Abstract
Outcomes for neurosurgery patients can be improved by enhancing intraoperative navigation and guidance. Current navigation systems do not accurately account for intraoperative brain deformation. So far, most studies of brain deformation have focused on brain shift, whereas this paper focuses on the brain deformation due to retraction. The heart of our system is a 3D nonrigid registration technique using a biomechanical model driven by the deformations of key surfaces tracked between two intraoperative images. The key surfaces, e.g., the whole-brain region boundary and the lips of the retraction cut, thus deform due to the combination of gravity and retractor deployment. The tissue discontinuity due to retraction is handled via the eXtended Finite Element Method (XFEM), which has the appealing feature of being able to handle arbitrarily shaped discontinuity without any remeshing. Our approach is shown to significantly improve the alignment of intraoperative MRI.
Introduction
The main goal of brain surgery is to remove as much of the lesion tissues as possible, while avoiding contact with eloquent areas and fiber tracts located in the white-matter tissues. Surgery is planned on the basis of preoperative images of multiple modalities, such as Computed Tomography (CT), structural and functional Magnetic Resonance Imaging (sMRI and fMRI), Diffusion Tensor Imaging (DTI), Positron Emission Tomography (PET), and Magneto-encephalography (MEG). Surgery is generally performed using an image guided navigation system that relates the 3D preoperative images to 3D patient coordinates, thereby allowing the surgeon to position his instruments in the patient's brain while navigating on the preoperative images. However, the brain deforms throughout the surgery, mostly as a result of leakage of cerebrospinal fluid (CSF) out of the skull cavity, modifications in cerebral perfusion, pharmacological modulation of the extracellular fluid, and surgical acts, such as cuts, retractions, and resections Citation[1]. Therefore, as the surgery progresses, the preoperative images become less representative of the actual brain, and navigation accuracy decreases.
These navigation errors could be reduced if one could acquire, throughout surgery, fresh images of the same modalities and quality as those acquired preoperatively. Acquisition of such images poses a major challenge, since, except at a handful of facilities, intraoperative MR (iMR) images are usually acquired using low-field MRI scanners that provide lower resolution and contrast than their preoperative counterparts. Moreover, even in those rare facilities that possess high-field intraoperative MRI scanners, the acquisition of intraoperative DTI images remains rather time-consuming. Finally, several useful imaging modalities, such as PET and MEG, cannot yet be acquired intraoperatively. The solution that is generally favored is to acquire reduced-quality intraoperative images at several critical points during surgery, to track the tissue deformation from one image to the next, to update (i.e., deform) the preoperative images accordingly, and to feed the resulting images to the navigation system Citation[2]. The deformation of preoperative images so that they conform to the intraoperative images falls into the domain of nonrigid registration.
One category of such techniques uses biomechanical models based on the Finite Element Method (FEM). Prior to surgery, a biomechanical brain model specific to the patient is built from preoperative images: the model consists of a volume mesh of finite elements (FEs), and of one or more mechanical behavior laws assigned to them. During surgery, a number of key anatomical landmarks are extracted and tracked through successive intraoperative images. When using the approach known as the forced-displacement method, the estimated displacements of these landmarks are directly applied to the biomechanical model and drive its deformation. The resulting displacement field of the biomechanical model is used to deform the preoperative images. Most of the mechanical conditions affecting the brain, such as the volume of CSF flowing out of the skull cavity, or the forces applied by a retractor tool, cannot yet be estimated in the operating room. The fact that an intraoperative image can provide knowledge of the current state of the brain after some deformation partly eliminates the need for a complete evaluation of these mechanical conditions. The nonrigid registration technique replaces them with the landmark displacements evaluated from successive intraoperative images.
Most studies of brain deformation based on biomechanical models have focused on shifts (in which the topology of the brain is not modified) occurring after the opening of the skull and dura Citation[3–12], and thus do not explicitly take into account any cuts or subsequent deformation. A good review of these different studies can be found in references Citation[9], Citation[13], Citation[14] and Citation[15]. In comparison, few studies have focused on brain deformations due to cuts, retractions and resections (the last two necessarily involving a cut). One of the main difficulties associated with a cut is the discontinuity it implies in the tissues, which changes the brain topology. Indeed, the finite element method (FEM) cannot handle discontinuities directly and requires one to realign the discontinuity with element boundaries based on mesh-adaptation Citation[16–18] or remeshing techniques Citation[19–22], which provide, from an initial mesh, a new mesh conforming to the discontinuity.
This paper presents a solution for updating multimodal preoperative images with respect to surgical brain deformations due to retraction that can be seen and quantified using iMR images. The landmarks tracked between the iMR images correspond to the whole-brain region boundary and the lips of the retraction cut, both deforming due to the combination of gravity and retractor deployment. In the field of fracture mechanics, which studies the growth and propagation of cracks in mechanical parts, several methods have been developed so as to avoid using FEM in combination with mesh adaptation or remeshing techniques Citation[23], Citation[24]. One of these methods is the eXtended Finite Element Method (XFEM or X-FEM), which first appeared in 1999 Citation[24] and has since been the subject of considerable research Citation[25]. XFEM works by allowing the displacement field to be discontinuous within some FEs of the mesh. The mesh does not have to conform to the discontinuities, so these can be arbitrarily located with respect to the underlying FE mesh. Because XFEM allows for an accurate representation of the discontinuities while avoiding mesh adaption or remeshing, and because of the similarity between cracks in mechanical parts and cuts in tissue, we proposed the use of XFEM for handling cuts, retraction, and resection in the updating of preoperative images. As explained earlier, the goal of this work is to incrementally update the preoperative images with each new pair of successive iMR images; as each iMR image is acquired, this new image and the preceding one are used to estimate the deformation of the brain. Typically, after brain shift, the patient undergoes successive resections with prior retraction, depending on the location of the tumor site. In the modeling of successive brain deformations, XFEM is thus appropriate to avoid successive remeshing. Some 2D results were previously presented for retraction modeling and successive resection modeling, both handled separately Citation[26]. This paper is the first complete and detailed account of the generalization to three dimensions of our 2D work on retraction modeling. In the future, 3D retraction followed by successive resections will be modeled. Different constitutive laws have already been used for modeling brain deformation. The simplest constitutive law is the linear elastic law Citation[7], Citation[10], which is used in this work, while the brain shift was also modeled using a hyperviscoelastic law characterized by a strain rate dependence Citation[11] or a poroelastic law characterized by a solid matrix and a fluid phase Citation[8].
