Abstract:
Plasma expander-like properties of albumin induced on hexa as well as dodecacPEGylation using Extension Arm Facilitated PEGylation platform make it an excellent resuscitation fluid. PEGylation induced changes in the structure, drug binding, and plasma expander-like properties of bovine serum albumin has been now investigated as a function of PEGylation. The molecular volume of albumin increases on PEGylation nearly linearly; in the beginning up to about six PEG chains are conjugated, then plateau off, while the viscosity and colloidal osmotic pressure change very little initially and then increase exponentially as a function of PEG chains conjugated. PEGylation has essentially no influence on the secondary structure or drug properties of albumin. Tryphtophyl fluorescence of albumin is quenched on PEGylation as a direct correlate of the changes in molecular radius of PEG-albumin. It is concluded that hexaPEGylated and dodecaPEGylated albumin belong to two different configurational states of PEG-albumin in terms of packing of PEG-chains on the molecular surface of the protein. The results suggest a transition of PEGylated albumin from the initial mushroom-like conformation to brush conformation as the PEGylation increases. The therapeutic efficacy of the two PEGylated species is needed to establish the optimum level of PEGylation to function as resuscitation fluids.
| ABBREVIATIONS | ||
| BSA | = | Bovine serum albumin |
| COP | = | Colloidal oncotic pressure |
| CLP | = | Cecal ligation and puncture |
| EAFP | = | Extended arm facilitated PEGylation |
| HES | = | Hy-droxyethyl starch |
| HAS | = | Human serum albumin |
| IT | = | 2-Iminothiolane |
| LPS | = | Lipopolysaccharides |
| PEG | = | Polyethylene glycol |
| 4-PDS | = | 4,4′-dithiopyridine |
INTRODUCTION
PEGylation has emerged as a unique technology to increase the therapeutic efficacy of proteins and peptides by reducing their antigenecity and lowering of their degradation by proteolysis in vivo. Recently, PEGylated Hbs and PEGylated albumins have emerged as oxygen- carrying and nonoxygen-carrying plasma expanders, respectively. This is a reflection of the recognition that PEGylation of proteins confers unique molecular properties to proteins that are also the solution properties of plasma expanders. These properties include a high hydrodynamic volume, viscosity, and colloidal osmotic pressure (COP) of the PEGylated proteins as compared to the parent protein.
Covalent modification of the proteins by functionalized PEG-chains is generally targeted to the surface amino groups. The PEGylation technology was first described by Abuchowski et al. [Citation1] and they generated PEGylate BSA. PEG-chains functionalized with cyanuric chloride had been used for the PEGylation studies. Since then, many functionalized PEG-chains targeted to the amino groups of proteins and peptides have been developed; the succinimidyl esters of monomethoxy PEG acids and the PEG aldehydes [Citation2,Citation3] represent the most widely used reagents. PEGylation of the proteins with multiple copies of small molecular weight functionalized PEG-chains has been the general approach to achieve the masking of the molecular surface of the proteins. However, this approach generally results in formation of heterogeneous products, particularly in terms of the apparent molecular sizes of the proteins as seen by size exclusion chromatography.
Advanced PEGylation or modification of the protein with one or a very limited number of larger molecular weight PEG-chains has been advanced as a better approach as compared to conjugation of multiple copies of PEG chains [Citation4] for production of more homogeneous products. This is possible as only a limited number of chemical reactions are involved in generating the PEGylated products and the reaction is targeted to the most reactive side chain functional groups of proteins. Other recommended improvements in the PEGylation technology involve the use of branched PEG-chains instead of the linear PEG-chains. Each of these approaches exposes unique advantages in specific proteins. Advanced PEGylation, particularly targeted to the reactive thiols of internal Cystine resides of proteins and PEG maleimides as the functionalized PEG derivatives, has turned out to be a very useful procedure to generate homogeneous PEGylated proteins and has attracted significant attention. Accordingly there has been renewed interest in combining the Advanced PEGylation strategy targeted to the Cys residues (with the site directed mutagenesis to engineer new Cys residues into proteins at desired sites) for PEGylation using PEG-maleimide.
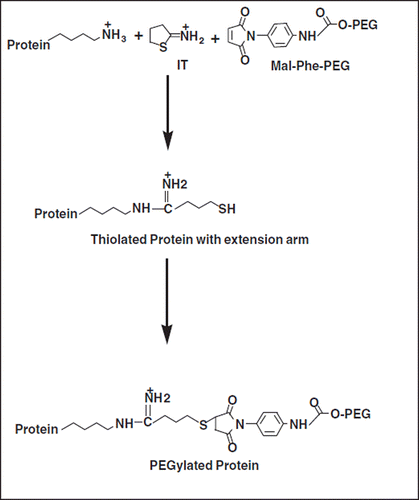
Extension Arm Facilitated PEGylation (EAFP) of proteins has been developed in our lab [Citation5] to facilitate the conjugation of multiple copies of PEG-chains to Hb. This approach takes advantage of the high reactivity and the stoichiometric aspects of the thiol maleimide reaction (). This novel EAFP approach was applied to Hb using 2-iminothiolane (IT) and maleidophenyl PEG 5K (represented schematically in ) and the approach has been used to generate vasoinactive hexaPEGylated Hb. Surprisingly, this product exhibited a high degree of molecular size homogeneity as reflected by SEC-chromatography [Citation5]. The thiol groups are engineered at the distal end of the extension arm, butyrmidyl chain placed on the reactive ϵ- amino groups of lysine residues of the protein. Accordingly the thiol groups are placed ∼1 nm away from the derivatized amino group. This can result in an increased accessibility of the new thiol groups introduced on the protein surface for PEGylation as compared to the parent amino group. Besides, the reactivity of all the new thiol groups thus introduced is likely to be comparable to one another, thereby leading to a reasonably uniform molecular size distribution for the PEGylated Hb generated. Additionally, this approach is also simple and cost-effective as the reaction of the maleimide PEG with thiols is nearly quantitative.
Figure 1. Schematic representation of Extension Arm Facilitated PEGylation of Proteins: Iminothiolane (IT) reacts with amino groups of proteins, adds an extension arm, and generates new thiol groups on reaction with protein amino groups. These nascent thiol groups at the distal end of the extension arm are modified by maleimide-PEG (Mal-Phe-PEG).
