Abstract
Context: Marine natural products are a rich source of potent, selective, and structurally novel compounds. Marine bacteria are considered the most promising source of biologically active compounds which can be applied to treat a wide range of diseases.
Objective: The current study was designed to establish the bases for a future marine exploration in the Ecuadorian coast based on the molecular identification of a marine bacterium and its potential use as an antibacterial or cytotoxic compounds source.
Materials and methods: Isolation and characterization of the marine bacterium were carried out through microbiological methods from desiccated sediment. Molecular identification was made by means of 16S rDNA analysis. MIC was measured by the microdilution broth method against six pathogenic bacteria: two Gram positive and four Gram negative strains. Cytotoxicity was evaluated by Crystal violet assay against breast adenocarcinoma (MCF7) and ductal carcinoma (T47D and ZR-75-30).
Results: Our present study has shown that EtOAc extract and fraction A1 obtained from marine Streptomyces sp. revealed the maximal antibacterial and cytotoxic activity. Enterococcus faecalis was found to be more sensitive strain (MIC 0.78 μg/ml) than the other five bacteria tested. ZR-75-30 and T47D cell lines were found to be more sensitive (IC50 value, 31.88 ± 0.05 and 68.35 ± 0.12 μg/ml) than adenocarcinoma MCF7 (IC50 value was 83.65 ± 0.06 μg/ml).
Discussion and conclusion: The results obtained herein indicate that EtOAc extract of Streptomyces sp. has shown a strong antibacterial activity as well as moderate cytotoxic activity which make it a good candidate for metabolite isolation.
Introduction
The acquired resistance of bacterial infections and various human cancers to conventional chemotherapy has renewed the interest of biomedical professionals and scientists alike to produce new specific types of molecules with novel mechanisms of action that help to diminish the attack of these pathogenic strains and to reduce the possibility of cancer reemerging diseases (Demain & Sanchez, Citation2009; Liu et al., Citation2010).
Many different approaches have been carried out to identify new substances with potent activities against pathogenic bacteria such as the Methicillin Resistant Staphylococcus aureus (MRSA) or the Vancomycin Resistant Enterococcus faecium or E. faecalis (VREF), which normally are acquired as nosocomial infections, however, no new compounds have reached clinical use yet (Kwon et al., Citation2006).
World Health Organization (WHO) explicitly emphasizes cancer as a leading cause of 7.6 million deaths worldwide in 2008. Currently, chemotherapy is so far the best treatment for human cancer; however, the high toxicity of anti-cancer chemical compounds underscores the urgent necessity to find, purify, and develop novel naturally produced biological compounds, with fewer side effects and/or with greater therapeutic efficiency (Olano et al., Citation2009).
Most microbial small molecule discovery efforts have focused on Actinomycetales from terrestrial environments but the rate of discovery has decreased considerably, thereby reinforcing the discovery efforts (Kwon et al., Citation2006). Marine-derived Actinomycetales are proving to be an important resource of such compounds leading to a discovery of many biologically active molecules with unique chemical structures (Nam et al., Citation2010).
Actinomycetales are an order of Actinobacteria, which are filamentous Gram-positive bacteria, characterized by their complex life cycle and are widely distributed in terrestrial and aquatic ecosystems. Among these, Streptomyces genus is one of the most diverse and shows the ability to produce clinically useful active compounds since the discovering of the streptomycin in 1943 (Waksman, Citation1953). To date, the Actinomycetales have produced more than 10 000 bioactive compounds, 7600 derived from Streptomyces (Bérdy, Citation2005).
The Ecuadorian coast has been poorly explored as a source of chemical diversity and few works have been conducted, focusing the research on medicinal plants rather than marine organisms. The aim of this work was to establish the bases for a future marine exploration in the Ecuadorian coast based on the molecular identification of a marine bacterium and its potential use as an antibacterial or cytotoxic compounds source.