The structure of the paper is as follows. In the next section, the state of the art in retraction modeling is presented. The basic principles of XFEM are then introduced. Our global system for updating preoperative images is described and details of our algorithms for retraction modeling are provided. After that, a patient case that illustrates our approach is considered, and our results are validated. Finally, conclusions are drawn and future work discussed.
State of the art
Retraction is usually performed when the target, e.g., the tumor, is deep inside the brain. The surgeon cuts through the brain tissues and inserts the blades of a retractor to spread the tissues out from the incision and create a free path towards the tumor. Ferrant et al. Citation[7] modeled retraction using a linear elastic biomechanical model. They deleted the FEs falling into the retraction path, which is visible on the iMR image acquired after retraction. This way of modeling retraction implied the removal of some tissues, which was not physically correct, and did not model explicitly the motion of tissues as they were spread out.
Miga et al. Citation[27] modeled retraction using a linear poroelastic biomechanical model. This model was subject to the effects of gravity, as well as to those arising from the deployment of the retractor. Optical images from the operating microscope and cortical features, such as surface vasculature, were used to estimate the position of the retractor. The tissue discontinuity was modeled by splitting the mesh along existing boundary edges to obtain the best possible representation of the cut by a jagged discontinuity. To model the opening of the path, the duplicated nodes were moved apart. The nodes located on the front side of the retractor (i.e., facing the opening) were moved to within a distance consistent with the optical images. The nodes on the back side were not allowed to move orthogonally to the retractor blade.
Platenik et al. Citation[28], Citation[29] attempted to validate the retraction modeling of Miga et al. Citation[27]. They performed in vivo incremental inter-hemispheric retraction (from 3 mm up to 10 mm) on four porcine subjects, using a controlled retractor mounted on the stereotactic frame attached to the subject. Intraoperative CT (iCT) scans were used to estimate the position of the retractor. Validation was performed using beads implanted in the porcine brains and tracked between successive iCT scans. This group also improved the model of Miga et al. by allowing tissue on the back side of the retractor to move with the retractor at the beginning of the retraction and then separate at larger retractions. This phenomenon was visible on iCT scans. They were able to capture up to 85% of brain motion. Lamprich and Miga Citation[31] also attempted to validate the retraction modeling of Miga and colleagues Citation[27] on ex vivo inter-hemispheric retraction porcine experimental images using iMR.
Lunn et al. Citation[31] used the data collected by Platenik et al. Citation[28] from these porcine experiments to test a different way of modeling retraction. The volume displacement field of the biomechanical model was computed as a weighted combination of basis solutions, computed prior to surgery with a linear poroelastic biomechanical model. The weight coefficients were computed with a minimization procedure using the implanted beads as control points, in order to reduce the error between the observed and computed displacements of the beads. They analyzed different basis solutions and evaluated the best combination. The best results were obtained with a combination of three basis solutions: gravity, retraction with normal displacements to the retractor blades, and pressure on the back of the retractor. They also analyzed the number and locations of control points necessary to obtain such results. While the results showed equal or decreased mean vector error compared to the results of Platenik et al., the method still calls for further study. Indeed, the method is, of course, sensitive to the type of basis solutions used, and does not necessarily provide better solutions with additional basis solutions. Furthermore, while it was feasible to use implanted beads for porcine subjects and track them in iCT scans, the use and tracking of control points in real patients is expected to be more challenging.
Sun et al. Citation[32] studied the capacity for capturing cortical motion during retraction, using stereo cameras mounted on the operating microscope. They registered stereo images acquired after craniotomy to a linear poroelastic biomechanical model of the patient's brain and tagged the nodes of the external surface of the biomechanical model that corresponded to the visible surface of the brain. From these nodes, they defined a subset that were visually hidden by the retractor on the stereo images acquired after insertion of the retractor and tagged them as the retractor nodes. They estimated the motion of the retractor based on stereo images acquired before and after the deployment of the retractor, using an iterative closest point (ICP) algorithm. The displacement of the retractor was transposed to displacements applied to the retractor nodes of the biomechanical model. The method was validated by computing, based on the ICP algorithm, the displacements of the tagged nodes minus the retractor nodes from stereo images, and comparing these displacements to the displacements of nodes resulting from the deformation of the biomechanical model. For the patient case studied, it was found that they could capture approximately 75% of the cortical motion.
The methods described above have all been developed using an FEM-based biomechanical model for intraoperative image registration. Surgical simulation is another research field that makes extensive use of FEM-based biomechanical models. The objective of a surgical simulator is to provide an interactive manipulation with force feedback of the anatomical part to be operated on using various surgical instruments. To enable modeling of a wide range of surgical procedures, a real-time interactive cutting method should be included in the simulator. Jerabkova and Kuhlen Citation[33] have applied nonlinear XFEM for simulating cuts, and have shown that XFEM is successful and efficient for such purposes.
Principles of XFEM
Before presenting the principles of XFEM, a few key properties of FEM will be introduced to make understanding of XFEM easier. FEM Citation[34] discretizes the solid of interest into a mesh, i.e., into a set of finite elements (FEs) interconnected by nodes, and approximates the displacement field u(x) by the FEM displacement field uFEM(x), defined aswhere x is the position of a point of the solid, I is the set of nodes, the φi(x)'s are the nodal shape functions (NSFs), and the ui's are the nodal degrees of freedom (DOFs). Each φi(x) is defined as being continuous on its compact support ωi, which corresponds to the union of the domains of the FEs connected to node i, with the property
where xj is the position of node j and δij the usual Kronecker signal. In our approach, linear NSFs are used.
FEM requires its displacement field uFEM(x) to be continuous over each FE. In contrast, XFEM handles a discontinuity by allowing the displacement field to be discontinuous within FEs Citation[24], Citation[35–37]. Arbitrarily shaped discontinuities can then be modeled without any remeshing. The XFEM displacement field generalizes the FEM displacement field (Equation 1) withThe first term corresponds to the FEM displacement field (Equation 1), where I is the set of nodes, the φi(x)'s are the FEM NSFs, and the ui's are the nodal FEM DOFs. The heart of XFEM is the “enrichment” that adds a number,
, of DOFs aji to each node i of the set J, which is the subset of nodes of I whose support is intersected by the discontinuity of interest. These DOFs are multiplied by the NSFs φi(x) and the discontinuous functions gj(x).