PEGylation induced plasma expander-like properties of Hb and albumin, namely enhanced molecular volume, increased viscosity and increase in COP makes this protein a unique resuscitation fluid [Citation7]. However, the cross correlation of PEGylation induced plasma expander-like properties of albumin as a function of the PEG-chains has not been delineated. Accordingly, in the present study PEGylation induced molecular properties of PEG albumin have been studied as a function of number of PEG-5K chains conjugated to the protein. The results presented here suggest a mushroom-like conformation of PEGylated albumin until about six copies of PEG chains are conjugated to a brush- conformation as the level of PEGylation increased further. Even at the level of dodecaPEGylation with PEG-5K, PEGylation has very little influence on its secondary structure of the protein. DodecaPEGylated-albumin adduct binds drugs targeted to its sites I and II, reflecting a flexibility of the conjugate and the accessibility of the molecular surfaces for drug binding These studies pose new questions as to the potential advantages mushroom conformation of PEGylated albumin versus the brush conformation in designing novel resuscitation fluids.
EXPERIMENTAL
Bovine serum albumin (Fraction V, essentially fatty acid free, purity, 99%), N-ethylmalemide, dansyl sarcosine, warfarin were obtained from Sigma, St. Louis, MO. MalPhePEG5K (Lot 01234) and 2-Iminothiolane (Lot 10222) were obtained from Bioaffinity Systems, Rockford, IL. Sodium chloride, disodium phosphate, dipotassium phosphate and potassium phosphate were of analytical grade products from Fisher Scientific. This preparation of bovine serum albumin contains small amounts of albumin dimer, and has been used without further purification.
PEGylation of Albumin
EAF-PEGylation of albumin was carried out in PBS pH 7.4 in cold room (40 C) at a protein concentration of 0.25mM in the presence of 2-iminothiolane and maleimido phenyl PEG-5K. The extent of conjugation of PEG maleimide to BSA (0.25 mM) has been studies as function of varying concentrations of either IT or of Mal-Phe-PEG-5K in PBS at 4°C overnight. The reactions were stopped by adding excess N-ethylmaleimide and incubating it further for an additional 30 minutes. After the incubation, the excess reagents were removed either by tangential flow filtration using a 70K membrane with Minim system (Pall Life Sciences, Ann Arbor, MI) or by filtering through Centricon-50. Complete removal of unreacted PEG reagent was confirmed by analyzing the retentate of the filtered products on Superose-12 column (Pharmacia), monitoring the refractive index (RID-10A, Shimadzu) of the eluate.
Estimation of Thiols
The number of thiol groups introduced on the amino groups of BSA by 2-iminothiolane was determined by the method previously described [Citation8]. Typically, an aliquot of 20 μL of the reaction mixture was removed and diluted with PBS to 0.75ml. An aliquot of 0.25 ml of 4-PDS (1mM) was added to the sample and incubated for 30 minutes at room temperature. The absorbance of the sample at 280 and 324 nm were used to determine the concentration of albumin and the reaction product of 4-PDS, 4-thiopyri-done, respectively. The number of thiols generated was calculated from the concentration of 4-thiopyridone.
SDS-PAGE Analysis of PEGylated Albumin
SDS-PAGE of protein samples was carried out using precast 4-12% Tris-glycine gels from Invitrogen. The gels were stained with Coomassie brilliant blue R-250 after the runs.
Dynamic Light Scattering
The molecular radii of the samples were determined using the principle of light scattering and the Dyna Pro instrument, Protein Solutions, Lakewood, NJ. Control and PEGylated samples were centrifuged at 13000 rpm prior to analysis. A protein concentration of 1mg/ml was used and the observations recorded at 22 ˚C.
NMR Analysis of PEGylated Albumin
Known amounts of Mal-Phe-PEG-5K were dissolved in D2O and Proton NMR spectra were recorded to determine the standard curve for free PEG. The spectra for PEGylated albumin was also recorded under similar conditions. The amount of Mal-Phe-PEG-5K associated with albumin was calculated from the standard curve. The protein contnent was estimated using Bradford reagent.
Fluorescence Spectroscopy
The fluorescence emission spectra of albumin and PEGylated albumins were determined in the range of 300-400 nm by exciting the samples at 295 nm at room temperature. The slit width was 3 and 5 nm for excitation and emission, respectively. A protein concentration of 1mg/ml was used. The quantum yield was determined at the emission maxima, at 340 mm.
Circular Dichroic Spectra
The far UVCD spectra of BSA and PEGylated BSA were recorded on a JASCO 720C spectropolarimeter (Tokyo, Japan) at 25°C using a 0.2 cm light path cuvette (310 μL). All the samples (0.4 mg/ml) were scanned in the range of 200-250 nm in PBS, pH 7.4. The CD results were expressed in terms of mean residue ellipticity.
COP Determination
The COP of albumin samples in PBS, pH 7.4 was determined at room temperature with a Wescor 4420 Colloidal Osmometer (Wescor, Logan, UT) using a 30 kDa cut off membrane. The instrument was calibrated using Osmocoll (Wescor) reference standards.
Viscosity Measurements
The viscosity of the samples in PBS, pH 7.4 was measured at 37°C using a cone and plate rheometer (Brookfield Engineering, Middleboro, MA) using a cone spindle CPE-40 (Brookfield Engineering) at a shear rate of 375 s-1.
Drug Binding Studies
The drug binding studies were carried out employing the fluorimetric method. The excitation and emission wavelengths used for warfarin were 320 and 380 nm and for dansylsarcosine 350 and 500 nm, respectively. The fluorimetric titrations for both the drugs were performed as described earlier [Citation9]. Scatchard plots were generated to determine the drug binding constants.
