Abstract
A new gut bacterial adhesion model has been developed. For this, a continuous-flow bioreactor packed with bacteria-coated beads was designed to simulate the gut lining and other features. In vitro model efficacy shows successful bacterial cell gut adhesions: bacterial adhesion was higher with mucin–alginate compared to controls. In feasibility study, adhesion of Lactobacillus fermentum NCIMB 5221 and Lactobacillus reuteri NCIMB 701359 was investigated for their metabolic activities for bile salt. Bile salt hydrolase (BSH)-active Lactobacillus reuteri exerted higher activity than non-BSH-active L. fermentum. This model has potential use in gut health, probiotic, bacterial cell gut adhesion and other delivery applications.
Introduction
Over the past decade, there has been a tremendous increase of interest for probiotic supplements. Probiotics are live microorganisms, which, when administered in adequate amounts, confer health benefits to the host (FAO/WHO, Citation2001). Lactobacillus sp. such as, L. reuteri and L. fermentum are common microorganisms used as probiotics (Martoni, Bhathena, Urbanska, & Prakash, Citation2008; Lee et al., Citation2011). Among many hypothesized mechanisms of action, it has been suggested that positive effects of probiotics might be induced through the remodeling of intestinal microbial communities and suppression of pathogens naturally present in the human gut microbiota such as, Escherichia coli (Prakash, Rodes, Coussa-Charley, & Tomaro-Duchesneau, Citation2011; DebRoy, Roberts, & Fratamico, Citation2011). Unfortunately, the study of the gut microbiotia presents methodological and ethical considerations which hamper its exploration. As a consequence, it is hardly possible to elucidate the effects of orally delivered probiotic bacteria on the bacterial microhabitats of the gastrointestinal tract in clinical trials. In order to overcome these limitations, in vitro and in vivo models are used (Rumney & Rowland, Citation1992).
In vivo models include various species of laboratory animals whose gut microbiota can be conventional, germ-free or selectively colonized. Germ-free laboratory animals refer to animals grown in a sterile environment lacking a gut microbiota. Those animals can be colonized with defined organisms, such as probiotic bacteria or with microbiota transferred from a laboratory animal or a human volunteer. Gnobiotic animals are particularly useful to study host-bacterial and bacterial-bacterial interactions (Prakash et al., Citation2011). Despite their usefulness, in vivo models present considerable technical issues which limit the investigation of the gut microbiota in its natural environment. On the opposite, in vitro models of the gut microbiota provide remarkable tools for controlled mechanistic studies. These are typically computer-controlled dynamic multi-compartmental reactors simulating the different parts of the gastrointestinal tract. Fecal inoculums from healthy volunteers, without any antibiotics history within the past 6 months, are often utilized to mimic the human gut microbiota (Prakash et al., Citation2011). Semi continuous and continuous batch culture systems provide excellent devices to study the effects of probiotics on metabolic and ecological variations of the gut microbiota over long periods (more than 2 weeks) (Martoni et al., Citation2008; Miller & Wolin, Citation1981; Manning, Federle, & Cerniglia, Citation1987; Afkhami et al., Citation2007; Molly, Vande, & Verstraete, Citation1993). However, those models do not differentiate the bacterial microhabitats of the intestines such as, the mucosal-associated and luminal bacterial communities.
An alternative to investigate the adhesion of bacterial cells to the gut lining is the use of gut adhesion models. Gut adhesion models are mostly static and involve the adhesion of different micro-organisms on microtiter plates using intestinal epithelial cell lines or immobilized mucus (Coconnier, Klaenhammer, Kerneis, Bernet, & Servin, Citation1992; Elo, Saxelin, & Salminen, Citation1991; Forestier, De Champs, Vatoux, & Joly, Citation2001; Kos et al., Citation2003; Laparra & Sanz, Citation2009; Matijasic, Narat, & Zoric, Citation2003; Ouwehand & Salminen, Citation2003; Tuomola & Salminen, Citation1998; Zarate, De Ambrosini, Chaia, & Gonzalez, Citation2002; Matsumoto, Tani, Ono, Ohishi, & Benno, Citation2002; Ouwehand et al., Citation2002; Cinquin, Le Blay, Fliss, & Lacroix, Citation2006; Macfarlane, Woodmansey, & Macfarlane, Citation2005; Probert & Gibson, Citation2004). However, those models allow for short duration studies lasting less than 24 hours. As, a consequence, the effect of probiotic bacteria on the bacterial communities of the gut lining are poorly understood. This study aimed at developing a novel continuous gut bacterial adhesion model to investigate adhesion of probiotic bacterial cells on the intestinal mucosa.
A series of experiment was performed to design, evaluate and test the novel in vitro gut adhesion model for probiotic studies on the intestinal mucosa. A continuous flow bioreactor was packed with bacteria-coated beads to simulate the gut lining. Adhesion efficacy of bacterial cells on glass, polystyrene, alginate and mucin–alginate packing beads were quantified. The material which demonstrated maximized bacterial cell adhesion was selected to design the model. The model was then evaluated for probiotic bacterial cell adhesion efficacy and activity using bile salt hydrolase (BSH)-active L. reuteri and nonactive BSH L. fermentum probiotic bacteria (Martoni et al., Citation2008). Finally, the model was tested for its feasibility to conduct probiotic studies on the intestinal mucosal-associated microbiota using potentially pathogenic E. coli and simulated human gut microbiota.