Materials and methods
Collection and characterization of MJG-3 strain
Different samples (20–50 g) were collected from Lat. 3°15′792 S and Long. 80°00′739 W to Lat. 03°17′711 S and Long. 80°01′924 W in Jambelí mangrove (Ecuador). These samples were collected from 1 to 3 m depth from marine sediment and put into sterile polythene bags. Water samples were collected in 15 ml Falcon tubes for pH measuring. A total of 20 sediment samples were collected and desiccated for 24 h at 32 °C. Stamps of pulverized samples were seeded on agar plates of a sea water-based medium (10 g starch, 4 g yeast extract, 2 g peptone, 1 g CaCO3, 40 mg Fe2(SO4)3·4H2O, 100 mg KB, 15 g Bacto Agar, per liter), acidic Gauze medium, Humic acid medium, and TCG medium, all of them containing 1% of polymyxin B and cycloheximide as inhibitors of undesired growth. The inverted plates were incubated at 30 °C and observed periodically. Gram staining was used to identify branched filamentous Gram-positive bacteria.
DNA isolation, PCR, sequencing, and phylogenetic analysis
The DNA was extracted with the PureLinkTM Genomic DNA mini kit (Invitrogen, Waltham, MA) for bacteria Gram positive, according to recommendations to the manufacturer. The partial 16S rDNA was amplified using the universal primers for bacteria 27F (AGAGTTTGATCMTGGCTCAG Lane, Citation1991) and 1492R (I) (GGTTACCTTGTTACGACTT, Turner et al., Citation1999) and the combination 341F (CCTACGGGAGGCAGCAG; Muyzer et al., Citation1993) and 907R (CCGTCAATTCMTTTGRTT; Lane, Citation1991). PCR conditions are similar as explained by Cruz et al. (Citation2011). Each PCR sample was tested by a 1% agarose gel electrophoresis containing GelRed nucleic acid stain (Biotum, Hayward, CA). The products were purified with the QI quick PCR purification kit (Qiagen, Valencia, CA). Sequencing was carried out in a 3500RUO Genetic Analyzer (Applied Biosystems, Waltham, MA). The sequences obtained in this study are available from GenBank JX463753 and JX463754.
Phylogenetic calculations were implemented in the programs RAxML v7.0.4 (Stamatakis, Citation2006). Maximum likelihood (ML) was based on 1000 bootstrap replicates (Pattengale et al., Citation2010) and GTRMIX as a DNA substitution model (Stamatakis, Citation2006).
Cultivation and extraction
The strain MJG-3 was cultured in ten 2.8 l Fernbach flasks each containing 1 l of a seawater based medium (10 g starch, 4 g yeast extract, 2 g peptone, 1 g CaCO3, 40 mg Fe2(SO4)3·4H2O, 100 mg KBr) and the flasks were shaken at 200 rpm at 30 °C for 7 d. After this time, sterilized XAD-7-HP (SIGMA) resin (20 g/l) was added to the flasks to adsorb the organic products, and the system shaken at 200 rpm for 2 h. The resin was filtered through cheesecloth, washed with deionized water, and the organic compounds absorbed eluted with acetone (Freel et al., Citation2011; Hu et al., Citation2011). The acetonic extract was dissolved in MeOH and the precipitated salts discarded yielding 5 g of methanolic extract. It was chromatographed using 50 g of silica gel 60 (230–400 mesh-MERCK) and EtOAc:MeOH as eluents (9:1, 7:3, 5:5, 3:7, and 1:9), collecting 1 l per proportion. TLC was carried out to compare the resulting fractions.
Antibacterial assay
Bacterial strains and culture conditions
The following six pathogenic bacteria (American Type and Culture Collection, ATCC) were included in this study: Staphylococcus aureus ATCC 25923, E. faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Proteus vulgaris ATCC 8427, and Klebsiella pneumoniae ATCC 9997. Tryptic Soy Broth (TSB-DIFCO, DIFCO, Sparks, MD) was employed for all bacteria except for E. faecalis where Brain Heart Infusion broth (BHI-DIFCO, DIFCO, Sparks, MD) was used. A cryogenic reserve of each bacterium kept at −80 °C was used to produce an overnight culture at 37 °C for 16 h.
Minimum inhibitory concentration (MIC) determination
MIC values were determined by the microdilution broth method using a final concentration of 5 × 105 CFU/ml. The MIC was defined as the lowest concentration of substance that completely inhibits growth of the organism in the microdilution wells as detected by the unaided eye (CLSI, M7-A7. 2006). DMSO solutions of the extract and fractions were prepared at a concentration of 20 μg/ml. The assays were carried-out in 96-well plates and two-fold serial dilution was employed to obtain decreasing concentrations since 1000–0.024 μg/ml. Incubation was at 37 °C for 24 h (Cos et al., Citation2006). Gentamicin was used a positive control with a MIC value of 0.40 μg/ml except for E. faecalis where ampicillin (MIC 1.56 μg/ml) was used.
