Abstract
The statistical atlas is a 3D medical image analysis tool to enable more patient-oriented and efficient diagnosis. The atlas includes information on geometry and its variation across populations. The comparison with information from other patients is very useful for objective quantitative diagnosis. The statistical atlas can also be used to solve other challenging problems such as image segmentation. As a key to the construction of statistical atlases, 3D registration remains an important yet unsolved problem in the medical image field due to the geometrical complexity of anatomical shapes and the computational complexity arising from the enormous size of volume data.
In this work we developed a two-level framework to efficiently solve 3D non-rigid registration, and applied the method to the problem of constructing statistical atlases of the femur. In contrast to a general multi-resolution framework, we employed an interpolation to propagate the matching instead of repeating the registration scheme in each resolution. The registration procedure is divided into two levels: a low-resolution solution to the correspondences and mapping of surface models using Chui and Rangarajan's thin-plate spline (TPS)-based algorithm, followed by an interpolation to achieve high-resolution matching. Next, principal component analysis (PCA) is used to build the statistical atlases. Experimental results show the shape variation learned from the atlases, and also demonstrate that our method significantly improves the efficiency of registration without decreasing the accuracy of the atlases.
Introduction
Statistical atlases are used as references in interpreting CT and MRI images Citation[1], and represent the shape or appearance of human anatomical structures Citation[2], Citation[3]. Some atlases are based on physical properties, such as elastic models Citation[4], Citation[5], “snakes” Citation[6], geometric splines Citation[7], and finite element-based models Citation[8]. Others are modeled from the statistical perspective: Szeliski Citation[9] introduced the statistical atlas to analyze shape variation between patients. To analyze shape and appearance variation, principal component analysis (PCA) is widely used Citation[10], Citation[11], Citation[12]. The two most common statistical atlases are the shape atlas Citation[13] and the appearance atlas Citation[14]. The shape atlas uses only geometric information such as landmarks, surfaces (boundaries of 3D objects) or crest lines Citation[3], while the appearance atlas uses both geometric features and the intensity of pixels or voxels.
As a key to the construction of atlases, 3D registration has been studied for many years in the context of computer vision, but remains a critical problem in the medical image field due to the geometrical complexity of anatomical shapes and the computational complexity arising from the enormous size of volume data. Three-dimensional registration has numerous clinical applications, such as statistical atlas construction for group study and statistical parameter analysis Citation[15], Citation[16], mapping anatomical atlases to individual patient images for disease analysis Citation[17], Citation[18], Citation[19], and image segmentation Citation[20], Citation[21].
Depending on the type of transformation involved, registration can be rigid or non-rigid. If the shape does not change between the two images, registration should be rigid, such as in intra-subject inter-modality (same patient; different imaging system) registrations, where images are captured at the same time. However, when we take into account the time, i.e., when two images are captured at different times, as in intra-subject registrations, most are non-rigid due to the shape variation of the anatomical structures caused by swelling, prostate probing, bone fractures, tumor growth, intestinal movements and so on. In addition, inter-subject (different patients) registrations are usually non-rigid because of the local anatomic difference between patients. So far, non-rigid (or deformable) registration remains a challenging problem Citation[22].
Non-rigid registration is used to find a non-rigid transformation from one 3D surface to the reference surface by minimizing the distance between two surfaces. In general, a non-rigid transformation is represented by a global rigid or affine transformation plus a local non-linear deformation, which can be represented by radial basis functions (RBFs) Citation[23], octree splines Citation[24], thin-plate splines Citation[25], Citation[15], geometric splines Citation[7], finite elements Citation[8], or free-form B-splines Citation[12], etc. To evaluate the results of registration, different similarity measurements are selected according to the different image features and imaging modalities. For example, the sum of squared distance (SSD) is usually used for geometric features Citation[26], but correlation coefficients (CC) Citation[27], Ratio Image Uniformity (RIU) Citation[28] and mutual information (MI) Citation[29] are used for intensity features. Registration can be simplified given known correspondences, e.g., when using markers Citation[30]; however, markers are not permissible or even available in many scenarios. Alternate estimations of correspondences and transformations are widely used for both rigid cases Citation[26] and non-rigid cases Citation[15], Citation[25], Citation[31]. Moreover, with the increase in data size and geometrical complexity, a multi-resolution strategy has been adopted for the registration framework Citation[19], Citation[32], Citation[33]. Sparse matrices are also used to handle the computational complexity Citation[34].
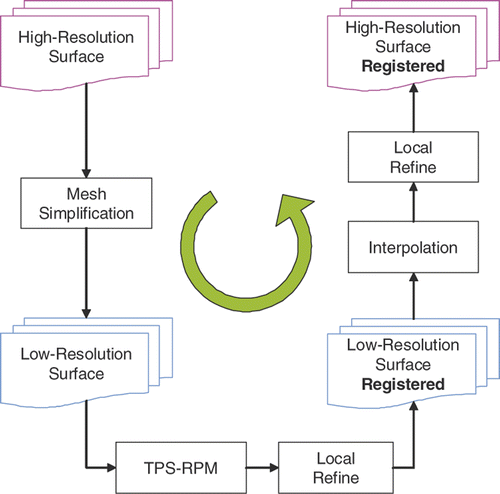
To build a femur statistical atlas given partial 3D surfaces, we developed a two-level approach inspired by Chui and Rangarajan's thin-plate spline-based algorithm Citation[25] and the previous multi-resolution work Citation[33]. Since Chui and Rangarajan's algorithm is unable to handle more than 2,000 3D points Citation[34], we broke down registration into a two-level process to deal with both the computational and geometrical complexity. We first applied the algorithm to the simplified low-resolution surfaces. To improve the efficiency, instead of successively matching each resolution from coarse to fine, we directly propagated the correspondences from low resolution to high resolution by interpolation. A local refine procedure was introduced for both low-resolution and high-resolution surfaces to improve matching. Finally, we applied PCA to the aligned surfaces to construct the femur atlas. shows a flowchart of our two-level framework.
