The highly conserved NOTCH signaling pathway has pleiotropic functions during multicellular animal development [Citation1]. NOTCH is a transmembrane receptor that mediates local cell–cell communication between adjacent cells. It is therefore not surprising that this pathway plays a major role in chronic lymphocytic leukemia (CLL): CLL cells are strongly dependent on microenvironmental support for their survival, and aberrant interactions with most of their bystander cells have been reported [Citation2].
In CLL, malignant B cells display activated NOTCH signaling, resulting in increased cell survival and leukemic transformation [Citation3]. After activation, the intracellular domain of the NOTCH protein translocates to the nucleus and acts as a transcription factor [Citation1]. So far, this NOTCH hyperactivity can be partially explained by simultaneous expression of the NOTCH1 and NOTCH2 receptors as well as of their ligands JAGGED-1 and JAGGED-2 [Citation3]. In addition, recently gain- of-function mutations in the NOTCH1 gene have been found in about 10% of patients with CLL (see references in [Citation4], this issue). Clinically, NOTCH1 mutations identify a patient subgroup with shorter progression-free and overall survival [Citation5].
Conspicuously, most of these patients carry mutations in the very C-terminal proline, glutamic acid, serine and threonine (PEST) degradation domain of the intracellular NOTCH1 protein (NICD), the activated form of NOTCH1 (, [Citation5]). In the vast majority (80%) of cases with mutation in the PEST domain, the same two base pairs are deleted (7544–7545delCT). The two base pair deletion causes a shift in the open reading frame of the NOTCH1 protein, thereby changing the subsequent three amino acids of the Cdc phosphodegron (CPD) consensus motif targeted by FBXW7, and introducing a premature STOP codon that deletes the following WSSSSP sequence (, [Citation1,Citation4,Citation5]).
Figure 1. The NOTCH1 intracellular domain cleavage product harbors different domains: the juxtamembrane portion (JM, orange), the Rbp-transcription factor associated domain (RAM, red), nuclear localization signals (NLS, white), ankyrin repeats (ANK, turquoise), the transactivation domain (TAD, blue) and the proline, glutamic acid, serine and threonine degradation domain (PEST, yellow). The PEST domain is enlarged below, and amino acid sequences of the Wt and mutated sequences are shown. The amino acids that correspond to the FBXW7 target sequence and to the WSSSSP motif are highlighted in orange and red. In the wild type sequence (Wt), both motifs are present. The two base pair deletion (delCT) results in a change of the amino acid sequence (PES> RVP, bold) and a premature STOP codon (asterisk). The novel single base pair substitution (C > T) reported by Sportoletti et al. in this issue leaves the FBXW7 motif unchanged, but its premature STOP codon (asterisk) deletes the WSSSSP sequence.
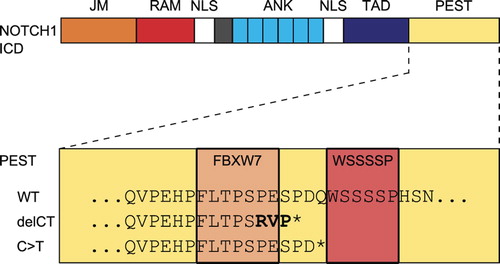
The CPD motif is recognized by the ubiquitin E3 ligase FBXW7 that ubiquitinates NICD for rapid degradation [Citation1,Citation4]. Similarly, phosphorylation of the serine residues in the WSSSSP sequence leads to NICD degradation and thereby shutdown of NOTCH signaling [Citation4]. As both the CPD and the WSSSSP motif are affected by the 7544–7545delCT, degradation of the NICD protein is markedly impaired and active NOTCH1 accumulates in the nucleus [Citation4].
In this issue, Sportoletti et al. report the discovery of a novel mutation in the NICD PEST domain in a single patient with CLL [Citation4]. Intriguingly, the single base pair substitution (7558C> T) they report in the PEST domain leads to a premature STOP codon and loss of the WSSSSP motif, but in contrast to the most common 7544–7545delCT, the CPD motif remains unchanged [Citation4]. So what is the functional difference between the two motifs?
The FBXW7 motif is targeted by the E3 ligase FBXW7 for ubiquitination, but the kinase(s) that phosphorylate the serine amino acids in the WSSSSP motif have not been identified yet [Citation6]. Interestingly, the homologous NOTCH4 lacks the WSSSSP sequence and degradation of its activated form is exclusively regulated by FBXW7, suggesting that FBXW7-dependent ubiquitination is sufficient for NICD degradation [Citation6]. The WSSSSP site is thus likely targeted by a distinct pathway and may have specific additional functions. It will therefore be of interest to dissect the role of the WSSSSP and the FBXW7 target site in CLL, especially since mutation of the WSSSSP sequence alone has been shown to be leukemogenic in T-cell acute lymphoblastic leukemia [Citation6]. The mutation identified by Sportoletti et al. might therefore point to an interesting duality in the regulation of NOTCH1 in CLL.
Supplementary Material
Download Zip (1.9 MB)Potential conflict of interest:
Disclosure forms provided by the authors are available with the full text of this article at www.informahealthcare.com/lal.
References
- Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development 2011;138:3593–3612.
- Burger JA, Ghia P, Rosenwald A, et al. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood 2009;114:3367–3375.
- Rosati E, Sabatini R, Rampino G, et al. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood 2009;113:856–865.
- Sportoletti P, Baldoni S, Del Papa B, et al. A novel NOTCH1 proline, glutamic acid, serine and threonine domain mutation in a case of chronic lymphocytic leukemia. Leuk Lymphoma 2013;54:1780–1782.
- Gianfelici V. Activation of the NOTCH1 pathway in chronic lymphocytic leukemia. Haematologica 2012;97:328–330.
- Chiang MY, Xu ML, Histen G, et al. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol Cell Biol 2006;26:6261–6271.