (2011). Clathrin-independent endocytosis: mechanisms and function. Curr Opin Cell Biol 23:413–20
- Santarella-Mellwig R, Franke J, Jaedicke A, et al. (2010). The compartmentalized bacteria of the planctomycetes-verrucomicrobia-chlamydiae superphylum have membrane coat-like proteins. PLoS Biol 19:e1000281
- Satir P, Guerra C, Bell AJ. (2007). Evolution and persistence of the cilium. Cell Motil Cytoskeleton 64:906–13
- Sato T, Atomi H. (2011). Novel metabolic pathways in Archaea. Curr Opin Microbiol 14:307–14
- Saudek V. (2012). Cystinosin, MPDU1, SWEETs and KDELR belong to a well-defined protein family with putative function of cargo receptors involved in vesicle trafficking. PLoS One 7:e30876
- Sawa M, Suetsugu S, Sugimoto A, et al. (2003). Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J Cell Sci 116:1505–18
- Schafer DA, Schroer TA. (1999). Actin-related proteins. Annu Rev Cell Dev Biol 15:341–63
- Schertzer JW, Whiteley M. (2012). A bilayer-couple model of bacterial outer membrane vesicle biogenesis. Mbio 3:e00297-11
- Schlacht A, Mowbrey K, Elias M, et al. (2013). Ancient complexity, opisthokont plasticity, and discovery of the 11th subfamily of Arf-GAP proteins. 14:636–49
- Schurko AM, Logsdon JM Jr. (2008). Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays 30:579–89
- Schwob E, Martin RP. (1992). New yeast actin-like gene required late in the cell cycle. Nature 355:179–82
- Sehring IM, Mansfeld J, Reiner C, et al. (2007). The actin multigene family of Paramecium tetraurelia. BMC Genomics 8:82
- Selkrig J, Mosbahi K, Webb CT, et al. (2012). Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol 19:506–10
- Serpeloni M, Vidal NM, Goldenberg S, et al. (2011). Comparative genomics of proteins involved in RNA nucleocytoplasmic export. BMC Evol Biol 11:7
- Shapiro L, McAdams HH, Losick R. (2009). Why and how bacteria localize proteins. Science 326:1225–8
- Shimomura Y, Takahashi Y, Kakuta Y, Fukuyama K. (2005). Crystal structure of Escherichia coli YfhJ protein, a member of the ISC machinery involved in assembly of iron-sulfur clusters. Proteins 60:566–9
- Siebers B, Schönheit P. (2005). Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol 8:695–705
- Simon DN, Wilson KL. (2011). The nucleoskeleton as a genome-associated dynamic network of networks. Nat Rev Mol Cell Biol 12:695–708
- Snider J, Houry WA. (2006). MoxR AAA+ ATPases: a novel family of molecular chaperones? J Struct Biol 156:200–9
- Song D, Lee FS. (2011). Mouse knock-out of IOP1 protein reveals its essential role in mammalian cytosolic iron-sulfur protein biogenesis. J Biol Chem 286:15797–805
- Spang A. (2012). The DSL1 Complex: The smallest but not the least CATCHR. Traffic 13:908–13
- Staehelin LA, Kang BH. (2008). Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol 147:1454–68
- Starr DA, Fridolfsson HN. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26:421–44
- Stechmann A, Baumgartner M, Silberman JD, Roger AJ. (2006). The glycolytic pathway of Trimastix pyriformis is an evolutionary mosaic. BMC Evol Biol 6:101
- Stehling O, Vashisht AA, Mascarenhas J, et al. (2012). MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337:195–9
- Suda Y, Nakano A. (2011). The yeast Golgi apparatus. Traffic 13:505–10
- Summons RE, Bradley AS, Jahnke LL, Waldbauer JR. (2006). Steroids, triterpenoids, and molecular oxygen. Philos Trans R Soc Lond B Biol Sci 361:951–68
- Sun M, Wartel M, Cascales E, et al. (2011). Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci USA 108:7559–64
- Tabak HF, van der Zand A, Braakman I. (2008). Peroxisomes: minted by the ER. Curr Opin Cell Bio. 20:393–400
