Abstract
Context: It is well known that marine fungi are an excellent source of biologically active secondary metabolites, and by 2011, it was reported that over 400 bioactive metabolites were derived from marine fungi.
Objective: This study establishes the basis for future research on antiproliferative compounds of marine endophytes inhabited in the Veracruz Reef System.
Materials and methods: Isolation of the 34 fungal strains was carried out by microbiological method from samples of sponges, corals, and other biological material from the Veracruz Reef System. The fungal biomass and broth were separated and extracted with a mixture of solvents MeOH:CHCl3. Characterization and molecular identification of the fungal strains were performed through microbiological methods and the analysis of the ITS-rDNA regions. Antiproliferative activity was tested at a dose of 250 μg/mL on human solid tumor cell lines HBL-100, HeLa, SW1573, T-47D, and WiDr by the SRB assay after 48 h-exposure to the fungal extracts.
Results: The extracts from five isolates showed an antiproliferative effect against one or more of the tested cell lines (percentage growth < 50%). The mycelial extract from the isolate LAEE 03 manifested the highest activity against the five cell lines (% PG of 17 HBL-100, 19 HeLa, 23 SW1573, -6 T-47D, and 10 WiDr) and the strain was identified as Curvularia trifolii (Kauffman) Boedijn (Pleosporaceae).
Discussion and conclusion: The results obtained indicate that the extract from a marine derived C. trifolii has the antiproliferative effect, thus suggesting that this organism is a good candidate for further analysis of its metabolites.
Introduction
It is widely known that fungi represent an excellent source of biologically active secondary metabolites. However, it was not until 1981 with the isolation of the antibiotic siccaine from a marine fungus that these organisms started to draw attention (Kupka et al., Citation1981). As stated in the literature (Gareth, Citation2011), the number of bioactive metabolites derived from marine fungi has increased yearly (from 1 in 1981; 100 in 2002; 272 in 2004; and >400 in 2011). The marine fungi are a source of bioactive natural products of diverse chemical nature such as alkaloids, macrolides, terpenes, and peptides among others (Zhang et al., Citation2009).
According to statistics (Bungi & Ireland, Citation2004), 85% of the reported compounds derived from marine fungi were obtained from endophytic fungi isolated from vegetal substrates, algae, mollusks, and marine invertebrates, among other organisms. An example of this was the isolation of sporolids A and B obtained from a microscopic fungus of genus Cladosporium isolated from an alga (Shigemori et al., Citation2004). Both the compounds manifested a cytotoxic activity against murine lymphoma cell line L1210. Similarly, varitrol obtained from the fungus Emericella variecolor Berk. & Broome showed cytotoxic activity against cancer cell lines from kidney and breast (Saleem et al., Citation2007).
Coral reefs are considered as one of the ecosystems with high biodiversity in coastal oceans, and as a source for isolation of endophytic microorganisms, including fungi (Golubic et al., Citation2005). The national park Veracruz Reef System (Sistema Arrecifal Veracruzano, SAV) is the second major reef system in Mexico, and it is formed by 23 reef structures (Ake-Castillo, Citation2011). It was declared a marine national park in 1992. Although it has been well studied for macroscopic organisms, so far there are only a few studies on endophytic fungi from this unique ecosystem. This work intends to establish the basis for future research on bioactive compounds in the Veracruz Reef System that based on the antiproliferative activity of marine endophytes.
Materials and methods
Collection and characterization of biological materials
Samples of corals, algae, and sponges were collected from La Blanca reef (Lat. 19.0865, Long. −95.9984), part of the (SAV), in two recollections carried out in the period from June to October 2011 by diving in shallow water (∼1–3 m) and were identified by Miguel Lozano (“Instituto de Ciencias Marinas y Ecología” Universidad Veracruzana, Mexico). The samples were cut into small pieces, placed in a sterilized tube with sea water, and refrigerated for transportation to the laboratory. The coral and algae samples collected were processed as follows: the tissue was rinsed under flowing water, and subsequently cut and disinfected with 4% sodium hypochlorite, then rinsed with sterile water and after that, placed in Petri plates with potato dextrose agar (PDA) supplied with 50% marine water and Petri plates with Marine Agar 2216; both media were supplied with the antibiotic chloramphenicol (0.2 mg/L). Petri plates were incubated at room temperature (25 ± 2°C) until marine samples developed mycelia (approximately for 14–21 d).
Strain identification
The identification of the strains was carried out by the observation of reproductive structures under a microscope and employing taxonomic keys (Barnett & Hunter, Citation1998; Lima & Furtado, Citation2007). For the microscopic observation, thin layer PDA cultures (microculture technique) were used. The mycelial cultures were observed after seven days of incubation at 25°C employing lactophenol. The microscopic morphology of the fungi was examined under an optic microscope (Carl Zeiss Microscopy GmbH, Jena, Germany).
Genetic identification of cultures
The active strains were genetically identified using nuclear ITS-rDNA sequences data. Isolation of the genomic DNA from the mycelium was performed by a standard protocol (Liu et al., Citation2000). The primers pair ITS1F-ITS4 was used to amplify nuclear rDNA-ITS regions by direct PCR technique. PCR products were first purified by the SiO2-coated magnetic beads (Sileks M, Moscow, Russia) and then were sequenced in both directions using forward and reverse primers ITS1F and ITS4. Sequencing reactions were performed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Waltham, MA). Newly obtained sequences were compared against nucleotide entries in the NCBI GenBank (http://www.ncbi.nlm.nih.gov/) by using search tool Blastn to confirm species identity. For phylogenetic analysis, sequences were aligned using ClustalW algorithm of MegAlign from MEGA6 software (Tamura et al., Citation2013). Phylogenetic tree of nucleotide alignments was constructed using Maximum Likelihood algorithm (Juke-Cantor) of the MEGA6 software. Bootstrap analysis was performed with 1000 replications.