Two-level framework
Mesh simplification: decreased resolution
Garland and Heckbert's quadric error metrics (QEM)-based mesh simplification Citation[35] method was used to compute low-resolution surfaces. QEM is based on iterative contraction of vertex pairs. The cost of contraction is represented by a quadric error, and the whole process is an iterative minimization of the quadric error. Since QEM provides a fast and simple way to guide the entire process with relatively minor storage costs, the simplification step is extremely fast.
An important parameter involved in the simplification process is the number of vertices on the low-resolution surfaces. To maintain the trade-off between accuracy and efficiency, an appropriate number is selected based on a series of leave-one-out experiments (for details see the Selection of simplification parameters section below).
Low-resolution non-rigid registration: point-to-point
We applied Chui and Rangarajan's non-rigid registration method Citation[25] to the simplified surfaces. With this method, fuzzy correspondences and a smoother optimization process can be achieved. A dual update strategy conjoined with a deterministic annealing technique is adopted to estimate the correspondences and transformation alternately. The non-rigid transformation is parameterized using thin-plate splines to generate a smooth spatial mapping.
Initial alignment
Given the particularity of the femur data, it is necessary to perform a pre-alignment to eliminate the effect caused by the missing shape. We obtained CT scans of 87 patients, all with healthy femurs, using the same scanner. As most surgeries are performed on the joints, only the top portion including the femoral head and the bottom portion including the condyles were scanned to build surfaces.
In , the bottom portion of the femur is used as an example. It can be seen that surface Y has more femur shaft than surface X. If we were to simply align both centers as in previous work Citation[25], experiments have shown that the registration process would be very slow and might not converge in several cases. The reason for this is that part of surface Y (bounded by blue in ) has no counterpart on surface X. Therefore, in order to improve upon the process, it was decided to estimate the pseudo-center of Y instead of the true center. The poses of the two surfaces were then estimated and aligned.
Figure 2. Illustration of the rigid transformation from surface X to surface Y. The upper row compares the translated X with Y. Black points on Y are used to compute the pseudo-center. The lower row compares the translated and rotated X with Y. Red axes represent the pose of X, green axes that of Y.
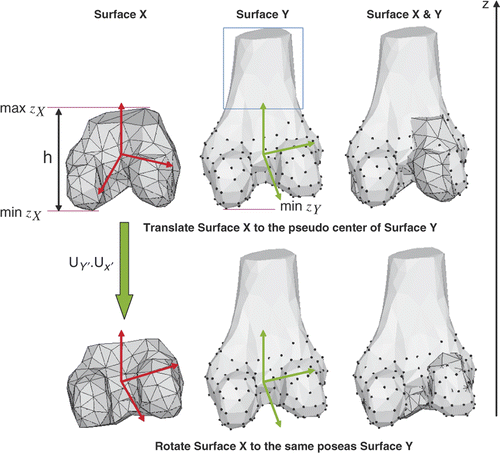
The height of X was used to estimate the pseudo-height of Y. Assuming the axis 𝒵 is in the scan direction from knee to hip, we selected those points bounded by the pseudo-height of Y (indicated in black in ) to estimate the pseudo-center κY ′ and the covariance matrices for the point set {pX} and {pY ′}:where NY ′ is the number of points {pY ′} on Y which satisfy (zY − min zY) < (max zX − min zX). We can solve for the principle axes by decomposing the covariance matrix using the moment analysis:
Each column of UX represents a principle axis of the point set {pX}, and UY ′ for {pY ′}. As shown in , three axes represent the pose of the point set: red for {pX} and green for {pY ′}. The rotation from coordinates of X to Y ′ is computed by UY ′ · UX′. We thus applied a rigid transformation [UY ′ · UX′|(κY ′ − κX)] to the point set {pX} and the two point sets were finally aligned. Experiments have shown that such alignment increases the rate of convergence from 78% to 95.2%.
Local refining procedure: point-to-surface
The result of the registration in the preceding sub-section can be further improved by minimizing the point-to-surface distance. This idea is illustrated by . xi is a vertex on the deformed surface X, whose corresponding vertex on surface Y is yi. The neighboring triangles which share the same vertex yi are S1, S2 and S3. We can compute the distance from xi to each neighboring triangle (the distance computed from the vertex xi to the plane where the triangle lies), i.e., d1, d2 and d3. If any of these is smaller than d0 = ‖xi − yi‖, we use the corresponding projected point to replace yi in order to achieve a smaller surface distance.
Figure 3. Illustration of the local refine procedure between the vertex and surface. xi is a vertex on the deformed surface X, whose correspondence on the surface Y is yi. The projection of xi to each triangle St sharing yi is denoted by . dt is the distance between xi and
(t = 1, 2, 3, …).