Evaluating the function uXFEM(x) at the position location xk of some node k that is enriched, i.e., for k ∈ J, one findsSince the last sum is generally not zero, one has
This means that, for nodes that are enriched, the FEM DOF uk does not represent the displacement of node k, and thus the XFEM displacement field uXFEM(x) does not interpolate the nodal displacements. This should be contrasted to the case of FEM, where the FEM DOF uk represents the displacement of node k. Equation 4 also implies that nodal displacements cannot be imposed by eliminating the DOFs uk, such as is done with FEM, but must be applied as a linear combination between several DOFs with, for instance, Lagrange multipliers for nodes that are enriched Citation[38].
The use of specific XFEM enrichment functions gj(x) for a node i ∈ J depends on the type of discontinuity, e.g., crack, hole, material interface, etc., to be modeled. Suppose that our goal is to model a crack, characterized by a discontinuity in the displacement field (as opposed to a material interface, for instance, characterized by a discontinuity in the derivative of the displacement field). When the crack fully intersects the support of the node, a simple choice is a piecewise-constant unit function that changes sign at the boundary across the crack, i.e., the Heaviside functionwhere x is again the position of a point of the solid, x* is the position of the point on the crack that is the closest to x, and en is the outward normal to the crack at x*, as shown in Citation[24].
Figure 1. Parameters related to the XFEM enrichment Heaviside function corresponding to a 3D crack surface.
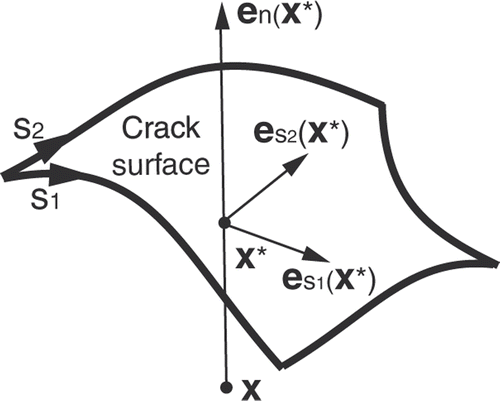
The support of any node in J is cut into two disjointed pieces by the crack. If, for a given node, one of the two pieces is very small compared to the other, then the Heaviside function is almost constant over the nodal support, which causes the XFEM problem to be ill-conditioned. Consequently, the node has to be removed from J, which is equivalent to moving the crack location to the boundary of the support. A criterion should be used to remove from J a node for which the ratio of the volume of the smallest piece to that of the full support is less than a given tolerance value, e.g., 10−4 Citation[35].
The XFEM equations that result from the minimization of the total potential energy remain sparse and symmetric as for FEM. Whereas FEM requires a remeshing and the duplication of the nodes along the crack to take into account any discontinuity, XFEM requires the identification of the nodes belonging to the set J and the addition of DOFs: any node whose support is not intersected by the discontinuity remains unaffected and thus possesses three DOFs; and any node whose support is intersected by the discontinuity is enriched with three Heaviside DOFs and thus possesses six DOFs. Because of the additional DOFs in XFEM, the number of equations to be solved is larger than for FEM.
Description of the baseline system
Our retraction modeling is part of a global framework for updating preoperative images using successive intraoperative MR (iMR) images acquired at various critical points during surgery. Images were acquired with the 0.5-Tesla GE Signa intraoperative scanner at the Brigham and Women's Hospital, Boston, MA. The iMR image size was 256 × 256 × 60 voxels, and the voxel size was 0.859 × 0.859 × 2.5 mm. In this paper, two simplifying assumptions are made. First, only retraction deformation is considered. As this occurred between the second and third iMR images for the patient case treated, brain shift deformation, which occurred between the first and second iMR images, is not taken into account; however, the modeling of retraction is not affected by this simplification. (The modeling of brain shift deformation – for a different case – was treated and illustrated in reference Citation[39].) Second, the second iMR image is used as a substitute for preoperative images. The biomechanical model is thus built based on structures visible in the second iMR image, as opposed to the preoperative images, and the structures used in the model are limited to those visible in the intraoperative image. Except for the rigid registration between the preoperative images, the biomechanical model, and the second iMR image, this simplified approach allows us to discuss, illustrate, and test all key aspects of the system.
The successive steps in the evaluation of brain retraction deformation consist of rigidly registering the iMR images and then segmenting them. The biomechanical model is built based on structures segmented in the second iMR image. A set of common anatomical surface landmarks, the whole-brain region boundary and the two lips of the cut, are segmented and tracked between the two iMR images. The biomechanical model is deformed, based on XFEM, in accordance with the displacement fields of these surface landmarks. The resulting volume displacement field of the biomechanical model is then used to warp the second iMR image. Finally, the method is validated by comparing images that are of the same modality and quality and thus show the same anatomical features (the original and deformed second iMR images are compared with the third iMR image). In the following sections, the successive processing steps just mentioned are described in detail.
Rigid registration of intraoperative images
Although the patient's head is supposedly fixed during the operation, one cannot totally rule out the possibility of slight head motion. To compensate for this potential source of rigid motion, the iMR images are rigidly registered using the point-based landmark transform available in VTK (http://www.vtk.org/), where the corresponding landmark points are manually selected in the two iMR images.