RESULTS
Extension Arm Facilitated PEGylation of Albumin
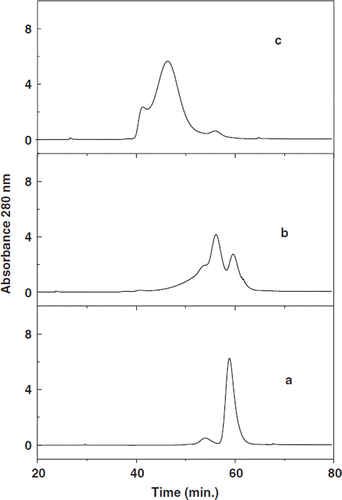
Size exclusion chromatography of BSA incubated with Mal-Phe-PEG 5K in the presence or absence of different concentrations of IT is shown in . The commercial preparations of BSA have two components (curve a); the major component represents the monomeric albumin, and the minor peak apparently represents dimer/oligomer of BSA known to be present in the commercial preparations. The samples of BSA (0.25 mM) incubated with 10 mM Mal-Phe-PEG-5K (a forty-fold molar excess) in the absence of IT shows three components (curve b): one coeluting with unmodified albumin, the second eluting earlier corresponding to the dimeric form of BSA. The third component is not resolved from the second peak and elutes earlier. Presumably this represents the species with two copies of PEG-chains. The reaction is apparently at the free Cys of albumin and also presumably at the ℵ-amino group, which exhibits a considerably lower pKa as compared to the ϵ-amino groups of Lys residues of albumin. Apparently, the early eluting peaks are albumin with PEG chains conjugated and resulting enhanced volume. The second peak eluting at the position of the unmodified dimeric species of albumin demonstrates the partial modification of albumin by MalPhe-PEG-5K, in spite of using a forty-fold molar excess of PEG maleimide. This may be due to the fact that the commercial albumin samples have reactive thiols partially blocked by the formation of disulfide bonds with cysteine/ glutathione derived from plasma [Citation10] and the reaction at the ℵ-amino group with maleimide is not quantitative. On the other hand, incubation of BSA with 10 mM Mal-PhePEG-5K in the presence of 5 mM IT generated much larger molecules and the major peak of this sample eluted earlier than BSA incubated with only the PEG reagent (curve c). This indicates that IT adds additional thiol groups on albumin that are modified by Mal-Phe-PEG-5K. No unmodified albumin was present in the chromatogram indicating the quantitative PEGylation of the protein. A control sample of BSA incubated with 5 mm IT for overnight had an elution pattern indistinguishable with that of the control BSA (data not shown), suggesting the absence of any extrinsic thiol catalyzed exchange reactions with thiolated BSA.
Figure 2. Size exclusion chromatography of PEGylated BSA: BSA (a) incubated at a protein concentration of 0.25 mM with either 10 mM Mal-Phe-PEG-5K (b) or with 5 mM IT and 10 mM Mal-Phe-PEG-5K (c) overnight at 4°C and the excess of reagents were removed by ultrafiltration. The products were subjected to size exclusion chromatography Superose 12 columns; two columns connected in series.
Influence of the Concentration of Iminothiolane and of PEG-maleimide on the Molecular Size of the PEGylated BSA
The extent of PEGylation of albumin by maleimide PEG is determined by the number of thiol groups added onto the protein; accordingly the thiolation of albumin has been studied as a function of IT concentration. The SDS-PAGE pattern of the PEGylated albumin is given in . BSA incubated with Mal-Phe-PEG-5K in the absence of IT displayed two bands, one corresponding to the unmodified albumin and the other slower-moving band related to PEGylated albumin (, Lane 4). These results are consistent with the size exclusion chromatography of this sample described above. PEGylated albumins generated in the presence of IT displayed slower mobility than albumin due to the enhanced molecular size. The molecular size increase was higher on increasing IT concentration, indicating the facilitation of PEGylation with thiolation. When the IT concentration is increased from 7.5 mM to 10 mM, the increase in the molecular size and number of PEG-chains conjugated appears to level off. Most of the PEGylated BSA bands appeared around the region expected for a globular protein with a molecular weight of 216 kDa. This anomalous migration may be due to the PEG chains attached.
Figure 3. Influence of concentration of 2-iminothiolane and of PEG maleimided on the EAF PEGylation as reflected by the SDS-PAGE analysis of PEGylated BSA. (A) Influence of the concentration of iminothiolane on EAF-PEGylation of BSA at a protein concentration of 0.25 mM) when incubated with 10 mm Mal-Phe PEG-5K. Lane-1, Marker; Lane-2, control BSA, Lane-3, BSA + 5 mM IT without PEG The samples in Lanes 4 to 9 represents the samples by reacting BSA with 10 mM Mal-Phe-PEG-5K in the presence of indicated concentrations of IT. .Lane-4, 10 mM Mal PEG 5K with out iminothiolane; Lane-5, 2.5mM IT; Lane-6, 5.0 mM IT; Lane-7, 7.5 mM IT; Lane-8, 10.0 mM IT; Lane-9, 12.5 mM IT; Lane-10 Marker. (B) Influence of concentration of Mal-Phe-PEG 5K on EAF-PEGylation of of BSA at a protein concentration of 0.25 mM when incubated with 5 mm 2-iminothiolane. Lane-1, marker; Lane-2, control BSA, Lanes 3 to 9 BSA incubated with 5 mM iminothiolane in the presence of indicated amounts of PEG maleimide. Lane-3, 1.25 mM; Lane-4, 2.5mM; Lane-5, 3.75 mM; Lane-6, 7.5mM; Lane-7, 10.0 mM; Lane-8, 12.5mM; Lane-9 marker.
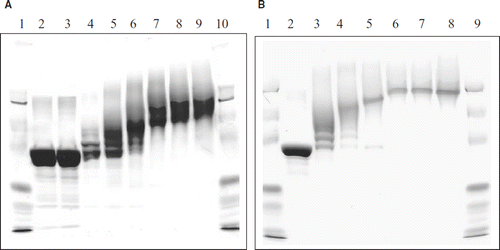
Influence of the concentration of Mal-Phe-PEG-5K at a fixed concentration of IT, 5 mM, on increasing the molecular size of the PEGylated albumin is shown in . The maximum increase in the molecular size of albumin is seen at a Mal-Phe-PEG-5K concentration of 7.5 mM and a further increase did not increase the molecular size of the PEGylated albumin further as reflected by SDA PAGE electrophoresis. These results indicate the complete modification of the thiol groups that were added on protein amino groups by 5 mM iminothiolane (∼ 20-fold molar excess over protein, BSA) is achieved in the concentration range of 7.5 mM to 10 mM of maleimide PEG (∼ 30- to 40-fold molar excess over BSA).