Materials and methods
Bacterial cultures and growth conditions
Lactobacillus fermentum NCIMB 5221 and Lactobacillus reuteri NCIMB 701359 were purchased from the National Collection of Industrial, Marine and food Bacteria (Aberdeen, Scotland). Escherichia coli ATCC 8739 was purchased from Cedarlane (Burlington, Canada). A 1% (v/v) bacterial inoculum of each bacterial strain was daily passaged in appropriate nutrient broth at 37°C in 5% CO2. Lactobacillus strains were grown in de Man, Rogosa and Sharpe (MRS) broth (Fisher Scientific, Ottawa, Canada). E. coli ATCC 8739 was grown in a nutrient broth comprising peptone (5 g/L), beef extract (1 g/L), yeast extract (2 g/L) and NaCl (5 g/L) (Sigma-Aldrich, Oakville, Canada).
Set up of the gut bacterial adhesion model
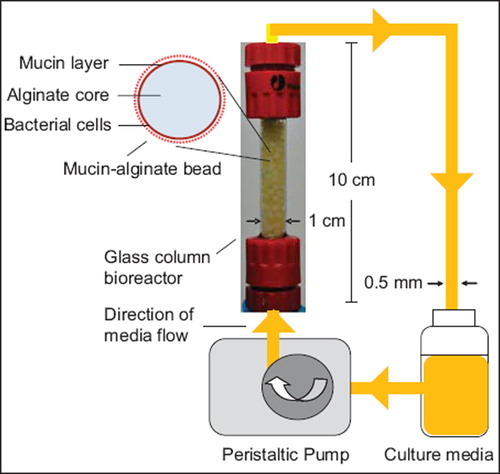
Culture media was circulated throughout a glass column bioreactor (1 × 10 cm) at a flow rate of 0.5 mL/minute using a Watson-Marlow peristaltic pump 323 E/D (Watson Marlow Pumps Group, Wilmington, USA). Bacteria-coated beads were transferred to the bioreactor under sterile conditions (). Culture media was daily replaced. The gut bacterial adhesion model was incubated at 37°C. Beads were extracted from the model at given time points for further analysis.
Figure 1. Schematic representation of the in vitro gut bacterial adhesion model using a continuous flow bioreactor packed with mucin–alginate beads. Mucin–alginate beads were composed of 2% (w/v) low-viscosity sodium alginate and 3% (w/v) mucin from porcine stomach. Beads were prepared by ionotropic gelation method. Beads were coated for 24 h with 1% (v/v) bacterial inoculum suspended in culture media, stirred at 50 rpm and incubated 24 h at 37°C in 5% CO2. Bacteria-coated beads were transferred to a glass column bioreactor under sterile conditions. Culture media was circulated throughout the system at a flowrate of 0.5 mL/min using a peristaltic pump. The whole system was incubated at 37oC.
Experimental design: development and applications of the gut bacterial adhesion model
A series of experiments was performed in order to design, evaluate and test the continuous in vitro gut adhesion model for probiotic studies. (1) The first experiment aimed at selecting an appropriate packing material for maximized bacterial cell adhesion. Effect of glass, polystyrene, alginate and mucin–alginate packing material on bacterial cell adhesion of L. fermentum NCIMB 5221 was investigated using plate counting and fluorescence microscopy. Culture media consisted of MRS broth supplemented with 0.2 M CaCl2. (2) Second, the gut bacterial adhesion model was evaluated for bacterial adhesion efficacy and activity. On time 0, the model was packed with beads coated with, respectively, BSH-active (L. reuteri NCIMB 701359) and nonactive BSH (L. fermentum NCIMB 5221) bacteria in distinct experiments. Culture media consisted of MRS broth supplemented with 0.2 M CaCl2. Bacterial cell adhesion on the beads was determined using plate counting. BSH-activity was monitored to evaluate the biological activity of the coated bacteria. (3) Third, the effect of daily administration of L. reuteri NCIMB 701359 on bacterial adhesion in the model packed with beads coated with the potentially pathogen E. coli ATCC 8739 was investigated using plate counting and scanning electron microscopy (SEM). On time 0, the model was packed with E. coli ATCC 8739-coated beads. Culture media (MRS broth supplemented with 0.2 M CaCl2) was inoculated daily with a 1% (v/v) inoculum of L. reuteri NCIMB 701359. A control test was performed without administration of L. reuteri NCIMB 701359. (4) Fourth, the model was tested for its feasibility to immobilize simulated human gut microbiota. Beads were coated with simulated human gut microbiota derived from an in vitro model of human colonic microbiotia (Rodes et al., Citation2011). Culture media consisted of a carbohydrate-rich media () supplemented with 20 mM CaCl2. Bacterial cell adhesion on the packed beads was determined using plate counting.
Table 1. Composition of the nutrient rich culture media circulating through the gut bacterial adhesion model.
Table 2. Comparison of the adhesion of simulated human gut Enterococci, Bifidobacteria, Lactobacilli and Clostridia in the gut bacterial adhesion model to available human colon data. On time 0, the gut bacterial adhesion model was packed with beads coated with simulated human gut microbiota. The model was incubated at 37 °C. Bacterial cell adhesion was determined using plate counting and expressed as log (CFU/cm2). All experiments were conducted in triplicates. Values are expressed as means ± SEM. Clinical data are presented as the range of the average composition of the human gut microbiota (Orrhage & Nord, Citation2000). There was not effect of time on bacterial cell adhesion of Enterococci, Bifidobacteria, Lactobacilli and Clostridia (p > 0.05).