Cytotoxicity assay
Cell cultures
Three cell lines: one of breast adenocarcinoma (MCF-7) and two of ductal carcinoma (T47D and ZR-75-30) from mammary gland were used. All cell lines were continuously grown on culture dishes in RPMI-1640 basal medium (GIBCO, Waltham, MA) supplemented with fetal bovine serum 10% (GIBCO, Waltham, MA), antibiotic-antimycotic 1% (GIBCO, Waltham, MA), l-glutamine 2 mmol/ml (GIBCO, Waltham, MA), sodium bicarbonate 0.2% (GIBCO, Waltham, MA), at 37 °C in a humidified atmosphere at 5% CO2 before using them in the colorimetric test.
Crystal violet assay
Cell inhibition was measured according to the Cell Adhesion Protocol Manual from BD Biosciences (Becton, Dickinson & Company, Citation2005) also known as Crystal violet assay, with some modifications as fixation with formaldehyde 4% and extraction of crystal violet with acetic acid 10% instead of Triton X-100. The assay was carried out in 96-well plates and cellular density of 3500 cells per well. After 24 h of incubation, the cells were treated with the extract and fractions at doses of 15, 30, 45, 60, and 75 μg/ml and incubated for 48 h. DMSO (0.5%) was used as a negative control and doxorubicin (DOX) as a positive control.
After 72 h of incubation, the absorbance at 620 nm was recorded in a Tecan microplate reader (Model Sunrise Tecan Austria GmbH, Grödig/Salzburg, Austria). The experiment was repeated three times with three replicates per dose for each cell line.
Statistical analysis
The recorded absorbances were switched to their equivalent percentages of inhibition (PI). GraphPad PRISM 5® statistical software (GraphPad Software Inc., San Diego, CA) was employed to obtain the IC50 values through a non-linear regression between the logarithms of the doses versus the PI.
Results and discussion
Collection, isolation, and molecular identification
From 20 sediments samples collected and desiccated, after cultivation and periodic examination, a total of 15 different kinds of Actinomycetales were isolated (data not shown) and further maintained in Petri dishes and cryopreserved at −80 °C in the presence of 10% DMSO. Sea-water samples showed pH 7.67–8.16 considering that the zone of collection was close enough to a shrimp farm. The MJG-3 strain, derived from the initial studies grown in acidic gauze medium, was isolated and selected for extraction. This was due to the production of a dark pigment in the sea water-based medium (), and the short generation time, which made it suitable for scale up of fermentation process and metabolic extraction.
Figure 1. MJG3 strain identified as Streptomyces sp. in agar sea-water-based medium exhibiting pigment production.

A fresh culture was used to extract the DNA from which two sequences of 790 bp each were obtained. The partial 16S rDNA analysis by Maximum likelihood (ML) reveals one clade supported with 99% ML within the Streptomyces genus ().
Figure 2. Phylogenetic tree of the partial 16S region includes new sequences to Streptomyces sp. and the most closely related sequences from GenBank (NCBI).
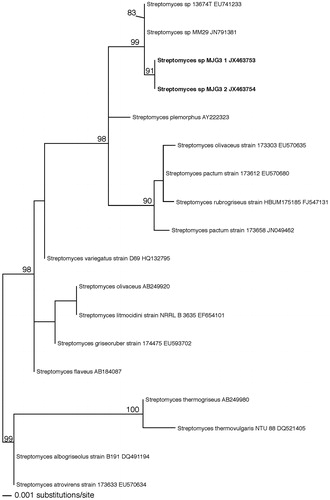
In , it is highlighted in bold the two new sequences of this study. All the values along the nodes correspond to ML and only values higher than 80% are shown in the tree.
Extraction and fractionation
The salt-free methanol extract (5 g) was Si-chromatographed using a specific mix of EtOAc and MeOH in decreasing proportions (9:1, 7:3, 5:5, 3:7, and 1:9). Each fraction was labeled as A1, A2, A3, A4, and A5, respectively. The start material was labeled as A0. Yields of each fraction are shown in .
Table 1. Minimal inhibitory concentration and half maximal inhibitory concentration values of extract and fractions of Streptomyces sp. yields of fractions are given in grams.