Segmentation of intraoperative images
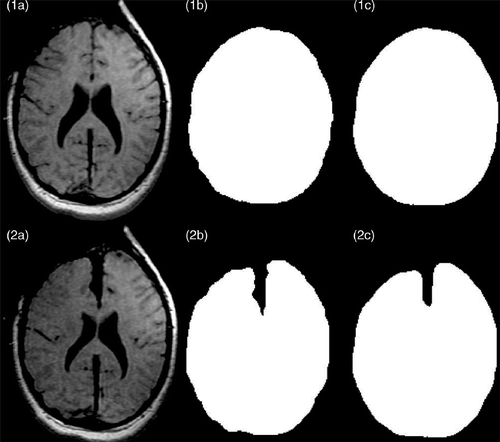
The segmentation of iMR images is necessary for both the building of the biomechanical model and the capture of intraoperative deformation between the two iMR images []. In the present application, the only region of interest is the whole-brain region, i.e., with the skull and external CSF masked out. The segmentation is performed in two steps. First, the whole-brain region is manually segmented out using 3D Slicer (http://www.slicer.org/) [] from the two iMR images. Second, the two segmented regions are smoothed out to minimize the dependence of the evaluation of surface landmark displacement fields on the roughness of manual segmentation. This smoothing is performed by building a surface mesh of the regions segmented manually in each iMR image, and by subsequently filling all voxels that fall inside the surface mesh with the numerical label of the segmented region []. By comparing , one sees that the smoothing causes the depth and the opening width of the path created by retraction to decrease. This problem will need to be fixed in the future.
Figure 2. Segmentation of intraoperative images. (1a) Original second iMR image, acquired before retraction. (2a) Original third iMR image, acquired after retraction. (1b) Whole-brain region segmented out manually from (1a) with 3D Slicer. (2b) Whole-brain region segmented out manually from (2a) with 3D Slicer. (1c) Smoothing of the segmented whole-brain region from (1b). (2c) Smoothing of the segmented whole-brain region from (2b). (1c) and (2c) are performed to minimize the dependence of the active surface algorithm on segmentation roughness and surface mesh resolution.
Building of the biomechanical model
As mentioned above, in the present work, the biomechanical model is built from the second iMR image rather than from the preoperative images. The biomechanical model is built in two steps. First, the volume mesh is built and the appropriate constitutive law is assigned to it. Second, the cut to be included in the biomechanical model is defined. This cut represents the discontinuity that is handled via XFEM, i.e., by enriching the appropriate nodes of the volume mesh with XFEM Heaviside DOFs.
To build the volume mesh, IsoSurf (http://www-math.mit.edu/~persson/mesh/) is first used to produce a surface mesh of the boundary of the segmented whole-brain region. This software takes as input a binary image, provided as stacks of slices and representing segmented regions, and meshes the boundary of these regions into surfaces of triangles. Gmsh (http://www.geuz.org/gmsh/) Citation[40], a 3D mesh generator, is then used to produce, based on the triangle surface mesh, a tetrahedron volume mesh of the whole-brain region. A linear elastic law is assigned to the whole-brain region, with Young's modulus E = 3000 Pa and Poisson ratio ν = 0.45 Citation[7]. Note that, since only displacements (and no forces) are applied to the homogeneous biomechanical model, its deformation is independent of the value of Young's modulus Citation[41].
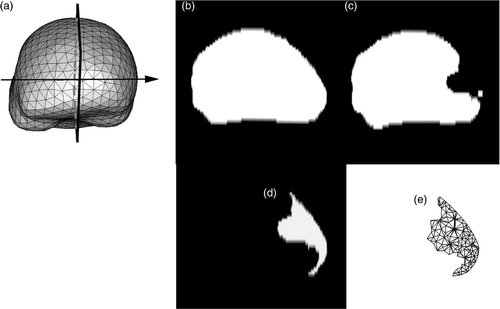
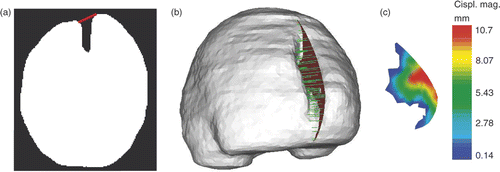
For modeling retraction, a representation of the cut in the biomechanical model is needed in order to enrich the appropriate nodes of the volume mesh with XFEM Heaviside DOFs. The cut is defined as a segmented region, and is ultimately represented by the surface mesh that is the intersection of the 3D volume mesh with this segmented region. For the patient case of interest, the mesh representing the cut is obtained as follows. First, the cut is assumed to be planar, and the cut plane, representing the location where the retractor was inserted, is taken as the plane between the two brain hemispheres (). Second, the 2D region that is the difference between the whole-brain regions segmented out from the two iMR images in the plane of the cut is determined (), where additional image processing has beforehand been performed on to remove small, isolated islands. The result is a 2D cut region, shown in . Third, the intersection of the cut with the volume mesh is computed. This is performed by first computing a distance map of the cut region and then using the filter VTKCutter, which allows one to compute the intersections of a volume mesh with an implicit function, such as a distance map. VTKCutter produces a triangle surface mesh, representing the cut mesh, composed of the set of points intersecting the volume mesh (). Finally, the nodes whose supports are intersected by the discontinuity, i.e., the cut mesh, are enriched with XFEM Heaviside DOFs. The volume mesh consists of 2,198 nodes which correspond to 6,594 FEM DOFs. Enrichment adds 561 XFEM Heaviside DOFs, giving a total of 7,155 DOFs.
Figure 3. Representation of the planar cut to be included in the biomechanical model. (a) Surface mesh of the biomechanical model showing the location of the plane where the retractor was inserted. This plane corresponds to the plane separating the two hemispheres. (b) Whole-brain region segmented out from the second iMR image, acquired before retraction, in the selected plane shown in (a). (c) Whole-brain region segmented out from the the third iMR image, acquired after retraction. (d) Cut defined as the difference between (b) and (c), where (c) has been processed beforehand to remove small, isolated islands. (e) Surface mesh representing the intersections of the cut with the volume mesh of the whole-brain region brain.
Evaluation of surface landmark displacement fields
To drive the deformation of the biomechanical model, the displacement fields of several surface landmarks are evaluated, namely those of the whole-brain region boundary and the two lips of the cut. The lips are assumed to be superimposed before the deployment of the retractor, and then spread out during its deployment.
Displacement field of whole-brain region boundary
To evaluate the displacement field of the whole-brain region boundary between two iMR images, an active surface algorithm Citation[42], Citation[43] is used. An active surface, i.e., a surface mesh with mechanical constraints such as elasticity, is initialized from the region boundary segmented in the second iMR image and then deformed under the influence of external forces computed from the corresponding region boundary in the second iMR image. The external forces attract the surface during an iterative process, in such a way that this surface deforms to cling to the region boundary to be matched. In our work, the external forces F(x) are computed using a gradient descent on a distance map of the region boundary, i.e.,where D(x) is the distance map of the region boundary to match and Smin is chosen to be +1 or −1 to ensure that the external forces point towards a position on the distance map with a smaller distance value. Further details on this specific active surface algorithm can be found in references Citation[7] and Citation[44].