Preparation of bacteria-coated beads
Packing materials tested included glass beads (3 mm, Fisher Scientific), polystyrene beads (Polyballs, 1/8” diameter, Polysciences, Niles, USA), alginate beads and mucin–alginate beads. Alginate-based beads were prepared as follows. 2% (w/v) low-viscosity sodium alginate (Sigma-Aldrich) dissolved in physiological solution (PS) (8.5 g/L NaCl) was added, dropwise from a height of 15 cm, to 0.2 M CaCl2 coating solution (Amsden & Turner, Citation1999). 3% (w/v) mucin from porcine stomach (Sigma-Aldrich) was added to the sodium alginate solution for the preparation of mucin–alginate beads. Beads were formed instantly due to ionotropic gelation and incubated overnight in 0.2 M CaCl2 at 37°C (Chang & Prakash, Citation2001). Beads were coated with 1% (v/v) bacterial inoculum suspended in culture media, stirred at 50 rpm and incubated 24 hours at 37°C in 5% CO2 for 24 hours. All solutions were heat- or filter- sterilized before use.
Evaluation of bacterial cell adhesion
Plate counting used for colony quantification
Removed beads were rinsed with PS to eliminate nonspecifically attached bacteria and vortexed in 1 mL of PS. Cell viability of Lactobacillus sp. was determined on MRS agar plates. In order to differentiate the viability of E. coli ATCC 8739 and L. reuteri NCIMB 701359, sorbitol-MacConkey (Becton Dickinson, Mississauga, Canada) and LAMVAB selective agar media were used (March & Ratnam, Citation1986; Hartemink & Rombouts, Citation1999). To quantify the viability of simulated gut microbiota genera, selective media used were enterococcus selective agar (Sigma-Aldrich) for Enterococci, LAMVAB for Lactobacilli, raffinose bifidobacterium agar for Bifidobacteria and tryptose sulphite cycloserin agar (Sigma-Aldrich) for Clostridia. Agar plates were streaked in triplicates at three different dilutions. Aerobic incubation was performed for 24 hours at 37°C in 5% CO2. Anaerobic incubation was performed in anaerobic jars with anaerobe atmosphere-generating bags (Oxoid, Nepean, Canada) for 48 hours at 37°C.
Fluorescence microscopy used for determination of live/dead bacter
A Viability Assay Kit for Bacteria Live & Dead Cells (Biotium, Hayward, USA) was used to confirm bacterial cell viability on the coated beads. Bacteria-coated beads were stained as follows: one volume of DMAO and two volumes of EtD-III were mixed thoroughly. Seven volumes of PS was added and mixed. 10 μL of the final fluorescent solution was added and incubated for 15 minutes at room temperature. Bacterial cells were observed using a fluorescent microscope (TE2000-U, Nikon, San Diego, USA). Live bacteria appeared green and dead bacteria were fluorescing red.
Microscopic observation using scanning electron microscopy
SEM was performed in order to visualize the bacteria coated on the beads using a Hitachi S-4700 FE scanning microscope. Removed beads were air-dried overnight and standard protocol for SEM was followed.
Evaluation of bile salt hydrolase activity
BSH activity was determined using a modified version of the spectrophotometric method described by Kumar et al. (Citation2006). Extracted beads were vortexed in 1 ml of PS supplemented with 30 µl protease inhibitor solution (Protease Inhibitor Cocktail Tablets, 25X stock solution, Sigma- Aldrich). The sample solution, maintained on ice, was sonicated 7 pulses at 50% power during 10 seconds using a Misonix XL2000 Ultrasonication System. The lysate was centrifuged at 20 000 × g for 10 minutes at 4°C. 20 mM GDCA (Sodium Glycodeoxycholic acid, Sigma-Aldrich), 10 mM dithiothreitol (DTT) and protease inhibitors were added to the mixture. pH was adjusted to 4.2 using 0.5M HCl. The cell-free bile salt hydrolysis reaction tube was incubated at 37°C. At time 0, 30, 60 and 90 minutes, 50 µl of the reaction mixture was removed and added to 50 µl of 15% (v/v) TCA solution to precipitate all proteins and cease the hydrolysis. The solution was centrifuged at 10 000 × g for 10 minutes at 4°C. 50 µl of the supernatant was added to 950 µl of ninhydrin solution (5:12:2 ratio by volume of 2% (v/v) ninhydrin:glycerol:0.5M sodium citrate pH 5.5). The mixture was boiled for 14 minutes and cooled at room temperature for 10 minutes. Absorbance was read at 570 nm against a standard curve of glycine concentrations using a PerkinElmer 1420 Multilabel Counter. BSH-activity was calculated as a rate of glycine release per colony-forming unit (CFU) (standardized to workable values by multiplying by 106).
Statistical analysis
All experiments were conducted in triplicates. Values were expressed as means ± SEM of triplicate experiments. D’Agostino and Pearson normality test was performed to assess Gaussian distribution of the data. Bartlett's test was performed to assess homogeneity of variances. Parametric analyses were performed using the unpaired Student's t-test and one-way ANOVA followed by Tukey's multiple comparison tests (Prism, Version 5.0, GraphPad Software, San Diego, USA). Nonparametric analyses were performed using the Mann–Whitney test or the Kruskal–Wallis test followed by Dunn's post-hoc tests (Prism, Version 5.0). The general linear model was used for the assessment of relationship between incubation time and adhesion of bacterial genera (Minitab, Version 16; Minitab Inc, Pennsylvania, USA). Statistical significance was set at p < 0.05.