Extent of PEGylation of Albumin as a Function of Concentration of PEG-maleimide
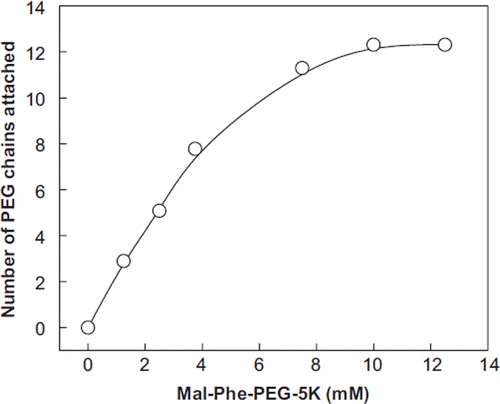
The number of PEG-5K chains conjugated to BSA (0.25 mM, ∼16 mg/ml) by EAF PEGylation in the presence of a 30-fold molar excess of 2-iminothiolane has been studied as a function of the concentration of maleimide PEG-5K. Approximately 12 copies of 2-iminthiolane are introduced onto BSA on an overnight reaction with a 30-fold molar excess of the reagent. PEGylated BSA samples from the above experiments were analyzed by NMR-spectroscopy for establishing the number of PEG chains conjugated to BSA as a function of maleimide PEG concentration (up to a concentration representing 50-fold molar excess, i.e. 12.5 mM). The number of PEG chains conjugated to each of BSA as a function of concentration of Mal-Phe-PEG-5K used for the modification is presented in . At a concentration of 7.5 mM Mal-Phe-PEG-5K (2.5-fold molar excess of the reagent over the number of the thiols introduced on the albumin), the number of PEG chains conjugated to the protein reached a value close to 12 and this value remained essentially unchanged even when the Mal-Phe-PEG-5K concentration is increased further, reflecting that thiolation may be a limiting factor. It may be noted that increasing the concentration of IT at this concentration of the protein (0.25 mM) does not significantly increase the apparent molecular size of PEG-BSA Thus dodecaPEGylation appears to be the maximum level of PEGylation that could be achieved under these experimental conditions. To increase the level of PEGylation, further lowering the concentration of albumin is a preferable approach since the increase in the viscosity of the reaction mixture increases as a function of PEGylation; at a given protein concentration the subsequent reactions become very slow, and the presence of more PEG reagent could lead to protein precipitation.
Figure 4. Correlation of the number of PEG-chains conjugated to BSA with the concentration of Mal-Phe-PEG-5K at a fixed concentration of 2-iminothilane. EAF PEGylation of BSA was carried out on the protein concentration of 0.25 mM in the presence of 7.5 mM iminothiolane. The number of copies PEG chains attached to albumin was determined by NMR analysis of the PEGylated BSA.
PEGylation Induced Colloidal Osmotic Pressure and Viscosity of dodecaPEGylated BSA
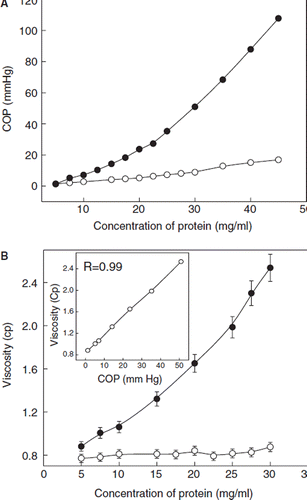
As noted above, under the present conditions 12 copies of the PEG-chains appear to be the maximum number of PEG-chains that could be introduced by EAF-PEGylation. The COP of the dodecaPEGylated BSA measured as a function of protein concentration is presented in . The COP of BSA showed a small and linear increase as a function of protein concentration. On the other hand, the COP of dodecaPEGylated BSA increased exponentially in the range of 0 to 4.5 g/dL. At this concentration, the COP of PEGylated albumin was about 10-old higher than that of BSA. The increase in the viscosity of PEGylated BSA also exhibited the same trend (). The influence of protein concentration on the viscosity of unmodified BSA was insignificant. At 3.0 g/dL, the viscosity of PEGylated BSA was about 3-fold higher than the control protein. As shown in the inset, the increase in the viscosity and COP as a function of protein concentration showed a direct correlation.
Figure 5. COP and viscosity and DodecaPEGylated BSA (filled circles) as a function of protein concentration. Open circles represent unmodified BSA and the closed circles represent dodecaPEGylated BSA. A: COP, B: Viscosity. Inset : Correlation between the viscosity and COP of PEGylated BSA. PEGylated BSA carrying the maximum number of PEG-5K chains (∼ 12 copies) was used in these studies.
Drug Binding Properties of PEGylated BSA
The drug binding capability of dodecaPEGylated BSA was probed with warfarin for site I and with dansylsar-cosine for site II. The protein-ligand association constants and the number of binding sites are displayed in . The warfarin association constant for PEGylated BSA was comparable to that of BSA. The number of binding sites for warfarin was also similar for both normal and PEGylated albumin. These results indicate that the PEGylation of albumin did not alter the affinity of albumin for binding warfarin. The dansylsarcosine binding constant for PEGylated BSA was 2.5-fold higher than that of BSA. Conversely, the number of binding sites was lower for PEGylated BSA than albumin. For BSA the binding sites were more than two and for PEGylated BSA it was less than two. Thus, PEGylation seems to inhibit the binding of dansylsarcosine to albumin at one or more high/low affinity sites. However, the same modification increased the affinity of albumin for dansylsarcosine to bind at some of the existing sites or at new sites.