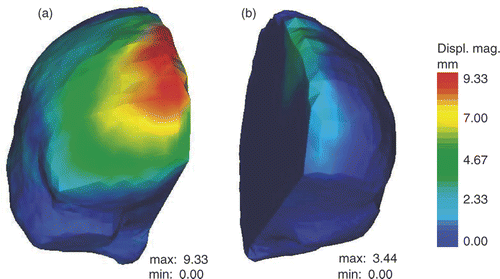
An active surface for the whole-brain region boundary built from the second iMR image would deform on the whole-brain region boundary of the third iMR image by entering into the free path created by the retraction. The resulting displacement field of the active surface algorithm would thus model a deformation that does not occur in reality. The deformation of the whole-brain region boundary is thus decoupled into two independent applications of the active surface algorithm, one for each of the two hemispheres. It can be clearly seen in that the two hemispheres deform differently. show the result of the active surface displacement fields of the two hemispheres. As expected, they deform differently; in particular, the magnitude of the displacement is much larger for the right hemisphere. (In accordance with radiological convention, the right side of the iMR images corresponds to the left side of the brain. Consequently, what we refer to as the left (right) brain hemisphere actually appears on the right (left) of the iMR images. The same applies to the left and right lips of the cut.)
Figure 4. Results of the active surface algorithm for the two hemispheres of the brain. (a) Initial active surface for right hemisphere with color levels corresponding to the magnitude of the displacement field. (b) Initial active surface for left hemisphere with same color coding. (In accordance with radiological convention, the right side of the iMR images corresponds to the left side of the brain, and vice versa.
Displacement field of cut lips
The displacement field of the cut lips is obtained by evaluating the displacement of each node of the cut mesh, as shown in , which represents the intersections of the cut with the volume mesh. In the following, these nodes are referred to as cut intersections.
First, the lips of the cut are assumed to be moved by the retractor along a direction that forms an angle with the plane of the cut, as shown in . A surface mesh of the whole-brain region segmented out from the third iMR image is then built. Finally, for each cut intersection, a vector is defined which starts at this intersection, forms the pre-defined angle with the cut plane (), and stops at its intersection with the surface mesh of the whole-brain region just mentioned. The length and direction of the vector represent the magnitude and direction of the displacement to be applied to the cut intersection of the lip. show the displacement field for the set of cut intersections for the right lip.
Figure 5. Evaluation of the displacement field of the cut for 3D retraction modeling. (a) Whole-brain region segmented out from the third iMR image, with segment (in red) defining the angle of the displacement field of the right lip. (b) Surface mesh built from the segmented third iMR image, cut mesh () (in red), and displacements of cut intersections (in green). (Displacements go from right to left in the figure.) (c) Cut mesh () with color levels corresponding to the magnitude of the displacement field of the right lip.
The displacement of each cut intersection n of the right lip is applied by imposing, based on Equations 3 and 6, the following conditions:
where
is the position of cut intersection n, An and Bn are the two nodes of the FE edge on which cut intersection n is located,
and
are the FEM NSFs of nodes An and Bn,
and
are the nodal FEM DOFs of nodes An and Bn, and
and
are the nodal XFEM Heaviside DOFs of nodes An and Bn. The Heaviside function H(x) is equal to +1 at each
, although it could also be taken as −1. The assignments of the values for H(x) + 1 and −1 can indeed be exchanged, depending which value is assigned on the left and right side of the cut. The conditions in Equation 8 are applied with Lagrange multipliers. A similar procedure is used for the left part of the brain.
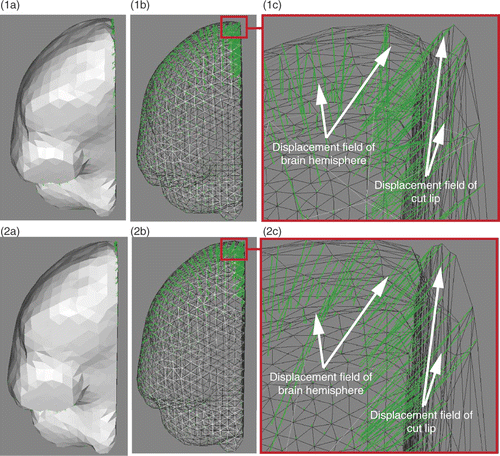
Once the displacement fields of both brain hemispheres and lips of the cut have been estimated, they are applied to the biomechanical model to drive its deformation. These displacement fields come from independent computations and are not necessarily consistent with one another, nor compatible with the volume mesh. jointly show the displacement fields of the right brain hemisphere and of the right lip of the cut. It can be clearly seen that, in the neighborhood of the external surface of the brain, the two displacement fields cross one another. The application of these two displacement fields to the biomechanical model would lead to element flipping. Before applying these displacement fields, they have to be smoothed. show the two displacement fields that have been jointly smoothed based on a weighted-distance average using ten neighbor nodes. The displacement fields are now consistent with one another, and compatible with the volume mesh. Furthermore, the direction of the displacement field of the right hemisphere () is clearly modified by the smoothing (). This modification of direction allows one to better capture the opening of the path created by retraction. It can thus be concluded that the use of an active surface algorithm for evaluating the displacement fields of the brain hemispheres is not sufficient, and that the evaluation of the displacement fields of the cut lips is required to correctly model retraction. A similar procedure is used for the left part of the brain. Note that the left and right displacement fields are smoothed separately because the deformation of the two sides of the brain is different and relatively uncoupled.
Figure 6. Comparison of unsmoothed (row 1) and jointly smoothed (row 2) displacement fields of right hemisphere and right lip of the cut. (1a) Surface meshes of right hemisphere and right lip of cut, and their surface displacement fields in green. (1b) Same as (1a), except that the surface meshes are left transparent. (1c) Zoom of (1b) around the external surface of the brain. (2a), (2b) and (2c) are, respectively, the same as (1a), (1b) and (1c), except that the displacement fields are jointly smoothed. (In each sub-figure, the direction of the displacement vectors is defined such that the vectors start at the surface mesh nodes.)