Antibacterial activity
To date only six antibacterial agents are in clinical development (phase III) and, just Difimicin developed by Optimer Pharmaceuticals is a new natural product for the treatment of Clostridium difficile-associated diarrhea (Donadio et al., Citation2010). Novel promises from S. platensis are platensimycin and platencin with MIC values of 0.1–0.32 μg/ml and 0.5–3.2 μg/ml against MRSA and VRE. Platensimycin also inhibits the highly pathogenic bacteria Mycobacterium tuberculosis (Martens & Demain, Citation2011).
Although daptomycin, a lipopeptide from Streptomyces roseus, represents a new natural product approved by FDA in 2003, until now, no new antibiotics have been approved, doing the research of natural products a relevant issue (Demain & Sanchez, Citation2009).
Marine-derived Streptomyces spp. continues affording new classes of chemical entities as reported by Zhang et al. (Citation2012) with the isolation of spiroindomicins A–D or the Lobophorins E and F isolated from the South of China. Lobophorin F exhibited strong antibacterial activity against S. aureus and E. faecalis with MIC values of 8 μg/ml (Niu et al., Citation2011).
The minimal inhibitory concentration (MIC) of the extract and fractions is shown in . The higher activity exhibited against E. faecalis demonstrates the selectiveness of the compounds present in the mixture. According to Cos et al. (Citation2006), good endpoints for antibacterial activity include records of IC50, IC90, MIC, and MBC values. For all anti-infective bioassays, the IC50 value of pure compounds should be below 25 μmol/l and, for mixtures, it should be below 100 μg/ml. Considering that MIC can be expressed as 99% inhibition, and comparing the MIC values of the extract and fraction A1 (0.78 μg/ml), we can conclude that the strong antibacterial activity exhibited results attractive for the isolation of chemical entities. A deep chromatographic research should carried out using the A1 (AcOET:MeOH 9:1) fraction as the starting material.
Cytotoxic activity
According to the College of American Pathologists (Citation2011) invasive ductal breast carcinoma is the most common invasive breast cancer representing 65–85% of all cases reported. The World Health Organization (WHO) estimates that deaths from cancer worldwide are projected to continue arising with 13.1 million deaths for 2030. In 2008, breast cancer has caused 458 000 deaths (Ferlay et al., Citation2010).
Natural sources continue rendering good alternatives for cancer treatment and marine Actinomycetales have proven to be a new source for anticancer drug discovery, leading to the isolation of salinosporamides from the recently discovered genus Salinispora (Danishefsky & Endo, 2005), the lobophorine F early mentioned (Niu et al., Citation2011), or the marinomycins from Marinispora (Kwon et al., Citation2006), all of them having significant cytotoxic activity.
Marine exploration (deep or shallow) arises as the promise for future development of drugs and the Actinomycetales have been recognized to contain non-ribosomal polyketide synthase (NRPK) and polyketide shyntase (PKs) pathways, the hallmarks of metabolic production (Manivasagan et al., Citation2013).
The IC50 values of the extract and fractions are shown in . A1 inhibits more strongly the ductal carcinoma cell line ZR-75-30 rather than T-47D or MCF7, indicating a selective mechanism of action. Although cytotoxic activity against ZR-75-30 is the most significant, the extract and fraction A1 show moderate activity against all the cell lines. According to US NCI plant screening program, a crude extract is generally considered to have in vitro cytotoxic activity, if the IC50 value is less than 30 μg/ml (Ponvinobala et al., Citation2012). Breast cancer cell (BCC) lines MCF7 and T-47D account for more than two-thirds of all abstracts reporting studies as concluded from a Medline-based survey, ZR-75-30 differentiates of them for having an amplified ERBB2 gene (estrogen receptor) (Lacroix & Leclercq, Citation2004); this indicates the correct choice to perform this kind of studies.
Acknowledgements
Special thanks is dedicated to John MacMillan, Principal Investigator of the Biochemistry Department at University of Texas Southwestern Medical Center at Dallas, for the extraordinary training given to the author, for Actinomycetales collection from shallow marine sediments, through which this project has started.
Our great thanks to Edward Ratoviski Ph.D. from Johns Hopkins (Baltimore) for his contribution in this work.
Declaration of interest
The authors report that they have no conflicts of interest. The present study was supported by Universidad Técnica Particular de Loja-Ecuador, through the project PROY IQA 0016.