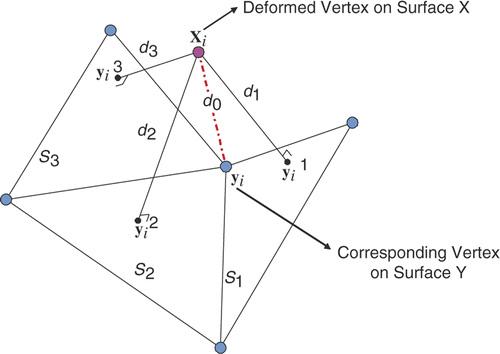
For those cases where different vertices on the surface X correspond to the same surface point on Y, we assign this corresponding surface point to the vertex on X with the smallest distance and mark it as unavailable to other vertices on X.
Low-resolution to high-resolution interpolation
Once we obtained the correspondences in low resolution (XL and YL), we propagated them to high resolution (XH and YH) by interpolation. The surface interpolation method is a derivative of methods known jointly as “moving least squares” Citation[36]. Radial basis functions (RBFs), finite elements, and multivariate splines, such as thin-plate splines (2D bivariate splines) and triharmonic thin-plate splines, are popular techniques used in surface interpolation. Carr et al. Citation[37] applied the multivariate splines method to RBFs by using splines as kernel functions. In the present work, we chose a Gaussian kernel due to its simple mathematical representation and fewer restrictions on nodes Citation[37]. More specifically, we used a linear affine function plus a series of RBFs to construct the interpolation function:where
is a vertex on the low-resolution surface XL whose correspondence on the low-resolution surface YL is
, i = 1, 2, …, N (N is the number of vertices on XL).
and
are both 3 × 1 vectors with three coordinates. c1 is a 3 × N coefficient matrix of RBFs.
is a symmetric RBF. We selected a Gaussian kernel φ(ui, uj) =exp(−‖ui − uj‖/0.5), as suggested by Pighin et al. Citation[38]. c2 and c3 are coefficients for the affine transformation. c2 is a 3×1 vector and c3 is a 3×3 matrix. Given N correspondences, there are N equations for each axis (𝒳, 𝒴 and 𝒵):
where c1k, c2k and c3k denote the kth row of c1, c2 and c3, respectively.
denotes the kth row of
, where k can be 1, 2 or 3, corresponding to the axes 𝒳, 𝒴, and 𝒵. There are 3N equations in total:
P is a 3N × (N + 4) matrix. To ensure smooth interpolation, additional orthogonality constraints
Citation[39] were added to Equation 5, where c1,i denotes the ith column of c1:
The least-squares solution for this linear system, Qc = w, is given by c = (QTQ)−1QTw.
Finally, the correspondence of a vertex in the high-resolution surface XH can be computed by using Equation 4:
, for j = 1, …, M (M is the number of vertices on XH). We also refined the resulted high-resolution surface by the procedure described in the preceding sub-section.
Femur atlas construction
Given the aligned surfaces, rigid pose alignments are applied to eliminate the effect of imaging poses Citation[40] prior to atlas construction. Suppose we have K aligned surfaces and each surface is represented by a 3M × 1 vector vi(i = 1, …, K), where M is the number of points on each surface and 3 denotes three coordinates 𝒳, 𝒴, and 𝒵. We compute the mean vector κ and covariance matrix Ψ, and then apply PCA to the data set:
Therefore, any surface model in the data set can be represented by a mean vector plus a linear combination of principal components (modes):where ηi is a K × 1 coefficient vector obtained by projecting vi onto each principal axis. New surface models not included in the data set can be generated by manipulating the coefficient ηi.
Selection of simplification parameters
In the two-level framework, the resolution of the simplified surfaces affects the final result. The smaller the number of points, the faster low-resolution registration is achieved, but results for high-resolution registration are less accurate. To maintain both accuracy and efficiency, an appropriate number is selected based on a series of leave-one-out experiments. For some extreme cases with symmetry shapes, fewer points may result in a perfect match in the low-resolution registration but a misalignment in the high-resolution registration. Therefore, not only the resolution parameter but also some landmarks are necessary to handle such ambiguity. Since the shape of the femur is already complex enough, we do not consider such extreme situations, and thus do not require any landmarks in our method.
Let denote the number of vertices on the low-resolution surfaces. For each surface vi(i = 1, …, K), K = 87, we used other K − 1 surfaces to construct the atlas using Equations 8 and 9. Let
denote the first S columns of the principal component matrix Ui, which constitutes 95% of the shape variation. Then, vi(i = 1, …, K) can be reconstructed by this atlas:
We compared the surface distance between the original surface vi and the reconstructed surface by computing the surface mean error and root mean square (RMS) error. We repeated this procedure for each surface and computed the average mean error and RMS error based on K leave-one-out experiments:
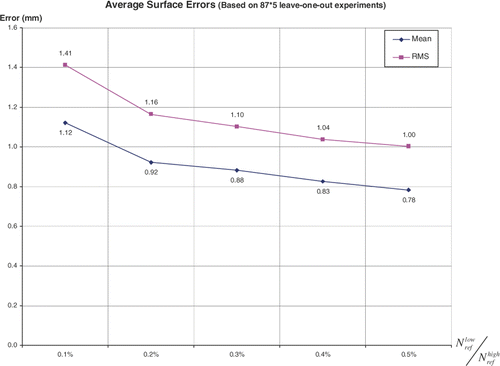
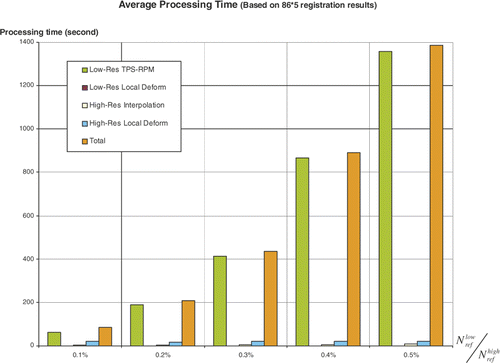
By tuning the number , we compared the error
and
and processing time
for the two-level registration as well. shows when
,
will be less than 1 mm, which is a practical number for clinical applications. shows that when
, the average processing time for the two-level registration will exceed 5 minutes (using a 2.4-GHz Pentium PC with 1 GB of RAM). Therefore,
was finally used in our algorithm to construct femur atlases and estimate the error distributions illustrated in .