- Tamura K, Fukao Y, Iwamoto M, et al. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22:4084–97
- Tetenbaum-Novatt J, Rout MP. (2010). The mechanism of nucleocytoplasmic transport through the nuclear pore complex. Cold Spring Harb Symp Quant Biol 75:567–84
- Thelander M, Olsson T, Ronne H. (2004). Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J 23:1900–10
- Thiergart T, Landan G, Schenk M, et al. (2012). An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol 4:466–85
- Tsaousis AD, Choudens SOD, Gentekaki E, et al. (2012). Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc Natl Acad Sci U S A 109:10426–31
- Tsaousis AD, Kunji ER, Goldberg AV, et al. (2008). A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453:533–6
- Van Der Giezen M. (2009). Hydrogenosomes and mitosomes: conservation and evolution of functions. J Euk Microbiol 56:221–31
- van Dam TPJ, Townsend MJ, Turk M, et al. (2013). Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci U S A 110:6943–8
- Vaughan S, Attwood T, Navarro M, et al. (2000). New tubulins in protozoal parasites. Curr Biol 10:R258–9
- Vaughan S, Wickstead B, Gull K, Addinall SG. (2004). Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol 58:19–29
- Vedovato M, Rossi V, Dacks JB, Filippini F. (2009). Comparative analysis of plant genomes allows the definition of the “Phytolongins”: a novel non-SNARE longin domain protein family. BMC Genomics 10:510
- Vothknecht UC, Otters S, Hennig R, Schneider D. (2012). Vipp1: a very important protein in plastids? J Exp Bot 63:1699–712
- Wanschers B, van de Vorstenbosch R, Wijers M, et al. (2008). Rab6 family proteins interact with the dynein light chain protein DYNLRB1. Cell Motil Cytoskeleton 65:183–96
- Weber K, Plessmann U, Ulrich W. (1989). Cytoplasmic intermediate filament proteins of invertebrates are closer to nuclear lamins than are vertebrate intermediate filament proteins; sequence characterization of two muscle proteins of a nematode. EMBO J 8:3221–7
- Whatley JM, John P, Whatley FR. (1979). From extracellular to intracellular: the establishment of mitochondria and chloroplasts. Proc R Soc Lond B 204:165–87
- Wickstead B, Gull K. (2007). Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8:1708–21
- Wickstead B, Gull K. (2011a). The evolution of the cytoskeleton. J Cell Biol 194:513–25
- Wickstead B, Gull K. (2011b). The evolutionary biology of dyneins. In: King S, ed. Dyneins: structure, biology and disease. Waltham (MA): Academic Press, 88–121
- Wickstead B, Gull K, Richards TA. (2010). Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol 10:110
- Wilkes DE, Watson HE, Mitchell DR, Asai DJ. (2008). Twenty-five dyneins in Tetrahymena: A re-examination of the multidynein hypothesis. Cell Motil Cytoskeleton 65:342–51
- Witman GB. (2003). Cell motility: deaf Drosophila keep the beat. Curr Biol 13:R796–8
- Woese CR, Fox GE. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA 74:5088–90
- Xu XM, Møller SG. (2011). Iron Sulfur Clusters: Biogenesis, Molecular Mechanisms, and Their Functional Significance. Antioxidants Redox Signal 15:271–307
- Xu XM, Meulia T, Meier I. (2007). Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol 17:1157–63
- Yang Z, Klionsky DJ. (2010). Eaten alive: a history of macroautophagy. Nature Cell Biol 12:814–22
- Yoshizawa AC, Kawashima S, Okuda S, et al. (2006). Extracting sequence motifs and the phylogenetic features of SNARE-dependent membrane traffic. Traffic 7:1104–18
- Yutin N, Wolf MY, Wolf YI, Koonin EV. (2009). The origins of phagocytosis and eukaryogenesis. Biol Direct 4:9
- Zhang Y, Franco M, Ducret A, Mignot T. (2010). A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol 8:e1000430