Table 1. Influence of dodecaPEGylation of bovine serum albumin on drug binding
Influence of PEGylation on the Secondary Structure of BSA
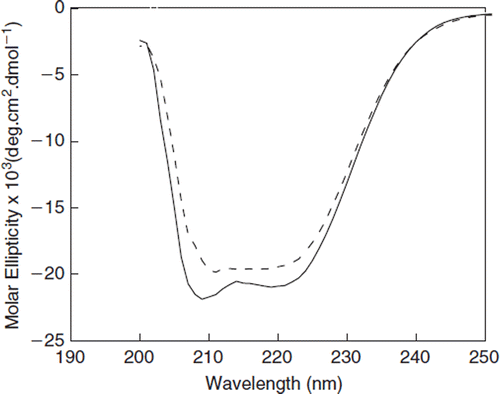
Circular dichroic spectra of dodecaPEGylated BSA in the far UV region has been compared with that of BSA in . As can be seen, only minor differences were observed between the spectra of dodecaPEGylated and unPEGylated BSA, indicating slight alterations in the secondary structure of BSA due to PEGylation.
Correlation of PEGylation Induced Colloidal Osmotic Pressure and Viscosity of BSA as a Function of Conjugation of PEG-chains
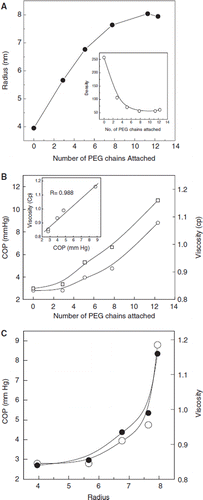
The COP and viscosity of albumin was also determined as a function of the number of PEG chains attached, at a constant protein concentration of 1 gm/dL. As shown in , the COP and viscosity of albumin increased exponentially as the number of PEG chains conjugated to the protein increased. Thus, the amount of PEG present on the surface of albumin makes the primary contribution towards the solution properties of the protein. There is a direct correlation between the COP and the viscosity (, inset) of the PEGylated albumin at different levels of PEGylation.
Figure 7. Correlation of the PEGylation induced molecular properties (molecular radius, viscosity and COP of BSA with the extent of PEGylation: (A) Increase in the molecular radius BSA as function of PEGylation. The molecular radius is determined by dynamic light scattering using DynaPro as described under methods. A concentration of 1mg/ml of the protein was used for all studies. The observations were the averages of three independent readings taken for 180 seconds each. The inset depicts the molecular density of albumin as a function of the molecular radius of the PEGylated protein. The inset shows the correlation of the density of PEGylated protein as a function of PEGylation. (B) Correlation of increase in the viscosity and COP of BSA as function of PEGylation. A protein concentration of 1% was used for both of these experiments. Inset shows correlation between viscosity and COP induced to BSA at a given level of PEGylation. (C) Correlation of COP and viscosity with the molecular radius.
Correlation of the Molecular Radius of the PEGylated BSA with the Number of PEG 5K Chains Conjugated
The molecular radius of PEGylated albumin generated as a function of Mal-phe-PEG5K concentration (at a protein and IT concentration of (0.25 mM, and 5 mM, a 20-fold molar excess) has been correlated with the number of PEG 5K chains conjugated to albumin (). The molecular radius increases linearly initially until about six copies of PEG 5K chains are added and then appears to plateau off when about 8 PEG 5K chains are conjugated. This indicates that additional PEG chains are accommodated within the hydrated PEG shell of the octaPEGylated albumin. The inset shows the decrease in the density (mass per volume) of albumin as a function of the molecular radius of PEGylated albumin. This correlation is biphasic, a linear correlation phase wherein the molecular density decreases as a function of the number of PEG-chains and a second phase wherein further conjugation of the PEG-chains to the PEGylated protein results only in a marginal change in the molecular density. The correlation of the increase in the molecular radius with the increase in the viscosity and COP of albumin is given in . These colligative properties of PEGylated albumin exhibit an exponential correlation with the molecular radius of PEG albumin.
Thus, of the three PEGylation induced molecular properties of PEG-BSA, the increase in the molecular radius is biphasic as a function of PEGylation. It is gradual after the initial reasonably linear increase. The PEGylation induced viscosity and COP increase exponentially as a function of PEGylation.
Correlation of Trp Fluorescence Changes (Tertiary Structure) of BSA with the Extent of PEGylation of BSA
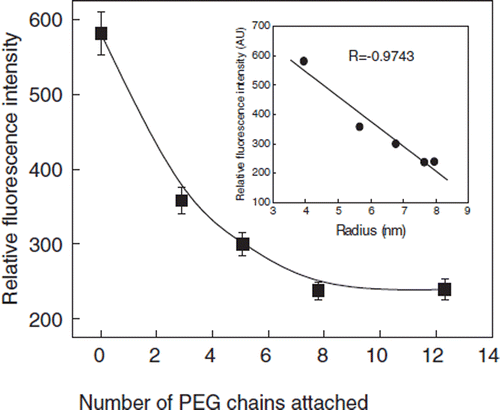
The influence of PEGylation of BSA on its Trp-fluorescence has been studied as a function of number of PEG-chains attached. The excitation of PEGylated BSA at 295 nm did not alter the maximum emission wavelength significantly. The fluorescence intensity of PEGylated BSA decreased with increase in the number of PEG chains conjugated to albumin (). The decrease in the florescence intensity is a biphasic phenomenon, a linear phase wherein the decrease in the intensity is directly correlated with the number of PEG-chains conjugated, and a second phase wherein the florescence intensity is not influenced by the increase in the number of the PEG chains conjugated to albumin. The decrease in the fluorescence intensity reached a plateau when about 8 PEG 5K chains are conjugated to albumin. A maximum of about 60% fluorescence quenching occurred at this point. This decrease in the florescence intensity as a function of number of PEG-chains conjugated appears to mimic the decrease in the molecular density of albumin as a function of the number of PEG-5K chains conjugated. There is a nearly linear correlation (, inset) between the quenching of the Trp fluorescence and molecular dimension of PEG-albumin adduct (structure of the PEG-shell).
Figure 8. Quenching of Tryptophyl Fluorescence of BSA as a function of PEGylation. The influence of the number of conjugated PEG chains on the fluorescence of BSA is presented. Protein samples (1 mg/ml) were excited at 295 nm and the emission was recorded at 340 nm. Inset: Correlation fluorescence intensity as a function of the molecular radius of PEGylated albumin.