Experiments and discussion
Over a period of several years, we obtained CT scans of 87 patients, all with healthy femurs, at the West Pennsylvania Hospital and Shadyside Hospital in Pittsburgh, Pennsylvania. The patients comprised 53 males and 34 females ranging in age from 39 to 78 years old; 43 left femurs and 44 right femurs were scanned, and the femur length ranged from 400 mm to 540 mm ( shows the data distribution in terms of age and femur length). The CT volumes were segmented to provide triangulated surface models using the Marching Cubes (MC) algorithm. All surface models were smoothed by the method proposed in reference Citation[41]. Given that most surgeries are performed on the joints, only the top portion including the femoral head and the bottom portion including the condyles were scanned to build surfaces. As a result, each set of femur data comprised two separated surfaces containing the femoral head and condyles, respectively.
Evaluation of two-level registration
show an example of how the surface distance is decreased at each step of the two-level framework. Here we use the bottom portion of the femur as an example, given its important relationship with the knee. In this case, there are two high-resolution surfaces XH (21,130 vertices, 42,256 triangles, 65.8 mm in the z-axis) and YH (26,652 vertices, 53,300 triangles, 105.9 mm in the z-axis) (Patient X was a 79-year-old female with femur length of 472.6 mm; Patient Y was a 53-year-old female with femur length of 477.6 mm). We first computed the point-to-surface distance from XH to YH Citation[42]:where ‖ · ‖2 is the Euclidean norm. The HSV (hue, saturation, value) color of each vertex on X H denotes the distance d(p, YH). We also computed the mean error dm(X H, Y H) and root mean square error dRMS(X H, YH) between X H and Y H based on Equation 13:
shows the high-resolution surfaces XH and YH. With respect to the bounding box diagonal of YH (158.5 mm), the mean error is 6.49% and the root mean square error is 7.70%.
shows the low-resolution surfaces XL (169 vertices, 334 triangles) and YL (213 vertices, 422 triangles) after simplification. With respect to the bounding box diagonal of YL (158.3 mm), the mean error is 6.53% and the root mean square error is 7.74%.
shows the deformed low-resolution surfaces XL(1) and YL after applying Chui and Rangarajan's non-rigid registration Citation[25] to XL. With respect to the bounding box diagonal of YL, the mean error is 1.68% and the root mean square error is 2.13%. The surface distance has been significantly decreased by applying Chui and Rangarajan's method.
shows the deformed low-resolution surfaces XL(2) and YL after applying a local refine process to XL(1). With respect to the bounding box diagonal of YL, the mean error is 0.68% and the root mean square error is 1.42%, which shows that the local point-to-surface refining procedure can further decrease the surface distance.
shows the interpolated high-resolution surfaces XH(1) and YH after applying the interpolation to XH. With respect to the bounding box diagonal of YH, the mean error is 1.65% and the root mean square error is 2.10%. The surface distance slightly increases by interpolation because only 0.80% of the vertices on XH(1) have correspondences computed from low-resolution registration; the other correspondences were obtained by interpolation.
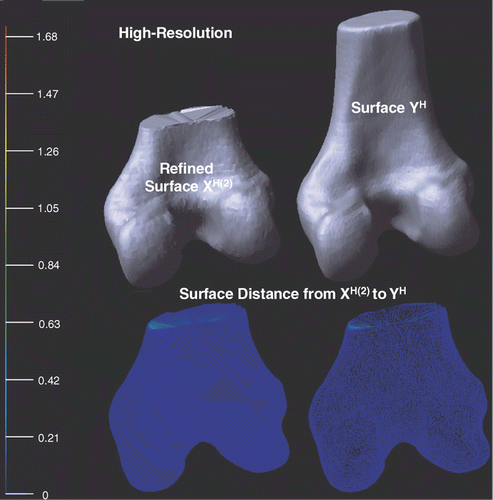
shows the deformed high-resolution surfaces XH(2) and YH after applying a local refining process to XH(1). With respect to the bounding box diagonal of YH, the mean error is 0.28% and the root mean square error is 1.26%, once again demonstrating that local point-to-surface refining procedure is helpful for decreasing surface distance.
Figure 7. Illustration of the high-resolution surfaces XH and YH. Point-to-surface distances from XH to YH are illustrated by an HSV color map: the color of each vertex on XH represents the distance d(p, YH), p ∈ XH. Both surface (left) and mesh (right) are shown on the bottom row.