XFEM-based biomechanical model deformation
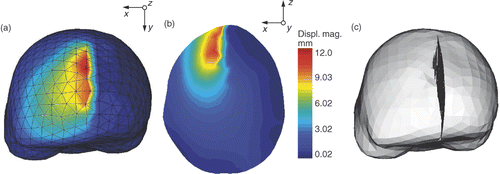
To deform the biomechanical model, one must transpose the smoothed displacement fields of the brain hemispheres and lips of the cut into displacements to be applied to the biomechanical model. The smoothed displacement fields of the lips are directly applied to cut intersections. The displacement of each cut intersection is applied by imposing the conditions described in Equation 8 with Lagrange multipliers. The displacement of each node of the external surface of the biomechanical model is given by the displacement of the closest node of both initial active surfaces, representing the brain hemispheres, or of the surface mesh representing the intersections of the cut with the volume mesh. The biomechanical model then deforms, based on XFEM. For this we use the FEM software tool Metafor (http://metafor.ltas.ulg.ac.be), developed in our mechanical engineering department, to which we have added an XFEM module. The initial stress state of the brain is unknown and is thus set to zero Citation[7], Citation[45]. The resulting volume displacement field of the biomechanical model is shown in .
Figure 7. Deformation of the biomechanical model based on XFEM for retraction modeling. (a) External surface mesh of the biomechanical model with color levels corresponding to the magnitude of the displacement field. (b) Slice of the biomechanical model with same color coding. (c) Final mesh resulting from the modeling of the retraction using XFEM. The tetrahedra that were added to display the opening of the cut lips are there only for visualization purposes. The edges of FEs cut by the discontinuity have actually been made discontinuous and their nodes moved apart.
Results of image warping and validation
The resulting volume displacement field of the biomechanical model () is used to warp the part of the second iMR image corresponding to the whole-brain region, i.e., with the skull and external CSF masked out. shows the second iMR image, and shows the result of rigidly registering the third iMR image to the second iMR image. shows the whole-brain region extracted from , while shows the result of the warping of .
Figure 8. Results of retraction modeling. (1a) Original second iMR image. (2a) Third iMR image, rigidly registered to (1a). (1b) Whole-brain region extracted from (1a). (2b) Deformation of (1b) using the volume displacement field of the biomechanical model, computed via XFEM. (1c) Juxtaposition of edges of whole-brain region of second iMR image (1a) (in green) and edges of whole-brain region of third iMR image (2a) (in red). (2c) Juxtaposition of edges of whole-brain region of second iMR image deformed for modeling retraction (2b) (in green) and edges of whole-brain region of third iMR image (2a) (in red). (The red edges in (1c) and (2c) are identical.)
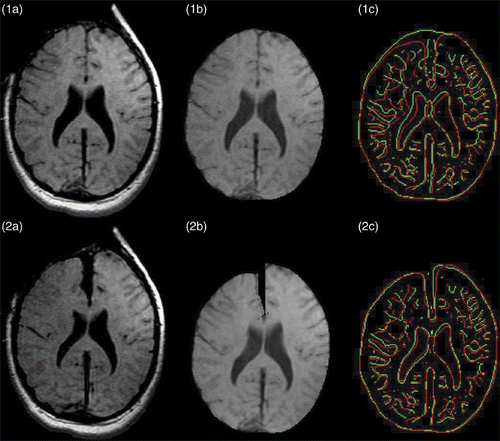
To evaluate the accuracy of our 3D nonrigid registration technique, the image similarity is compared for the two iMR images rigidly registered, with the similarity between the images resulting from the warping of the second iMR image and the third iMR image. This gives an estimate of how well one is able to capture, and compensate for, the local deformations between the two iMR images. To estimate qualitatively the similarity between two images, the edges extracted from these images using the Canny edge detector itkCannyEdgeDetectionImageFilter, available in ITK (http://www.itk.org/), are compared. This allows us to visually, and also locally, estimate the alignment of the iMR images. shows the juxtaposition of edges extracted from the whole-brain region of the two iMR images rigidly registered, while shows the juxtaposition of the edges extracted from the result of the warping of the second iMR image and the whole-brain region of the third iMR image. Visual comparison of shows that the edges match much better in the latter, which qualitatively indicates the benefit of the nonrigid registration.
To quantitatively estimate the similarity of the two edge maps, the modified Hausdorff distance is computed between the sets of edge points, i.e., voxels representing the edges, in these two images. The modified Hausdorff distance ℋ(A, B) [47] between two sets of points A and B is defined aswhere h(A, B) is the directed Hausdorff distance. The directed Hausdorff distance is a measure of the distance from the point set A to the point set B, and is defined as
where Na is the number of points in set A, and d(a, B) is the distance of point a ∈ A to the closest point in B, i.e., d(a, B) = minb∈B‖a − b‖, where ‖a − b‖ is the Euclidean distance. The directed Hausdorff distance h(A, B) thus computes the average distance of points of A to points of B. The averaging minimizes the effects of outlier points, e.g., due to image noise. The value of the modified Hausdorff distance ℋ(A, B) increases with the difference between the two sets of edge points. The advantage of using a criterion such as the modified Hausdorff distance is that it quantifies (numerically) the alignment of the iMR images; however, the quantification remains global and cannot reflect the fact that some local regions could be better non-rigidly registered than others. The qualitative and quantitative evaluations of the non-rigid registration, based on the visual comparison of edges and the modified Hausdorff distance, are thus complementary.
The modified Hausdorff distance decreases from 1.36 mm for the set of edges extracted from the two iMR images rigidly registered () to 1.10 mm for the iMR images nonrigidly registered (). This indicates that the nonrigid registration improves the alignment of the two iMR images, although the opening width of the path created by retraction is not totally captured, as shown in in comparison to . We expect this is due to the segmentation, as explained above. In addition, the need to jointly smooth (on each side of the brain, respectively) the displacement fields of the hemispheres and the lips of the cut, as explained above, tends to globally decrease the magnitude of the displacement fields, and thus to decrease the opening width of the path.