Results
Design of an in vitro gut bacterial adhesion model
Set up of the gut bacterial adhesion model
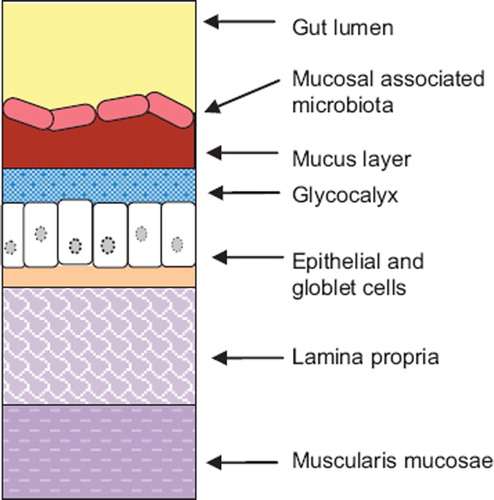
An in vitro gut bacterial adhesion model was set up () to simulate the gut lining. A glass column bioreactor was packed with bacteria-coated beads to allow for (i) maximized bacterial cell adhesion by increasing the surface area to volume ratio, and (ii) multiple continuous sampling. Culture media was circulated through the model using a peristaltic pump. The media was daily replaced. Bacteria adhered on the surface of the packing material beads simulated the mucosal-associated gut microbiota, as represented on (Johansson et al., Citation2011). Spaces between beads simulated the gut lumen which contains nutritive solution and the luminal gut microbiota.
Figure 2. Schematic representation of the anatomy of the gut lining. The gut lining is composed of a series of layer. The innermost layer consists of the muscularis mucosae. It is separated from the epithelial layer by a supportive tissue: the lamina propria. Epithelial cells are covered by the glycocalyx and a mucus viscoelastic gel layer composed mainly of mucin glycoproteins. Bacterial cells cover the mucus layer and form a complex microbial ecosystem interacting with its environment: the mucosal associated gut microbiota.
Selection of packing material for maximized bacterial cell adhesion
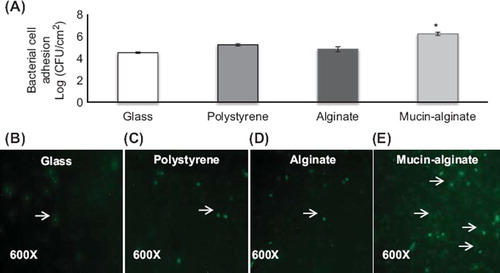
Glass, polystyrene, alginate, and mucin–alginate materials were tested for bacterial cell adhesion efficacy. Adhesion of L. fermentum NCIMB 5221 on the different surfaces was assessed after 24 hours incubation using plate counting and fluorescence microscopy. Bacterial cell adhesion averaged 4.52 ± 0.05 log (CFU/cm2) on glass, 5.24 ± 0.06 log (CFU/cm2) on polystyrene, 4.84 ± 0.13 log (CFU/cm2) on alginate beads and 6.23 ± 0.09 log (CFU/cm2) on mucin–alginate beads (). Mucin–alginate beads demonstrated increased bacterial cell adhesion compared to glass, polystyrene and alginate beads (p < 0.05). A fluorescence viability assay was performed to confirm bacterial cell viability on the surface of the beads. Fluorescence images () demonstrated higher density of viable bacteria observed on the mucin–alginate surface compared to glass, polystyrene and alginate materials. Thus, mucin–alginate beads were selected to pack the column bioreactor of the bacterial gut adhesion model.
Figure 3. Effect of glass, polystyrene, alginate and mucin–alginate packing material on bacterial cell adhesion of L. fermentum NCIMB 5221. (A) L. fermentum NCIMB 5221 cell adhesion on glass, polystyrene, alginate and mucin–alginate packing material was measured by plate counting (log (CFU/cm2)). (B-D) Live/dead fluorescence assay images of L. fermentum NCIMB 5221 on (B) glass, (C) polystyrene, (D) alginate and (E) mucin–alginate packing materials. Viable cells fluoresced green (as indicated with an arrow). Original magnification was 600X. A 1% (v/v) bacterial inoculum of L. fermentum NCIMB 5221 was added to, respectively, glass, polystyrene, alginate and mucin–alginate beads suspended in MRS broth supplemented with 0.2 M CaCl2. Beads in solution were incubated 24 h at 37°C in 5% CO2 and stirred at 50 rpm. All experiments were conducted in triplicates. Values are expressed as means ± SEM. *Indicates statistical significance (p < 0.05).