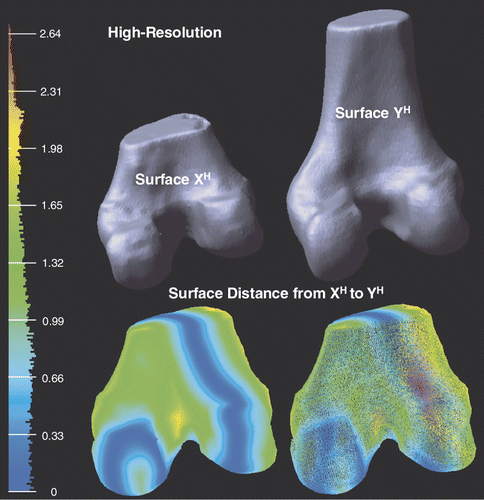
Figure 8. Illustration of the low-resolution surfaces XL and YL after applying the mesh simplification (see Mesh simplification sub-section) to both XH and YH. Point-to-surface distances from XL to YL are illustrated by an HSV color map: the color of each vertex on XL represents the distance d(p, YL), p ∈ XL. Both surface (left) and mesh (right) display modes are shown on the bottom row.
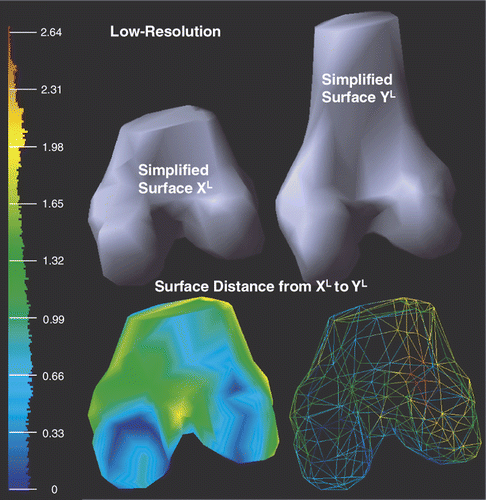
Figure 9. Illustration of the deformed low-resolution surfaces XL(1) and YL after applying Chui and Rangarajan's non-rigid registration method (see Low-resolution non-rigid registration sub-section) to XL. Point-to-surface distances from XL(1) to YL are illustrated by an HSV color map: the color of each vertex on XL(1) represents the distance d(p, YL), p ∈ XL(1). Both surface (left) and mesh (right) display modes are shown on the bottom row.
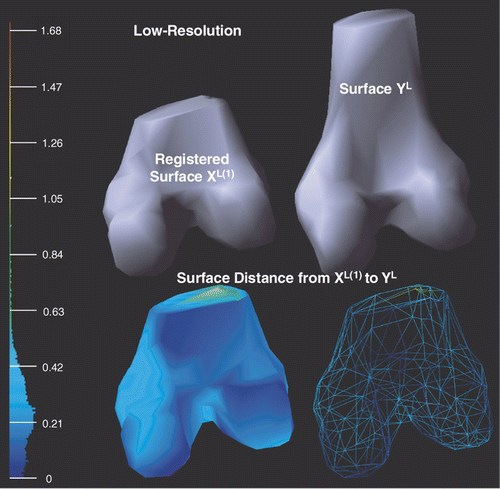
Figure 10. Illustration of the deformed low-resolution surfaces XL(2) and YL after applying the local refinement (see Local refining procedure sub-section) to XL(1). Point-to-surface distances from XL(2) to YL are illustrated by an HSV color map: the color of each vertex on XL(2) represents the distance d(p, YL), p ∈ XL(2). Both surface (left) and mesh (right) display modes are shown on the bottom row.
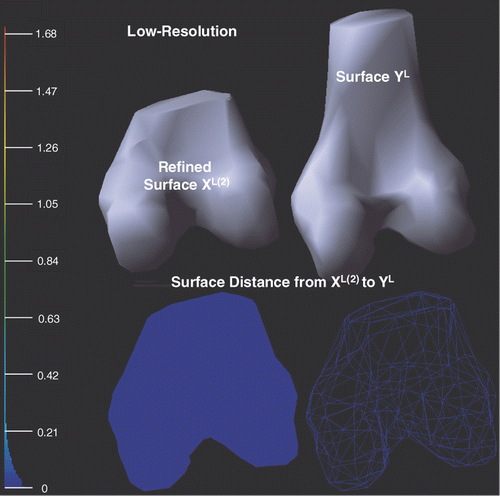
Figure 11. Illustration of the deformed high-resolution surfaces XH(1) and YH after applying the interpolation (see Low-resolution to high-resolution interpolation sub-section) to XH. Point-to-surface distances from XH(1) to YH are illustrated by an HSV color map: the color of each vertex on XH(1) represents the distance d(p, YH), p ∈ XH(1). Both surface (left) and mesh (right) display modes are shown on the bottom row.
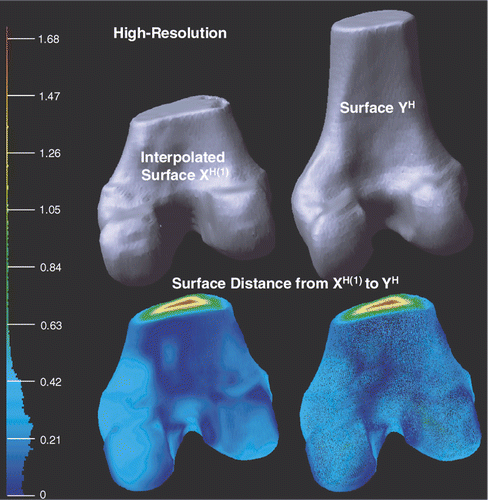
Figure 12. Illustration of the deformed high-resolution surfaces XH(2) and YH after applying the local refinement (see Local refine procedure sub-section) to XH(1). Point-to-surface distances from XH(2) to YH are illustrated by an HSV color map: the color of each vertex on XH(2) represents the distance d(p, YH), p ∈ XH(2). Both surface (left) and mesh (right) display modes are shown on the bottom row.