References
- Becton, Dickinson and Company. (2005). Cell adhesion protocol manual, Available from: http://www.bdbiosciences.com/documents/Cell_Adhesion_protocol_manual.pdf [last accessed 20 May 2013]
- Bérdy J. (2005). Bioactive microbial metabolites. Jpn J Antibiot 58:1–26
- Clinical and Laboratory Standards Institute. (2006). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; approved standard. 7th edn. Method M7-A7, Vol. 26, No. 2 p 1–45. Wayne Ed
- College of American Phatologists. (2011). Breast Cancer. Invasive Ductal Carcinoma, Available from: http://www.cap.org/apps/docs/reference/myBiopsy/BreastInvasiveDuctalCarcinoma.pdf [last accessed 26 Apr 2013]
- Cos P, Vlietinck AJ, Berghe DV, et al. (2006). Anti-infective potential of natural products: How to develop a stronger in vitro ‘proof-of-concept’. J Ethnopharmacol 106:290–302
- Cruz D, Suárez JP, Kottke I, et al. (2011). Defining species in Tulasnella by correlation morphology and nrDNA ITS-5.8s sequence data of basidiomata from a tropical Andean forest. Mycol Prog 10:229–38
- Danishefsky SJ, Endo A. (2005). Total synthesis of Salinosporamide A. J Am Chem Soc 127:8298–9
- Demain AL, Sanchez S. (2009). Microbial drug discovery: 80 years of progress. Jpn J Antibiot 62:5–16
- Donadio S, Maffioli S, Monciardini P, et al. (2010). Antibiotic discovery in the twenty-first century: Current trends and future perspectives. Jpn J Antibiot 63:423–30
- Ferlay J, Shin H, Bray F, et al. (2010). Estimates of worldwide burden cancer in 2008: GLOBOCAN 2008. Int J Cancer 127:2893–917
- Freel KC, Jam S, Fenical W, et al. (2011). Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl Environ Microbiol 77:7261–70
- Hu Y, Espindola APDM, Stewart NA, et al. (2011). Chromomycin SA analogs from a marine-derived Streptomyces sp. Bioorg Med Chem 19:5183–9
- Kwon HC, Kauffman CA, Jensen P, et al. (2006). Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J Am Chem Soc 128:1622–32
- Lacroix M, Leclercq G. (2004). Relevance of breast cancer cell lines as models for breast tumours: An update. Breast Cancer Res Treat 83:249–89
- Lane DJ. (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, eds. Nucleic Acid Techniques in Bacterial Systematics. New York, NY: John Wiley and Sons, 115–75
- Liu X, Ashforth E, Ren B, et al. (2010). Bioprospecting microbial natural product libraries from the marine environment for drug discovery. Jpn J Antibiot 63:415–22
- Manivasagan P, Venkatesan J, Sivakumar K, et al. (2013). Marine actinobacterial metabolites: Current status and future perspectives. Microbiol Res 168:311–32
- Martens E, Demain AL. (2011). Platensimycin and platencin: Promising antibiotics for future application in human medicine. Jpn J Antibiot 64:705–10
- Muyzer G, De Waal EC, Uittierlinden AG. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
- Nam SJ, Gaudêncio SP, Kauffman CA, et al. (2010). Fijiolides A and B, inhibitors of TNF-alpha-induced NF kappa B activation, from a marine-derived sediment bacterium of the genus Nocardiopsis. J Nat Prod 73:1080–6
- Niu S, Li S, Chen Y, et al. (2011). Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. Jpn J Antibiot 64:711–16
- Olano C, Méndez C, Salas JA. (2009). Antitumor compounds from marine Actinomycetales. Marine Drugs 7:210–48
- Pattengale ND, Alipour M, Bininda-Emonds O, et al. (2010). How many bootstrap replicates are necessary? J Comput Biol 17:337–54.
- Ponvinobala K, Kanchana G, Rubalakshmi G. (2012). In vitro anticancer activity of hydro-alcohol extract of leaves of Andrographis neesiana against PC-3 and MCF-7 cell lines. Int J Pharm Sci 4:396–9
- Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–90
- Turner S, Pryer KM, Miao VPW, et al. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46:327–38
- Waksman SA. (1953). Streptomycin: Background, isolation, properties, and utilization. Science 118:259–66
- Zhang W, Liu Z, Li S, et al. (2012). Spiroindimicins A-D: New bisindole alkaloids from a deep-sea-derived Actinomycete. Org Lett 14:3364–7