Evaluation of the gut bacterial adhesion model: bacterial cell adhesion and activity
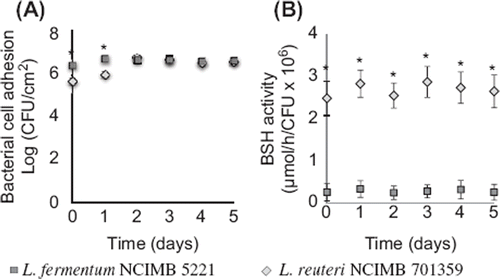
In order to determine bacterial cell adhesion efficacy and activity in the gut bacterial adhesion model, viability and BSH-activity of L. reuteri NCIMB 701359 (BSH-active) and L. fermentum NCIMB 5221 (nonactive BSH) bacteria, coated on packed mucin–alginate beads, were determined over 5 days in distinct experiments. Bacterial cell adhesion averaged 6.32 ± 0.25 log (CFU/cm2) for L. reuteri NCIMB 701359 and 6.58 ± 0.06 log (CFU/cm2) for L. fermentum NCIMB 5221 (). Bacterial cell adhesion of L. reuteri NCIMB 701359 and L. fermentum NCIMB 5221 demonstrated no significant statistical difference after 1 day of incubation in the model (p > 0.05). BSH-activity of L. reuteri NCIMB 701359 in the gut bacterial adhesion model was continuously high, with an average rate of 2.70 ± 0.20 µmol/h/CFU × 106 over 5 days (). On the opposite, BSH production of L. fermentum NCIMB 5221 averaged 0.28 ± 0.09 µmol/h/CFU × 106 over 5 days. BSH production was significantly higher with L. reuteri NCIMB 701359 as compared to L. fermentum NCIMB 5221 (p < 0.05).
Figure 4. Bacterial cell adhesion and BSH-activity of L. reuteri NCIMB 701359 (BSH-active) and L. fermentum NCIMB 5221 (nonactive BSH) coated on mucin–alginate beads packed in the gut bacterial adhesion model. (A) Bacterial cell adhesion was determined using plate counting and expressed as log (CFU/cm2). (B) BSH-activity was determined using spectrophotometry and expressed as the rate of glycine release per CFU (µmol/h/CFU 3 106). On day 0, the gut bacterial adhesion model was packed with mucin–alginate beads coated with, respectively, L. reuteri NCIMB 701359 and L. fermentum NCIMB 5221 in distinct studies. The model was incubated at 37°C. Culture media was replaced daily. All experiments were conducted in triplicates. Values are expressed as means ± SEM. *Indicates statistical (p < 0.05).
Applications of the bacterial gut adhesion model
Effect of a probiotic bacteria on the colonization of potentially pathogenic bacteria
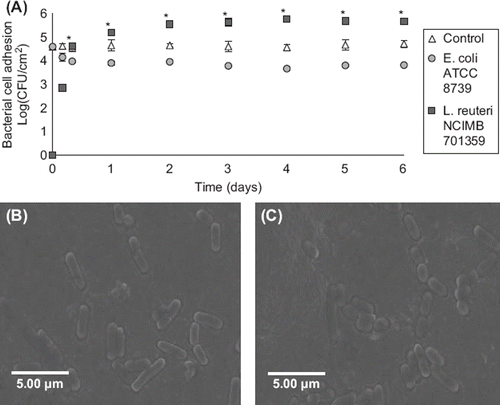
The effect of daily administration of L. reuteri NCIMB 701359 on bacterial cell adhesion in the gut model packed with potentially pathogenic E. coli ATCC 8739-coated beads was investigated over a 6-day period. Bacterial cell adhesion of L. reuteri NCIMB 701359 on mucin–alginate beads rapidly increased to reach a plateau phase on day (5.65 ± 0.05 log (CFU/cm2) (). Adhesion of L. reuteri NCIMB 701359 significantly exceeded adhesion of E. coli ATCC 8739 after 8 hours incubation in the gut bacterial adhesion model (p < 0.05). The effect was maintained during the duration of the experiment (p < 0.05). Bacterial cell adhesion of potentially pathogenic E. coli ATCC 8739 decreased by 9.67 ± 1.88% within 4 hours when exposed to L. reuteri NCIMB 701359l. Lower adhesion of E. coli ATCC 8739 was maintained over a 6-day period at 3.84 ± 0.06 log (CFU/cm2), but the effect was not significant compared to control. SEM images confirmed the adhesion of bacterial cells on the surface of mucin–alginate beads in the gut bacterial adhesion model during the course of the experiment ().
Figure 5. Effect of daily administration of L. reuteri NCIMB 701359 on bacterial cell adhesion in a gut bacterial adhesion model packed with mucin–alginate beads coated with E. coli ATCC 8739. (A) Bacterial cell adhesion was determined using plate counting and expressed as log (CFU/cm2). (B-C) Scanning electron microscope (SEM) images of mucin–alginate beads coated with E. coli ATCC 8739: (B) Control after 1 day incubation and (C) 1 day incubation after administration of L. reuteri NCIMB 701359. On time 0, the model was packed with mucin–alginate beads coated with E. coli ATCC 8739. Culture media was inoculated daily with a 1% (v/v) inoculum of L. reuteri NCIMB 701359. The model was incubated at 37°C. In the control, there was no administration of L. reuteri NCIMB 701359. All experiments were conducted in triplicates. Values are expressed as means ± SEM. *Indicates statistical significance (p < 0.05).