DISCUSSION
The extension arm facilitated PEGylation developed initially for the PEGylation of Hb, represents the reaction of the protein with a small molecule with hetero bifunctional reagent that introduces a thiol [Citation11–15] group as the targeted site for conjugating PEG-chains. This increases the accessibility of the functional groups of Hb and/or of red blood cells to the macromolecular PEG-reagents. In the case of Hb, the engineered extension arms appears to buffer (reduce) the influence of PEG-chains on the inter dimeric interactions of the molecule. The present study extends the study of the EAF-PEGylation to albumin, using the 2-iminothiolane and the thiolating agent, particularly to correlate the PEGylation induced plasma expander-like properties of proteins as a function of extent of PEGylation. The results presented here first confirm that the thiol groups, generated in situ on albumin, do not facilitate inter or intra molecular thiol disulfide exchange reaction. Accordingly, a wider application of the EAF PEGylation to other therapeutic proteins/peptides with internal disulfide bonds is envisioned.
The development of novel PEGylation strategies and the application of PEGylated proteins as therapeutic proteins is a subject of considerable interest in protein peptide therapeutics [Citation16–18]. The increased solubility, decreased proteolysis and the surface coverage to hide the molecules from the antibodies and being recognized by the immune system as an alien material have been the major PEG-induced molecular properties that have advanced the development of PEGylation proteins/peptides a new class of therapeutic agents. However, the development of PEG Hb as the oxygen carrying plasma expander is based on the observation that PEGylation induces the plasma expander-like properties to Hb. The plasma expander like properties PEG-albumin adduct include, but are not limited to, the high hydrodynamic volume, high viscosity and high COP. This has prompted the development of PEG-albumin as the non-oxygen carrying plasma expander. We have used the same extension arm facilitated PEGylation platform that was designed for the development of PEG-Hb and applied here to develop PEGylated albumin. The correlation between the PEGylation induced increase in hydrodynamic volume, viscosity and COP has not been investigated so far, and this has been attempted here.
The surface decoration of bovine serum albumin with PEG-chains was carried out using cyanuric chloride activated-PEG-5K [Citation1]. This procedure has been used more recently to generate PEGylated human serum albumin with multiple copies of PEG-5K chains [Citation6] and the PEGylated albumins have been advanced as plasma expander to overcome the leakage of the albumin in situations of capillary leak [Citation1,Citation6].
The molecular size homogeneity of PEGylated albumin that we have generated by EAF-PEGylation is in contrast to the PEGylated albumin generated by cyanuric chloride functionalized [Citation6]. The molecular size homogeneity is also true of hexaPEGylated uncrosslinked Hb generated by EAF PEGylation. A prototype molecule of the hexaPEGylated Hb generated at Einstein has been developed by Sangart as oxygen therapeutic and is in clinical trial under the trade name Hemospan [Citation13].
The molecular size homogeneity of hexaPEGylated albumin and hexaPEGylated Hb generated by EAF PEGylation is, apparently, a consequence of multiple structural factors. The accessibility of the thiol groups (of the thiolated protein) located at the distal end of the extension arm engineered into protein is expected to be higher compared to the original amino groups to which the extension arms are attached. Accordingly, the macromolecular PEG reagents can react with it much more readily. Besides, reactivity of all nascent thiol groups generated through iminothiolane mediated thiolation is anticipated to be comparable to each other, is expected to play their respective roles in determining the molecular size homogeneity of the PEG-protein conjugate [Citation19]. Chemical reactivity of the PEG reagents could also play their own role in the final homogeneity of the PEG-protein adducts. PEG-reagents like cyanuric hydrochloride functionalized PEG exhibit some potential to introduce crosslink, besides their primary role of monofunctional modification of proteins. The influence of the viscosity of the reaction mixture on the generation of new events of PEG reagent with the PEGylated protein also needs to be considered in understanding the molecular size homogeneity seen in the EAF-PEGylation studies.
The calculated molecular mass of dodecaPEGylated BSA is comparable to that of BSA dimer. However, the BSA dimer elutes much earlier than the hexaPEGylated albumin, reflecting the very high level of increase in the molecular volume on conjugation of PEG-chains to the proteins. The molecular radius of dodecaPEGylated is nearly twice that of albumin, as estimated by dynamic light scattering. This reflects highly disordered conformation of the PEG chains conjugated onto the surface of the protein.
The flexible conformation of the PEG-chains conjugated onto the surface of albumin is consistent with accessibility of the molecular surface for binding of small molecular weight drugs. The drugs warfarin and dansylsarcosine bind to BSA at more than one site [Citation24–28]. In the present studies warfarin binds to BSA at about two sites. One appears to bind at site I and another may be bound at a discrete site or diffused site [Citation29]. The binding constant and the number of warfarin binding sites for PEGylated albumin were comparable to that of albumin. The binding of dansylsarcosine to PEGylated albumin was different as compared to albumin. While this drug bound to albumin at more than two sites, it bound to PEGylated albumin at less than two sites. Dansylsarcosine binds to albumin at site II that is located in IIIA domain. In addition to this site, it also binds to albumin at several other sites that are not characterized. PEGylation of albumin may have inhibited the binding of dansylsarcosine to some of these sites, including site II. Interestingly, the dansylsarcosine binding constant for PEGylated albumin is higher than that for albumin, apparently a consequence of the PEG-shell surrounding the protein core.
The dodecaPEGylation of albumin caused only minimal changes in the secondary structure of albumin as reflected by the far UV CD spectroscopy; PEG-chains exert limited influence on PEGylation on the overall helical structure of the protein. On the other hand, a significant level of fluorescence quenching has occurred as a result of PEGylation. The microenvironment of Trp, i.e. exposure of the Trp residue to the solvent phase, is influenced by PEGylation without significantly altering the packing of the secondary structural units of albumin. The PEG shell surrounding the protein hydration shell of albumin seems to impact the microenvironment of Trp. The quenching is a direct correlate of the molecular radius of PEG-albumin adducts. Similar results have been reported [Citation22,Citation23] for BSA in the presence of surfactants.
Resuscitating capability of the dodecaPEGylated BSA under hypovolemic conditions has been tested earlier in hamster models with induced endotoximia [Citation20]. PEGylated BSA improved the micro vascular functions more effectively than dextran 70. In hemorrhagic shock hamster models PEGylated BSA restored systemic and micro vascular parameters for a much longer period than HES [Citation21]. The PEGylated albumin seems to provide both early and long-term sustained systemic and microvascular recovery much better than the other plasma expanders that are currently in use.