Comparison to other registration methods
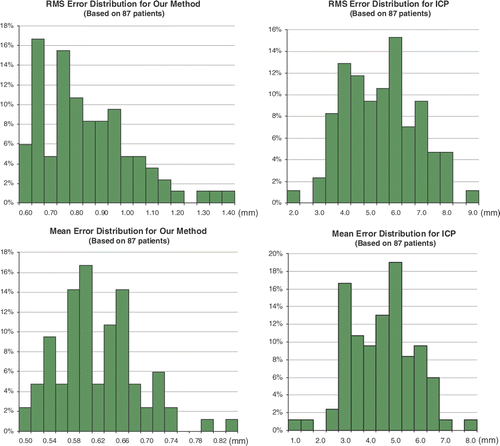
We compared our registration algorithm with the classical iterative closet point (ICP) method since the latter is widely used in many medical image registration problems. Given 87 training surfaces, the distributions of the RMS error and mean error of the two approaches are shown in . For the RMS error, the distribution is centered between 0.6 and 1.0 mm (max: 1.4 mm) with our method, but with ICP the range is between 3.1 and 6.5 mm (max: 8.5 mm). For the mean error, the distribution is centered between 0.5 and 0.7 mm (max: 0.9 mm) with our method, but with ICP the range is 2.5 to 6.5 mm (max: 7.5 mm). From this it is apparent that the two-level non-rigid registration has a better performance than ICP.
Since Chui and Rangarajan's thin-plate spline (TPS)-based method cannot deal with the complexity of our data, it is not possible to compare the two methods directly. Alternatively, it can be seen from that our method only requires 5 min or less to match any size of surfaces with less than 200,000 vertices. In contrast, the method of Chui and Rangarajan takes 5 min for 350 vertices, 10 min for 460 vertices, 20 min for 610 vertices, and so on. It is therefore clear that our algorithm represents a significant improvement in efficiency. To maintain a good level of accuracy for practical applications, a best simplification parameter was selected based on a series of leave-one-out experiments (see details in Selection of simplification parameters section).
Evaluation of atlases
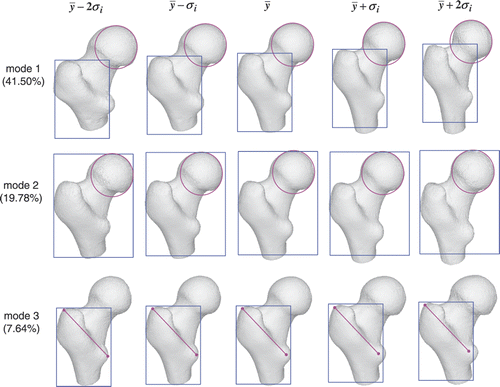
We built three atlases for the femoral head, the condyles and the entire femur. shows the shape variation in the first three modes of the femoral head atlas. The pink and blue shapes are used to highlight the local variation. For example, the size of the femoral head (denoted by the pink circle) gradually reduces along the first mode, but remains almost unchanged along the second mode. The greater trochanter (denoted by the blue rectangle) becomes longer but progressively narrower along the first mode, whereas it remains the same length but becomes progressively wider along the third mode.
Figure 14. Femoral head atlas: illustration of the shape variation in the first, second and third modes. The first mode encodes 41.50% of the shape variation, while the second and third modes encode 19.78% and 7.64% of the shape variation, respectively. The color shapes highlight the local shape variation.
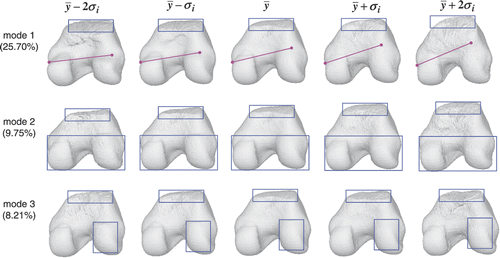
shows the shape variation in the first three modes of the condyle atlas. We noticed that the top of the condyles (denoted by the blue rectangles) changed considerably along each mode. This is because each scan was cut differently. The top area can only be estimated from the reference surface without any refining from the original surfaces. The lateral epicondyle (on the right) becomes progressively larger along the first mode, compared to the medial epicondyle (on the left). This is similar to the trend seen in the third mode but the change is smaller; the condyles become progressively longer but narrower along the second mode.
Figure 15. Condyle atlas: illustration of the shape variation in the first, second and third modes. The first mode encodes 25.70% of the shape variation, while the second and third modes encode 9.75% and 8.21% of the shape variation, respectively. The color shapes highlight the local shape variation.
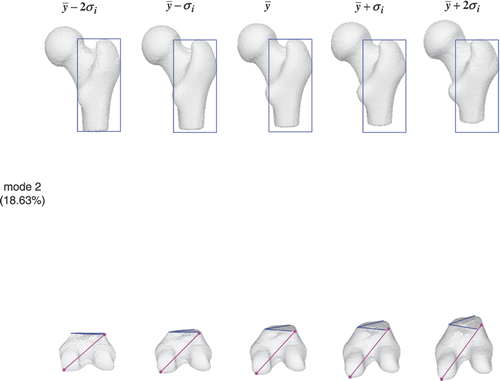
and show the shape variation in the first two modes of the entire femur atlas. For the first mode, the femoral head and greater trochanter exhibit a similar change to that shown in , while the condyles become progressively longer and narrower as in . The atlas of the entire femur behaves in a manner similar to that of the individual portion in the first mode. For the second mode, the greater trochanter becomes shorter but remains the same as in , while the condyles become much longer but keep the same length as in . This means that the second dominant mode in the individual atlas may not be the second dominant mode in the conjunction case.