Immobilization of simulated human gut microbiota in the gut bacterial adhesion model
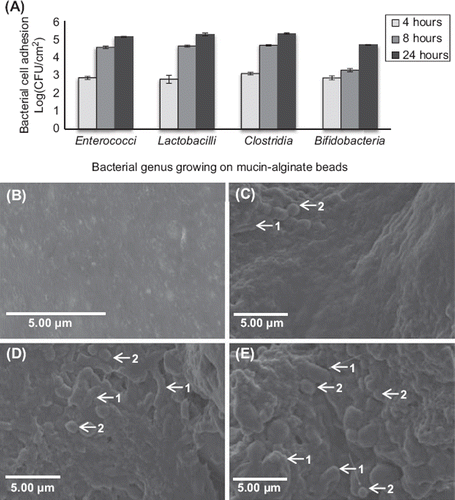
The gut bacterial adhesion model was tested for its feasibility to immobilize simulated human gut microbiota. In a first step, bacterial adhesion of simulated human gut microbiota on mucine–alginate beads was monitored during the coating procedure. Plate counting results demonstrated that bacterial adhesion of Enterococci, Bifidobacteria, Lactobacilli and Clostridia on mucin-alginate beads increased in a time dependent manner over 24 hours (). Precisely, bacterial cell adhesion on the surface of mucin–alginate beads after 4 hours incubation was low for all genera, ranging from 2.77 ± 0.13 log (CFU/cm2) for Lactobacilli to (3.09 ± 0.04 log (CFU/cm2)) for Clostridia. After 8 hours, bacterial immobilization was highly increased for Enterococci (4.56 ± 0.04 log (CFU/cm2)), Lactobacilli (4.62 ± 0.03 log (CFU/cm2)) and Clostridia (4.66 ± 0.02 log (CFU/cm2)). On the opposite, adhesion of Bifidobacteria remained relatively low (3.28 ± 0.05 log (CFU/cm2)) but highly increased to (4.69 ± 0.02 log (CFU/cm2)) after 24 hours incubation. SEM observation confirmed the growth of diverse populations of bacteria including rounded-shaped cocci and elongated-shaped bacilli (). The mucin–alginate scaffold was totally covered by bacterial cells after 8 hours of incubation.
Figure 6. Effect of coating duration on the adhesion of simulated human gut microbiota on mucin–alginate beads. (A) Bacterial cell adhesion during the bead coating process was determined using plate counting and expressed as log (CFU.cm‐ 2). (B-E) Scanning electron microscope (SEM) images of mucin–alginate beads coated with simulated human gut microbiata after: (B) 0 hour, (C) 4 hour, (D) 8 hour and (E) 24 hour incubation. Elongated-shaped bacilli are labeled 1 and rounded-shaped cocci are labeled 2. On time 0, mucin–alginate beads were coated with a 1% (v/v) bacterial inoculum of simulated human gut microbiota. All experiments were conducted in triplicates. Values are expressed as means ± SEM.
In a second step, bacterial cell adhesion of simulated human gut microbiota coated on mucin–alginate beads was evaluated in the gut bacterial adhesion model over a period of 16 days. There was not effect of time on bacterial cell adhesion of Enterococci, Bifidobacteria, Lactobacilli and Clostridia (p > 0.05) indicating quantitative stability of the bacterial populations in the adhesion model (). Bacterial cells adhesion averaged 5.20 ± 0.09 log (CFU/cm2) for Enterococci, 4.38 ± 0.05 log (CFU/cm2) for Bifidobacteria. 5.59 ± 0.13 log (CFU/cm2) for Lactobacilli and 5.38 ± 0.05 log (CFU/cm2) for Clostridia which corresponded well with clinical data (Orrhage & Nord, Citation2000).
Discussion
The gut mucosa constitutes the interface between the gut microbiota and host tissues. It is composed predominantly of intestinal cells, a mucus layer produced by goblet cells, and the mucosal-associated gut microbiota. Interaction between these components compose a dynamic equilibrium involved in the functioning of the immune system and maintaining human health (DebRoy et al., Citation2011; Johansson et al., Citation2011; Perdigon, Locascio, Medici, Pesce de Ruiz, & Oliver, Citation2003). Despite the growing interest for probiotic bacteria and their effect on the gastrointestinal system, very little is known about biofilms formation on the gut lining. Biofilms are defined as microbial accretions that adhere to surfaces and represent a significant mode of growth for bacteria (Hall-Stoodley, Costerton, & Stoodley, Citation2004). As a consequence, in vitro models have been developed to investigate the adhesion of bacterial cells on the gut lining (Cinquin et al., Citation2006; Macfarlane et al., Citation2005; Probert & Gibson, Citation2004). However, those models may not be sufficient to describe the adhesion capacity of bacteria onto the gut mucosa during long term experiments, and often ignore the relations between luminal and mucosal microbial populations. In this study, a novel gut bacterial adhesion model was developed to investigate mucosal adhesion of bacterial cells for probiotic applications.