The PEGylation induced increase in the molecular radius, viscosity and COP of albumin has been studied as a function of PEGylation. The increase in the viscosity and COP of albumin as a function of PEGylation is exponential, thus by using a higher level of PEGylation with albumin a lower concentration of the protein could be used. The PEGylation induced viscosity and COP at different levels of PEGylation show a direct correlation. On the other hand, the increase in the molecular radius of albumin as a function of PEGylation is not a direct correlate of the number of the PEG-chains conjugate. In the initial stages of up to six to eight copies of PEG-5K chains, the correlation is reasonably linear, and the increase in the molecular radius of the octaPEGylated albumin as new PEG-chains are introduced is smaller. As seen from the inset in , the overall molecular densities of PEG albumin adduct decrease as a function of the number of PEG-chains. This suggests the ordering of the PEG-chains in the PEGylated albumin is not influenced much in the early stages and results in a lowering of the overall molecular density. As the number of PEG-chains on the molecular surface is increased, the propensity of the newly added PEG-chains to lower the density of the PEG-albumin adduct is lowered, the PEG-chains start getting packed within a reasonably finite molecular volume in such a way that molecular density of the PEG albumin adducts remain reasonably constant. In this connection, it is of interest to note that during the PEGylation of albumin with cyanuric chloride –PEG lowers the S-value of albumin significantly in the initial phase of modification wherein only about 5 % of the amino groups are PEGylated, about three groups, and on increasing the PEGylation further the change in the S-values is marginal.
The non-linear correlation of the extent of PEGylation of albumin with the increase in the molecular radius, particularly the biphasic pattern, is interesting. This is schematically depicted in a model () by comparing the distribution of PEG-chains in the hexaPEGylated and dodecaPEGylated albumin. The initial attachment of few PEG chains leads to a significant increase in the hydrodynamic volume of the protein (). The initial few chains conjugated are presumably distributed on the surface, leading to a “mushroom” type of conformation [Citation30], i.e. a low density PEG-shell and limited interactions between the PEG-chains. The addition of more PEG chains is achieved without significantly increasing the molecular volume of the PEG-albumin conjugate. This leads to a packing of the PEG-chains with a higher density within the PEG-shell. Accordingly, the density of the PEG chains is increased in the PEG-shell, i.e. PEG-chains in the shell begin a transition to the “brush conformation.” The new PEG chains are accommodated on the molecular surface with a very small increase in the hydrodynamic volume of PEGylated protein (). The exponential increase in the viscosity and COP of the conjugate as a function of molecular radius of PEG-chains is also consistent with the transition of the PEGylated albumin to a brush conformation once the molecular surface of albumin is covered by about six to eight PEG-chains.
Figure 9. Schematic representation of the structure of the PEGylated BSA depicting the protein core and the PEG-shell. (A) OctaPEGylated BSA and (B) DodecaPEGylated BSA. The color code is as follows: BSA (dark blue), protein core (yellow), PEG chains (dark brown), PEG shell (light brown), water (light blue). Green line represents the boundary of the protein core and dark blue line represents the increased volume after PEGylation assuming a globular shape.
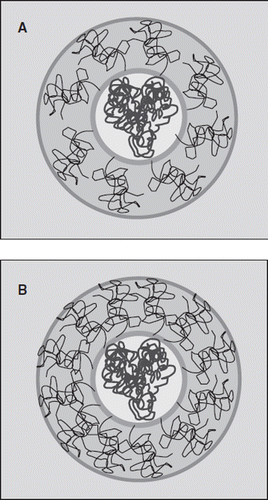
The present study exposes new information about the correlation of the PEGylation induced plasma expander-like properties of albumin as a function of PEGylation by PEG-5K. The results suggest that the packing of the PEG-chains on the molecular surface of albumin becomes denser with a limited increase in the hydrodynamic volume when PEGylated beyond the stage of hexaPEGylation, the viscosity of and the COP of the PEGylated Hb increase exponentially as a function of conjugated PEG-5K chains. We suggest that once PEG albumin is surface decorated with about six PEG-chains it represents the limit of the early phase of PEGylation wherein the molecular dimensions are enhanced without significant increase in viscosity and COP. After this stage PEG-albumin molecule transitions from a mushroom to a brush or brush-like conformation. Accordingly, we speculate that the hexaPEGylated albumin and dodecaPEGylated albumin generated by EAF PEGylation using EAF PEGylation and PEG-5K maleimide represent two very distinct configurational states of PEGylated albumin. In view of differences in the COP of hexa and dodecaPEGylated Hb, the hexaPEGylated albumin is used at 2.5 gm% and dodecaPEGylated albumin at 4 gm%; both have provided excellent functional capillary densities. The potential significance of this difference in the conformational states of PEGylated albumins and their impact in terms of microvascular responses in vivo needs to be delineated to expose any of the advantages and/or disadvantages of these for harnessing the therapeutic efficacy of PEGylated albumin as plasma expanders for specific clinical applications.
Acknowledgements
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
This paper was first published online as an Early Online article on 10 November 2009.
References
- Abuchowski, A. Es, van T., Palczuk, N. C., Davis, F. F. (1977). Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol. J Biol Chem. 252:3578–3581.
- Fella, C., Walker, G.F., Ogris, M., Wagner, E. (2008). Amine-reactive pyridylhydrazone-based PEG reagents for pH-reversible PEI polyplex shielding. Eur J Pharm Sci. 34:309–20.
- Lee, B.K., Kwon, J.S., Kim, H. J., Yamamoto, S., Lee, E.K. (2007). Solid-phase PEGylation of recombinant interferon alpha-2a for site-specific modification: process performance, characterization, and in vitro bioactivity. Bioconjug. Chem. 18:1728–34.
- Nacharaju, P., Boctor, F. N., Manjula, B.N., Acharya, S.A. (2005). Surface decoration of red blood cells with maleimidophenyl-polyethylene glycol facilitated by thiolation with IT: an approach to mask A, B, and D antigens to generate universal red blood cells. Transfusion45:374–383.