Figure 16. Femur atlas: 27.67% of the shape variation is encoded in the first mode. The color shapes highlight the local shape variation.
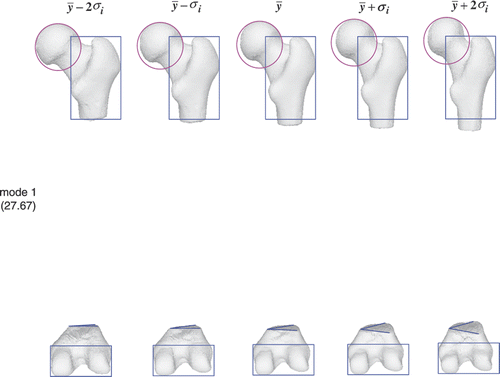
Figure 17. Femur atlas: 18.63% of the shape variation is encoded in the second mode. The color shapes highlight the local shape variation.
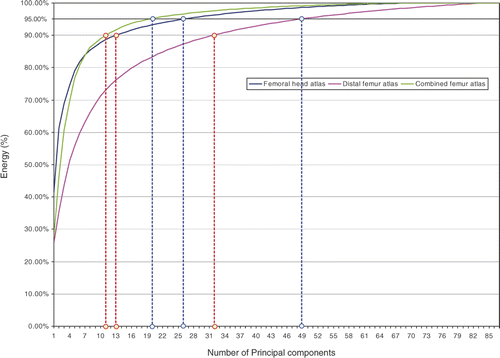
shows the shape variation encoded in the modes of each atlas. To keep 95% of the shape variation we only need 26 modes for the femoral head atlas, 49 modes for the condyle atlas, and 20 modes for the entire femur atlas. The condyle atlas needs more modes to retain most of the shape variation because the training surfaces of the bottom portion of the femur change shape considerably (i.e., each surface contains a different length of femur shaft).
Conclusion
In this paper we have developed a two-level framework to efficiently tackle non-rigid registration challenges caused by geometrical and computational complexity. Our work is mainly inspired by the method of Chui and Rangarajan Citation[25], but performs much faster and requires less memory. Experiments have demonstrated that our method significantly improves the efficiency of registration without decreasing the accuracy of the atlases. Shape variation learned from both femur atlases can be used for clinical studies and diagnosis in the future. Moreover, femur atlases can be used as a statistical prior for shape reconstruction from other imaging modalities such as endoscopy, and also for the segmentation of 3D surfaces from DICOM images.
References
- McInerney T, Terzopoulos D. Deformable models in medical image analysis: a survey. Med Image Anal 1996; 1(2)91–108
- Subsol G, Thirion JP, Ayache N. First steps towards automatic building of anatomical atlases. INRIA, Technical Report 2216, March, 1994
- Subsol G, Thirion JP, Ayache N. A general scheme for automatically building three-dimensional morphometric anatomical atlases: application to a skull atlas. INRIA, Technical Report 2586, May, 1995
- Broit C. Optimal registration of deformed images. PhD dissertation, Department of Computer and Information Science, University of Pennsylvania, Philadelphia, PA 1981
- Bajcsy R, Kovacic S. Multiresolution elastic matching. Proceedings of CVGIP 1989; 46: 1–21
- Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vision 1988; 1(4)321–331
- Farin G. Curves and Surfaces for CAGD. Academic Press, New York 1993
- Park J, Mataxas D, Young A, Axel L. Deformable models with parameter functions for cardiac motion analysis from tagged MRI data. IEEE Trans Med Imaging 1996; 15: 278–289
- Szeliski R. Bayesian modeling of uncertainty in low-level vision. Int J Comput Vision 1990; 5: 271–301
- Cootes TF, Beeston C, Edwards GJ, Taylor CJ. A unified framework for atlas matching using active appearance models. Proceedings of the 16th International Conference on Information Processing in Medical Imaging (IPMI '99). Visegrád, Hungary 28 June–2 July, 1999; 322–333
- Meller S, Kalender WA. Building a statistical shape model of the pelvis. In: Lemke HU, Vannier MW, Inamura K, Farman AG, Doi K, Reiber JHC, editors. Proceedings of the 18th International Congress and Exhibition on Computer Assisted Radiology and Surgery (CARS 2004), Chicago, IL, June 2004. Amsterdam: Elsevier; 2004. pp 561–566.