The aim of the study was threefold. (i) The first experiment aimed at designing the gut bacterial adhesion model for maximized adhesion of bacterial cells. A continuous-flow bioreactor was packed with bacteria-coated beads to simulate the gut lining. Plate counting and fluorescence assay demonstrated that mucin–alginate material provided a statistically superior platform for bacterial cell adhesion of L. fermentum, a commonly used probiotic bacteria (Lee et al., Citation2011). Thus, the model was further packed with mucin–alginate beads. Bacteria adhered on the surface of the packing material beads simulated the mucosal-associated gut microbiota. Spaces between beads simulated the intestinal lumen. The continuous flow of nutritive media was representative of the intestinal transit. (ii) The second experiment aimed at evaluating the model for bacterial cell adhesion efficacy and activity using BSH-active (L. reuteri) and nonactive BSH (L. fermentum) probiotic bacteria (Martoni et al., Citation2008). BSH-active bacteria have been suggested to lower cholesterol levels through the enzymatic deconjugation of bile salts (Martoni et al., Citation2008; Gilliland, Nelson, & Maxwell, Citation1985; Grunewald, Citation1982). Results showed that L. reuteri and L. fermentum demonstrated high adhesion capacities in the continuous gut adhesion model over a 5-day period. Biological activity of the BSH-active bacteria was significantly maintained through the course of the experiment, as opposed to the nonactive BSH bacteria. Therefore, this novel gut bacterial adhesion model allows to maintain bacterial cell adhesion and activity during long-term experiments. (iii) The last experiments aimed at testing the gut bacterial adhesion model for its feasibility to conduct probiotic studies on the intestinal mucosal-associated microbiota. The effect of a probiotic daily administration (L. reuteri) on bacterial adhesion in the model packed with potentially pathogenic E. coli-coated beads was investigated during 6 days. Plate counting and SEM results showed that adhesion of L. reuteri significantly increased to exceed adhesion of E. coli after 8 hours (DebRoy et al., Citation2011). On the opposite, adhesion of potentially pathogenic E. coli decreased within 4 hours, but the effect was not significant compared to control. Previous publications have reported that probiotic bacteria can reduce the viability of pathogenic bacteria. However, these studies were performed over very short incubation times, typically in the range of 1–1.5 hours (Candela et al., Citation2008; Collado, Meriluoto, & Salminen, Citation2007; Lee, Puong, Ouwehand, & Salminen, Citation2003). Finally, the novel gut bacterial adhesion model was tested for its feasibility to immobilize simulated human gut microbiota. Bacterial cell adhesion of Enterococci, Bifidobacteria, Lactobacilli and Clostridia corresponded well with clinical data. Viability of the bacterial populations in the model was somewhat lower but this is in accordance with previous results (Possemiers, Verthe, Uyttendaele, & Verstraete, Citation2004). In addition, there was no significant effect of time on bacterial adhesion of Enterococci, Bifidobacteria, Lactobacilli and Clostridia. We conclude that this gut bacterial adhesion model allows to maintain bacterial adhesion of simulated human gut microbiota over long time periods. The model is suitable to investigate the effect of probiotic bacteria on mucosal and luminal simulated human gut microbiota.
In this study, an in vitro gut bacterial adhesion model was designed to investigate mucosal adhesion of bacterial cells. A continuous flow bioreactor packed with mucin–alginate beads was set up to mimic the mucosal and luminal components of the gut lining. Findings demonstrated that the model provides an appropriate device to perform studies on the mucosal adhesion of bacterial cells. This novel gut bacterial adhesion model can be used as an alternative device to investigate mucosal adhesion of bacterial cells for probiotic, prebiotic and antibiotic applications.
Acknowledgements
This work was supported by a research grant from the Canadian Institutes of Health Research (CIHR) MOP # 64308 to S. Prakash. L. Rodes acknowledges the Doctoral Training Award from the Fonds de Recherche Santé Québec (FRSQ). A. Paul acknowledges the Alexander Graham Bell Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council (NSERC) and the FRSQ Postdoctoral Training Award. S. Abbasi acknowledges the McGill Faculty of Medicine Internal Studentship-GG Harris Fellowship. C. Tomaro-Duchesneau acknowledges the Alexander Graham Bell Canada Graduate Scholarship from NSERC.
Declaration of interest
The author reports no conflicts of interest in this work. The authors alone are responsible for the content and writing of the article.
References
- Afkhami F, Ouyang W, Chen H, Lawuyi B, Lim T, Prakash S. 2007. Impact of orally administered microcapsules on gastrointestinal microbial flora: in-vitro investigation using computer controlled dynamic human gastrointestinal model. Artif Cells Blood Substit Immobil Biotechnol. 35:359–375.
- Amsden B, Turner N. 1999. Diffusion characteristics of calcium alginate gels. Biotechnol Bioeng. 65:605–610.
- Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P, Rizzello F, . 2008. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol. 125:286–292.
- Chang TM, Prakash S. 2001. Procedures for microencapsulation of enzymes, cells and genetically engineered microorganisms. Mol Biotechnol. 17:249–260.
- Cinquin C, Le Blay G, Fliss I, Lacroix C. 2006. Comparative effects of exopolysaccharides from lactic acid bacteria and fructo-oligosaccharides on infant gut microbiota tested in an in vitro colonic model with immobilized cells. Fems Microbiol Ecol. 57: 226–238.
- Coconnier MH, Klaenhammer TR, Kerneis S, Bernet MF, Servin AL. 1992. Protein-Mediated Adhesion of Lactobacillus-Acidophilus Bg2Fo4 on Human Enterocyte and Mucus-Secreting Cell-Lines in Culture. App Environ Microbiol. 58:2034–2039.
- Collado MC, Meriluoto J, Salminen S. 2007. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett Appl Microbiol. 45:454–460.
- DebRoy C, Roberts E, Fratamico PM. 2011. Detection of O antigens in Escherichia coli. Anim Health Res Rev. 12:169–185.
- Elo S, Saxelin M, Salminen S. 1991. Attachment of Lactobacillus-Casei Strain Gg to Human Colon-Carcinoma Cell-Line Caco-2 - Comparison with Other Dairy Strains. Lett Appl Microbiol. 13:154–156.
- FAO/WHO 2001. Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria. Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Basel, Switzerland: World Heatlh Organization; 2001 Oct.
- Forestier C, De Champs C, Vatoux C, Joly B. 2001. Probiotic activities of Lactobacillus casei rhamnosus: in vitro adherence to intestinal cells and antimicrobial properties. Res Microbiol. 152:167–173.