Conclusions
We have developed a method using intraoperative images for evaluating brain deformation due to retraction. The nonrigid registration technique was based on a linear elastic biomechanical model driven by the deformations of key surfaces, extracted and tracked from two successive iMR images. The cut was defined directly based on the two iMR images, and was then included in the biomechanical model using XFEM. The iMR images used in our experiments were acquired with a 0.5-Tesla GE Signa intraoperative scanner. To facilitate the evaluation of our results, the iMR image acquired just before retraction was updated as a substitute preoperative image.
In this work, all performed in three dimensions, the whole-brain region boundary and the cut were chosen as surface landmarks. The deformation of the whole-brain region boundary was decoupled in two independent computations of the active surface algorithm for both hemispheres, and this was shown to be beneficial since they deformed very differently. Also shown was the importance of evaluating the displacement fields of the cut lips to correctly capture the opening of the path created by retraction. While the opening width of the path was not totally captured, the nonrigid registration was shown to improve the alignment of the two iMR images.
To further improve our retraction modeling, future work may be considered in four main areas. First, a way of preventing the opening width of the path from decreasing should be determined. The segmentation method, and the evaluation of the surface displacement fields, should thus be improved to ensure that they do not imply this effect. Second, a linear formulation of XFEM was used, which applies to small deformations. Nevertheless, a nonlinear formulation of XFEM Citation[33] should be implemented for surgery cases involving large deformations of the brain. Third, the fact that iMR images are used could be further exploited. Indeed, these images provide volume information (rather than surface information only), are of good quality in comparison to other intraoperative modalities, and possess a field of view that includes the full volume of brain tissues (for the 0.5-T GE Signa scanner). These images thus allow one to evaluate what, and how, new structures of the brain could be used. Some regions, e.g., the lateral ventricles region, could be extracted from the two iMR images and used as surface landmarks to drive the deformation of the biomechanical model. Fourth, as mentioned in the introduction, retraction followed by successive resections should be modeled to enable updating of preoperative images for any kind of deformation encountered.
Declaration of interest: This research was supported in part by the Fonds Léon Fredericq, Liège, Belgium; the Fonds National de la Recherche Scientifique (F.N.R.S.), Brussels, Belgium; a research grant from CIMIT; and NIH grants R01 RR021885, R01 GM074068 and R01 EB008015.
References
- Nabavi A, Black PM, Gering DT, Westin CF, Mehta V, Pergolizzi RS, Ferrant M, Warfield SK, Hata N, Schwartz RB, et al. Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery 2001; 48(4)787–798
- Nimsky C, Ganslandt O, Hastreiter P, Fahlbusch R. Intraoperative compensation for brain shift. Surgical Neurol 2001; 56(6)357–365
- Paulsen KD, Miga MI, Kennedy FE, Hoopens PJ, Hartov A, Roberts DW. A computational model for tracking subsurface tissue deformation during stereotactic neurosurgery. IEEE Trans Biomed Eng 1999; 46(2)213–225
- Miga MI, Paulsen KD, Hoopes PJ, Kennedy FE, Hartov A, Roberts DW. In vivo quantification of a homogeneous brain deformation model for updating preoperative images during surgery. IEEE Trans Biomed Eng 2000; 47(2)266–273
- Hagemann A, Rohr K, Stiehl HS. Coupling of fluid and elastic models for biomechanical simulations of brain deformations using FEM. Med Image Anal 2002; 6(4)375–388
- Skrinjar O, Nabavi A, Duncan J. Model-driven brain shift compensation. Med Image Anal 2002; 6(4)361–373
- Ferrant M, Nabavi A, Macq B, Kikinis R, Warfield SK. Serial registration of intra-operative MR images of the brain. Med Image Anal 2002; 6(4)337–359
- Lunn KE, Paulsen KD, Roberts DW, Kennedy FE, Hartov A, West JD. Displacement estimation with co-registered ultrasound for image guided neurosurgery: A quantitative in vivo porcine study. IEEE Trans Med Imaging 2003; 22(11)1358–1368
- Warfield SK, Haker SJ, Talos IF, Kemper CA, Weisenfeld N, Mewes AU, Goldberg-Zimring D, Zou KH, Westin CF, Wells WM, et al. Capturing intraoperative deformations: research experience at Brigham and Women's Hospital. Med Image Anal 2005; 9(2)145–162
- Clatz O, Delingette H, Talos I-F, Golby AJ, Kikinis R, Jolesz FA, Ayache N, Warfield SK. Robust non-rigid registration to capture brain shift from intra-operative MRI. IEEE Trans Med Imaging 2005; 24(11)1417–1427
- Wittek A, Miller K, Kikinis R, Warfield SK. Patient-specific model of brain deformation: Application to medical image registration. J Biomechanics 2007; 40(4)919–929
- Songbai J, Hartov A, Robert D, Paulsen K. Data assimilation using a gradient descent method for estimation of intraoperative brain deformation. Med Image Anal 2009; 13(5)744–756
- Kyriacou SK, Mohamed A, Miller K, Neff S. Brain mechanics for neurosurgery: Modeling issues. Biomechanics and Modeling in Mechanobiology 2002; 1(2)151–164
- Cohen-Adad J, Paul P, Morandi X, Jannin P. Knowledge modeling in image guided neurosurgery: application in understanding intra-operative brain shift. In: Cleary KR, Galloway RL Jr. Proceedings of SPIE Medical Imaging 2006: Visualization, Image-Guided Procedures and Display, San Diego, CA, February 2006. Proc SPIE 2006;6141:709–716
- Miller K, Wittek A, Joldes G, Horton A, Dutta-Roy T, Berger J, Morriss L. Modelling brain deformations for computer-integrated neurosurgery. Int J Numer Methods Biomed Eng 2009; 26(1)117–138
- Nienhuys HW, van der Stappen FA. A surgery simulation supporting cuts and finite element deformation. In: Niessen WJ, Viergever MA, editors. Proceedings of the 4th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2001), Utrecht, The Netherlands, October 2001. Lecture Notes in Computer Science 2208. Berlin: Springer; 2001. pp 153–160