- Rueckert D, Frangi AF, Schnabel JA. Automatic construction of 3-D statistical deformation models of the brain using nonrigid registration. IEEE Trans Med Imaging 2003; 22(8)1014–1025
- Cootes TF, Taylor CJ, Cooper D, Graham J. Active shape models: their training and application. Computer Vision and Image Understanding 1995; 61(1)38–59
- Cootes TF, Taylor CJ. Statistical models of appearance for medical image analysis and computer vision. Proceedings of SPIE Image Processing 2001; 4322: 238–248
- Chui H, Rangarajan A, Zhang J, Leonard CM. Unsupervised learning of an atlas from unlabeled point-sets. IEEE Trans Pattern Anal Machine Intel 2004; 2: 160–172
- Hill DLG, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Phys Med Biol 2001; 46(3)1–45
- Chen M. 3-D deformable registration using a statistical atlas with applications in medicine. PhD dissertation, Robotics Institute, School of Computer Science, Carnegie Mellon University, Pittsburgh, PA 1999
- Fleute M, Lavallée S, Desbat L. Integrated approach for matching statistical shape models with intra-operative 2D and 3D data. In: Dohi T, Kikinis R, editors. Proceedings of the 5th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2002), Tokyo, September 2002. Part II. Lecture Notes in Computer Science 2489. Berlin: Springer; 2002. pp 364–372.
- Jaume S, Macq B, Warfield SK. Labeling the brain surface using a deformable multiresolution mesh. In: Dohi T, Kikinis R, editors. Proceedings of the 5th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2002), Tokyo, Septermber 2002. Part I. Lecture Notes in Computer Science 2488. Berlin: Springer; 2002. pp 451–458.
- Rohlfing T, Russakoff DB, Maurer CR. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans Med Imaging 2004; 23(8)983–994
- Straka M, Cruz AL, Dimitrov LI, Sramek M, Fleischmann D, Groller E. Bone segmentation in CT-angiography data using a probabilistic atlas. Proceedings of the Vision, Modeling and Visualization Conference 2003 (VMV 2003), T Ertl, MunichGermany November, 2003; 505–512
- Dawant BM. Non-rigid registration of medical images: purpose and methods, a short survey. In: Proceedings of the IEEE International Symposium on Biomedical Imaging, Washington, DC, July 2002. pp 465–468.
- Yu H. Automatic rigid and deformable medical image registration. PhD dissertation, Department of Mechanical Engineering, Worcester Polytechnic Institute, Worcester, MA 2005
- Szeliski R, Lavallée S. Matching 3-D anatomical surfaces with non-rigid deformations using octree-splines. Int J Comput Vision 1996; 18: 171–186
- Chui H, Rangarajan A. A new point matching algorithm for non-rigid registration. Computer Vision and Image Understanding 2003; 89(2-3)114–141
- Besl PJ, McKay ND. A method for registration of 3-D shapes. IEEE Trans Pattern Anal Machine Intel 1992; 14(2)239–256
- Kim J, Fessler JA. Intensity-based image registration using robust correlation coefficients. IEEE Trans Pattern Anal Machine Intel 2004; 23(11)1430–1444
- Woods RP, Mazziotta JC, Cherry SR. MRI-PET registration with automated algorithm. J Comput Assist Tomography 1994; 17: 536–546
- Wells WM, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Med Image Anal 1996; 1(1)35–51
- Maurer CR, Fitzpatrick JM, Wang MY, Galloway RL, Maciunas RJ, Allen GG. Registration of head volume images using implantable fiducial markers. IEEE Trans Pattern Anal Machine Intel 1997; 16: 447–462
- Glaunes J, Trouvé A, Younes L. Diffeomorphic matching of distributions: a new approach for unlabelled point-sets and sub-manifolds matching. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR 2004). Washington, DC 27 June–2 July, 2004; 712–718
- Ellingsen LM, Prince JL. Deformable registration of CT pelvis images using Mjolnir. Proceedings of SPIE Medical Imaging 2006; 6144: 329–337
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging 2002; 21(11)1421–1439
- Papademetris X, Jackowski AP, Schultz RT, Staib LH, Duncan JS. Non-rigid brain registration using extended robust point matching for composite multisubject fMRI analysis. In: Ellis RE, Peters TM, editors. Proceedings of the 6th International Conference on Medical Image Analysis and Computer-Assisted Intervention (MICCAI 2003), Montreal, Canada, Novermber 2003. Part II. Lecture Notes in Computer Science 2879. Berlin: Springer; 2003. pp 788–795.
- Garland M, Heckbert PS. Surface simplification using quadric error metrics. Proceedings of SIGGRAPH 1997; 209–216
- Yngve G, Turk G. Robust creation of implicit surfaces from polygonal meshes. IEEE Trans Visualization Comput Graphics 2002; 8(4)346–359
- Carr JC, Fright WR, Beatson RK. Surface interpolation with radial basis functions for medical imaging. IEEE Trans Med Imaging 1997; 16(1)96–107
- Pighin F, Hecker J, Lischinski D, Szeliski R, Salesin DH. Synthesizing realistic facial expressions from photographs. In: Proceedings of SIGGRAPH 1998. pp 75–84
- Carr JC, Beatson RK, Cherrie JB, Mitchell TJ, Fright WR, McCallum BC, Evans TR. Reconstruction and representation of 3D objects with radial basis functions. Proceedings of SIGGRAPH 2001; 67–76
- Goryn D, Hein S. On the estimation of rigid body rotation from noisy data. IEEE Trans Pattern Analysis Machine Intel 1995; 17(2)1219–1220
- Desbrun M, Meyer M, Schroder P, Barr AH. Implicit fairing of irregular meshes using diffusion and curvature flow. In: Proceedings of SIGGRAPH 1999. pp 317–324
- Aspert N, Santa-Cruz D, Ebrahimi T. MESH: measuring errors between surfaces using the Hausdorff distance. Proceedings of the IEEE International Conference on Multimedia and Expo (ICME 2002). LausanneSwitzerland August, 2002; 705–708