- Acharya, S. A., Friedman, J.M., Manjula, B.N., Intaglietta, M., Tsai, A.G., Winslow, R. M., Malavalli, A., Vandegriff, K., Smith, P.K. (2005). Enhanced molecular volume of conservatively pegylated Hb: (SPPEG5K)6-HbA is non-hypertensive. Artif Cells Blood Substit Immobil Biotechnol. 33:239–255.
- Assaly, R.A., Azizi, M., Kennedy, D. J., Amauro, C., Zaher, A., Houts, F. W. Habib, R. H., Shapiro, J. I., Dignam, J. D. (2004). Plasma expansion by polyethylene-glycol-modified albumin. Clin Sci (Lond). 107:263–272.
- Hu, T., Prabhakaran, M., Acharya, S. A., Manjula, B. N. (2005). Influence of the chemistry of conjugation of poly(ethylene glycol) to Hb on the oxygen-binding and solution properties of the PEG-Hb conjugate. Biochem J. 392:555–64.
- Ampulski, R. S., Ayers, V.E., Morell, S. A. (1969). Determination of the reactive sulfhydryl groups in heme proteins with 4,4’-dipyridinedisulfide. Anal Biochem. 32:163–169.
- Maes, V., Engelborghs, Y., Hoebeke, Maras, Y., Vercruysse, A. (1982). Fluorimetric analysis of the binding of warfarin to human serum albumin. Equilibrium and kinetic study. Mol Pharmacol. 21:100–107.
- Carlsson, J., Svenson A. (1974). Preparation of bovine mercaptalbumin by means of covalent chromatography. FEBS Lett. 42:183–186.
- Li, D, Manjula, B.N., Acharya, A.S. (2006). Extension arm facilitated PEGylation of hemoglobin: correlation of the properties with the extent of PEGylation. Protein J. 25:263–274.
- Acharya, A.S., Manjula, B.N., Smith, P.K. (1996). Hemoglobin crosslinkers. US Patent 5585484.
- Manjula, B. N., Tsai, A. G., Intaglietta, M., Tsai, C. H., Ho, C., Smith, P. K., Perumalsamy, K., Kanika, N. D., Friedman, J. M., Acharya, S. A. (2005). Conjugation of multiple copies of polyethylene glycol to hemoglobin facilitated through thiolation: influence on hemoglobin structure and function. Protein J. 24:133–146.
- Nacharaju, P., Manjula, B.N., Acharya, S. A. (2007). Thiolation mediated pegylation platform to generate functional universal red blood cells. Artif Cells Blood Substit Immobil Biotechnol. 35:107–118.
- Ananda, K., Acharya, S. A. (2008). Role of extension arm in PEG-Hb conjugates on the stability of the tetramer: non-conservative EAF maleimide thio-PEG mediated PEGylation. Artif Cells Blood Substit Immobil Biotechnol. 36:499–512.
- Winslow, R.M. (1999). New transfusion strategies: red cell substitutes. Annu Rev Med. 50:337–353.
- Nacharaju, P., Friedman, J. M., Prabhakaran, M., Acharya, S. A., Manjula, B. N. (2007). Combining the influence of two low O2 affinity-inducing chemical modifications of the central cavity of hemoglobin. Biochemistry. 46:4554–4564.
- Nho, K., Linberg, R., Johnson, M., Gilbert, C., Shorr, R. (1994). PEG-hemoglobin: an efficient oxygen-delivery system in the rat exchange transfusion and hypovolemic shock models. Artif Cells Blood Substit Immobil Biotechnol. 22:795–803.
- Prabhakaran, M., Manjula, B. N., Acharya, S. A. (2006). Molecular modeling studies of surface decoration of hemoglobin by maleimide PEG. Artif Cells Blood Substit Immobil Biotechnol. 34:381–93.
- Hangai-Hoger, N., Nacharaju, P., Manjula, B. N., Cabrales, P., Tsai, A. G., Acharya, S. A. Intaglietta, M. (2006). Microvascular effects following treatment with polyethylene glycol-albumin in lipopolysaccharide-induced endotoxemia. Crit Care Med. 34:108–117.
- Cabrales, P., Tsai, A. G., Winslow, R. M., Intaglietta, M. (2005). Extreme hemodilution with PEG-hemoglobin vs. PEG-albumin. Am J Physiol Heart Circ Physiol. 289:H2392–2400.
- Gelamo, E. L., Silva, C. H., Imasato, H., Tabak, M. (2002). Interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants: spectroscopy and modelling. Biochim Biophys Acta. 1594:84–99.
- Gelamo, E.L., Tabak, M. (2000). Spectroscopic studies on the interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants. Spectrochim Acta A Mol Biomol Spectrosc. 56A:2255–2271.
- Kosa, T., Maruyama, T., Otagiri, M. (1997). Species differences of serum albumins: I. Drug binding sites. Pharm Res. 14:1607–1612.
- He, X.M., Carter, D.C. (1992). Atomic structure and chemistry of human serum albumin. Nature 358:209–215.
- Oester, Y. T., Keresztes-Nagy, S., Mais, R. F., Becktel, J., Zaroslinski, J. F. (1976). Effect of temperature on binding of warfarin by human serum albumin. J Pharm Sci. 65:1673–1677.
- Chakrabarti, S. K. (1978). Influence of heavy metals on the in vitro interaction between human serum albumin and warfarin. Biochem Pharmacol. 27:2957–2959.
- Wilting, J., van der Giesen, W. F., Janssen, L. H., Weideman, M. M., Otagiri, M., Perrin, J. H. (1980). The effect of albumin conformation on the binding of warfarin to human serum albumin. The dependence of the binding of warfarin to human serum albumin on the hydrogen, calcium, and chloride ion concentrations as studied by circular dichroism, fluorescence, and equilibrium dialysis. J Biol Chem. 255:3032–3037.
- Eckenhoff, R. G. (1996). Amino acid resolution of halothane binding sites in serum albumin. J Biol Chem. 271:15521–15526.
- Tirosh, O., Barenholz, Y., Katzhendler, J., Priev, A. (1998). Hydration of polyethylene glycol-grafted liposomes. Biophys J. 74:1371–1379.