- Gilliland SE, Nelson CR, Maxwell C. 1985. Assimilation of cholesterol by Lactobacillus acidophilus. Appl Environ Microbiol. 49:377–381.
- Grunewald KK 1982. Serum-cholesterol levels in rats fed skim milk fermented by Lactobacillus-acidophilus. J Food Sci. 47: 2078–2079.
- Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2:95–108.
- Hartemink R, Rombouts FM. 1999. Comparison of media for the detection of bifidobacteria, lactobacilli and total anaerobes from faecal samples. J Microbiol Methods. 36:181–192.
- Johansson ME, Ambort D, Pelaseyed T, Schutte A, Gustafsson JK, Ermund A, Subramani DB, . 2011. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci. 68: 3635–3641.
- Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosic S. 2003. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 94:981–987.
- Kumar RS, Brannigan JA, Prabhune AA, Pundle AV, Dodson GG, Dodson EJ, Suresh CG. 2006. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J Biol Chem. 281:32516–32525.
- Laparra JM, Sanz Y. 2009. Comparison of in vitro models to study bacterial adhesion to the intestinal epithelium. Lett Appl Microbiol. 49:695–701.
- Lee YK, Puong KY, Ouwehand AC, Salminen S. 2003. Displacement of bacterial pathogens from mucus and Caco-2 cell surface by lactobacilli. J Med Microbiol. 52:925–930.
- Lee J, Yun HS, Cho KW, Oh S, Kim SH, Chun T, Kim B, . 2011. Evaluation of probiotic characteristics of newly isolated Lactobacillus spp.: immune modulation and longevity. Int J Food Microbiol. 148:80–86.
- Macfarlane S, Woodmansey EJ, Macfarlane GT. 2005. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl Environ Microbiol. 71:7483–7492.
- Manning BW, Federle TW, Cerniglia CE. 1987. Use of a semicontinuous culture system as a model for determining the role of human intestinal microflora in the metabolism of xenobiotics. J Microbiol Meth. 6:81–94.
- March SB, Ratnam S. 1986. Sorbitol-MacConkey medium for detection of Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol. 23:869–872.
- Matijasic BB, Narat M, Zoric M. 2003. Adhesion of two Lactobacillus gasseri probiotic strains on Caco-2 cells. Food Tech Biotechnol. 41:83–88.
- Matsumoto M, Tani H, Ono H, Ohishi H, Benno Y. 2002. Adhesive property of Bifidobacterium lactis LKM512 and predominant bacteria of intestinal microflora to human intestinal mucin. Curr Microbiol. 44:212–215.
- Martoni C, Bhathena J, Urbanska AM, Prakash S. 2008. Microencapsulated bile salt hydrolase producing Lactobacillus reuteri for oral targeted delivery in the gastrointestinal tract. Appl Microbiol Biotechnol. 81:225–233.
- Miller TL, Wolin MJ. 1981. Fermentation by the human large intestine microbial community in an in vitro semicontinuous culture system. Appl Environ Microbiol. 42:400–407.
- Molly K, Vande WM, Verstraete W. 1993. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol. 39:254–258.
- Orrhage K, Nord CE. 2000. Bifidobacteria and lactobacilli in human health. Drugs Exp Clin Res. 26:95–111.
- Ouwehand AC, Salminen S, Tolkko S, Roberts P, Ovaska J, Salminen E. 2002. Resected human colonic tissue: new model for characterizing adhesion of lactic acid bacteria. Clin Diagn Lab Immunol. 9:184–186.
- Ouwehand AC, Salminen S. 2003. In vitro adhesion assays for probiotics and their in vivo relevance: a review. Microb Ecol Health Dis. 15:175–184.
- Ouwehand AC, Kirjavainen PV, Gronlund M-M, Isolauri E, Salminen SJ. 1999. Adhesion of probiotic micro-organisms to intestinal mucus. Int Dairy J. 9:623–630.
- Perdigon G, Locascio M, Medici M, Pesce de Ruiz HA, Oliver G. 2003. Interaction of bifidobacteria with the gut and their influence in the immune function. Biocell. 27:1–9.
- Possemiers S, Verthe K, Uyttendaele S, Verstraete W. 2004. PCR- DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 49:495–507.
- Prakash S, Rodes L, Coussa-Charley M, Tomaro-Duchesneau C. 2011. Gut microbiota: next frontier in understanding human health and development of biotherapeutics. Biologics. 5:71–86. Epub 2011 Jul 11:71–86.
- Probert HM, Gibson GR. 2004. Development of a fermentation system to model sessile bacterial populations in the human colon. Biofilms. 1:13–19.
- Rodes L, Paul A, Coussa-Charley M, Al-Salami H, Tomaro-Duchesneau C, Fakhoury M, Prakash S. 2011. Transit Time Affects the Community Stability of Lactobacillus and Bifidobacterium Species in an In Vitro Model of Human Colonic Microbiotia. Artif Cells Blood Substit Immobil Biotechnol. 39:351–356.
- Rumney CJ, Rowland IR. 1992. In vivo and in vitro models of the human colonic flora. Crit Rev Food Sci Nutr. 31:299–331.
- Tuomola EM, Salminen SJ. 1998. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int J Food Microbiol. 41:45–51.
- Zarate G, De Ambrosini VM, Chaia AP, Gonzalez S. 2002. Some factors affecting the adherence of probiotic Propionibacterium acidipropionici CRL 1198 to intestinal epithelial cells. Can J Microbiol. 48:449–457.