- Serby D, Harders M, Székely G. A new approach to cutting into finite element models. In: Niessen WJ and Viergever MA, editors. Proceedings of the 4th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2001), Utrecht, The Netherlands, October 2001. Lecture Notes in Computer Science 2208. Berlin: Springer; 2001. pp 425–433
- Steinemann D, Harders MA, Gross M, Székely G. Hybrid cutting of deformable solids. In: Proceedings of the IEEE Computer Society Conference on Virtual Reality. Alexandria, VA, March 2006. pp 35–42
- Mor A, Kanade T. Modifying soft tissue models: Progressive cutting with minimal new element creation. In: Delp S, DiGioia AM, Jaramaz B, editors. Proceedings of the 3rd International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2000), Pittsburgh, PA, October 2000. Lecture Notes in Computer Science 1935. Berlin: Springer; 2000. pp 598–607
- Ganovelli F, Cignoni P, Montani C, Scopigno R. A multiresolution model for soft objects supporting interactive cuts and lacerations. Computer Graphics Forum 2000; 19(3)271–282
- Nienhuys HW. Cutting in deformable objects. PhD thesis, Institute for Information and Computing Sciences, University of Utrecht, The Netherlands, 2003
- Bielser D, Glardon P, Teschner M, Gross MH. A state machine for real-time cutting of tetrahedral meshes. Graphical Models 2004; 66(6)398–417
- Duflot M, Nguyen-Dang H. A meshless method with enriched weight functions for fatigue crack growth. Int J Numer Methods Eng 2004; 59: 1945–1961
- Moës N, Dolbow J, Belytschko T. A finite element method for crack growth without remeshing. Int J Numer Methods Eng 1999; 46: 131–150
- Abdelaziz Y, Hamouine A. A survey of the extended finite element. Computers and Structures 2008; 86(11–12)1141–1151
- Vigneron LM, Duflot MP, Robe PA, Warfield SK, Verly JG. 2D XFEM-based modeling of retraction and successive resections for preoperative image update. Comput Aided Surg 2009; 14(1–3)1–20
- Miga MI, Roberts DW, Kennedy FE, Platenik LA, Hartov A, Lunn KE, Paulsen KD. Modeling of retraction and resection for intraoperative updating of images. Neurosurgery 2001; 49(1)75–84
- Platenik LA, Miga MI, Roberts DW, Lunn KE, Kennedy FE, Hartov A, Paulsen KD. In vivo quantification of retraction deformation modeling for update image-guidance during neurosurgery. IEEE Trans Biomed Eng 2002; 49(8)823–835
- Platenik LA, Miga MI, Roberts DW, Kennedy FE, Hartov A, Lunn KE, Paulsen KD. Comparison of an incremental versus single-step retraction model for intraoperative compensation. Proceedings of SPIE Medical Imaging 2001: Visualization, Image-Guided Procedures and Display, San Diego, CA, February 2001. Proc SPIE 2001;4319:358–365
- Lamprich BK, Miga MI. Analysis of model-updated MR images to correct for brain deformation due to tissue retraction. Proceedings SPIE Medical Imaging 2003: Visualization, Image-Guided Procedures and Display, San Diego, CA, February 2003. Proc SPIE 2003;5029:552–560
- Lunn KE, Paulsen KD, Roberts DW, Kennedy FE, Hartov A, Platenik LA. Nonrigid brain registration: synthesizing full volume deformation fields from model basis solutions constrained by partial volume intraoperative data. Computer Vision and Image Understanding 2003; 89: 299–317
- Sun H, Kennedy FE, Carlson EJ, Hartov A, Roberts DW, Paulsen KD. Modeling of brain tissue retraction using intraoperative data. In: Barillot C, Haynor DR, Hellier P, editors. Proceedings of the 7th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2004), Saint-Malo, France, September 2004. Part I. Lecture Notes in Computer Science 3216. Berlin: Springer; 2004. pp 225–233
- Jerabkova L, Kuhlen T. Stable cutting of deformable objects in virtual environments using xfem. IEEE Computer Graphics and Applications 2009; 29(2)61–71
- Zienkiewicz OC, Taylor RL. The Finite Element Method. Butterworth-Heinemann, Oxford 2000
- Sukumar N, Prévost JH. Modeling quasi-static crack growth with the extended finite element method. Part I: Computer implementation. Int J Solids Structures 2003; 40(26)7513–7537
- Dolbow JE. An extended finite element method with discontinuous enrichment for applied mechanics. PhD thesis, Northwestern University, Evanston, IL; 1999
- Sukumar N, Moës N, Belytschko T, Moran B. Extended finite element method for three-dimensional crack modelling. Int J Numer Methods Eng 2000; 48(11)1549–1570
- Felippa CA. Introduction to Finite Element Method, http://www.colorado.edu/engineering/cas/courses.d/IFEM.d/
- Vigneron LM, Boman RC, Ponthot J-P, Robe PA, Warfield SK, Verly JG. Enhanced FEM-based modeling of brain shift deformation in image-guided neurosurgery. J Comput Appl Math 2010; 234: 2046–2053
- Geuzaine C, Remacle J-F. Gmsh: A three-dimensional finite element mesh generator with built-in pre- and post-processing facilities. Int J Numer Methods Eng 2009; 79(11)1309–1331
- Miller K, Wittek A. Neuroimage registration as displacement – zero traction problem of solid mechanics. Presentation at Workshop on Computational Biomechanics for Medicine at the 9th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2006), Copenhagen, Denmark, October 2006
- Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. Int J Comput Vision 1988; 1(4)321–331
- Xu C, Pham DL, Prince JL. Medical image segmentation using deformable models. In: Sonka M, Fitzpatrick JM, editors. SPIE Handbook of Medical Imaging. Vol 2: Medical Image Processing and Analysis. SPIE; 2000. pp 129–174
- Ferrant M. Physics-based deformable modeling of volumes and surfaces for medical image registration, segmentation and visualization. PhD thesis, Université catholique de Louvain, Louvain-la-Neuve, Belgium; 2001
- Warfield SK, Talos F, Kemper C, Cosman E, Tei A, Ferrant M, Macq B, Wells WM, Black PM, Jolesz FA, Kikinis R. Augmenting intraoperative MRI with preoperative fMRI and DTI by biomechanical simulation of brain deformation. Proceedings of SPIE Medical Imaging 2003: Visualization, Image-Guided Procedures and Display, San Diego, CA, February 2003. Proc SPIE 2003;5029:79–86
- Dubuisson M-P, Jain AK. A modified Hausdorff distance for object matching. In: Proceedings of the 12th IAPR International Conference on Pattern Recognition, Jerusalem, Israel, October 1994. pp. 566–568