Culture and extraction
A culture medium composed of yeast extract (4 g/L), soluble starch (10 g/L), peptone (2 g/L), and sea water (75%) was employed to culture the fungal strains. For each strain, a total volume of 500 mL was distributed in 250 mL Erlenmeyer flasks (50 mL of medium in each) and the flasks were inoculated with a small piece of agar with the fungal strain (approximately 1 cm2) and were put into an orbital shaker for 14 d at 25 ± 2°C. After that time, the culture broth and the biomass were separated by vacuum filtration. Both biomass and culture broth were frozen and lyophilized. When dry, the biomass and the broth were extracted with a mixture of solvents MeOH:CHCl3 1:1 for 7 d. The extraction was repeated several times. The extracts were then filtered and concentrated in a rotatory evaporator; the lyophilized extracts were employed in the chemosensitivity testing assay.
Cell lines and culture
The human solid tumor cell lines HBL-100, HeLa, SW1573, T-47D, and WiDr were used in this study. These cell lines were a kind gift from Prof. G. J. Peters (VU Medical Center, Amsterdam, The Netherlands). Cells were maintained in 25 cm2 culture flasks in RPMI 1640 medium supplemented with 5% heat inactivated fetal calf serum and 2 mM l-glutamine in a 37°C, 5% CO2, 95% humidified air incubator. Exponentially growing cells were trypsinized and re-suspended in antibiotic containing medium (100 units penicillin G and 0.1 mg/mL of streptomycin). Single cell suspensions were counted using Orflow’s MoxiZ automated cell counter (Ketchum, ID) and dilutions were made to give the appropriate cell densities for inoculation into 96-well microtiter plates. Cells were inoculated in a volume of 100 μL per well at densities of 10 000 (HBL-100, HeLa and SW1573), 15 000 (T-47D), and 20 000 (WiDr) cells per well, based on their doubling times.
Chemosensitivity testing
Dry extracts were initially dissolved in DMSO at 400 times to the desired final maximum test concentration, i.e., 10 mg/mL and diluted in the culture medium until they reached an assay concentration of 250 μg/mL. Control cells were exposed to an equivalent concentration of DMSO (0.25% v/v, negative control). The drug treatment was started on the first day after plating. Drug treatment incubation time was 48 h, after that cells were precipitated with 25 μL ice-cold TCA (50% w/v) and fixed for 60 min at 4°C. Then the SRB assay was performed (Skehan et al., Citation1990). The optical density (OD) of each well was measured at 492 nm, using BioTek’s PowerWave XS Absorbance Microplate Reader (BioTek, Winooski, VT). Values were corrected for background OD from wells containing only the medium. The percentage growth (PG) was calculated with reference to untreated control cells (C) based on the difference in OD at the start (T0) and end (T) time points of drug exposure, according to NCI formulas (Monks et al., Citation1991). Briefly, if T is greater than or equal to T0 the calculation is 100 × [(T−T0)/(C−T0)]. If T is less than T0, that denotes cell killing, the calculation is 100 × [(T−T0)/(T0)]. With these calculations, a PG value of 0 corresponds to the amount of cells present at the start point of drug exposure, while negative PG values denote net cell kill.
Results and discussion
Sampling and isolation of the fungal strains
The specimens collected in the SAV include eight genera of corals [Montastraea Vaugham & Wells (Montastreaidae), Diploria Milne-Edwards Haime (Mussidae), Millepora Linneaus (Milleporidae), Acropora Oken (Acroporidae), Siderastrea Blainville (Siderastreidae), Pseudopterogorgia Gmelin (Gorgoniidae), Plexaura Lamouroux (Plexauridae), Plexaurella Kölliker (Plexauridae), and Pseudoplexaura Hottuya (Plexauridae)], four genera of marine sponges [Aphimedon Duchassing & Michelotti (Niphatidae), Chondrilla Schmidt (Chondriilidae), Agelas Wilson (Agelasidae), and Aplysina Higgin (Aplysinidae)], one genus of red alga (Hypnea sp.) and a zoanthus. From the whole sample collection, 34 fungal strains were isolated, of which 24 were isolated from corals, eight strains were obtained from marine sponges, and one strain was from the alga and one from the zoanthus as shown in .
Table 1. List of samples, isolation source and morphological identification.
Morphological identification
For taxonomic identification, the microculture technique was employed with the aim of observing microscopical morphology. After 7 d of incubation on PDA at 25°C, the microcultures were examined under the microscope and, with the use of a taxonomic key, fungi from 11 genera were identified as follows: Acremonium Link (Hypocreaceae), Curvularia Boedijn (Pleosporaceae), Fusarium Link (Nectriaceae), Aspergillus Micheli (Trichocomaceae), Alternaria Nees (Pleosporaceae), Khuskia Huds (Incertae sedis), Epicoccum Link (Pleosporaceae), Sarocladium W. Gams & D. Hawksw (Incertae sedis), Trichoderma Pers (Hypocreacea), Cladosporium Link (Cladosporiaceae), and Monilia Bonordi (Sclerotiniaceae). The characteristic structures observed were microconidia, macroconidia, and chlamydospores for the genus Fusarium. Dark brown geniculate and transversely septate conidia with an enlarged central cell and with a pale terminal cell were characteristics for the genus Curvularia. In the case of the genus Acremonium, long and straight phialides producing unicellular conidia and aggregated in the apex of the phialide were identified. In the genus Monilia subglobose chains of conidia were observed, while in Khuskia black and globose spores were found, slightly flattened characteristics for the genus. In Alternaria, conidia were with transversal and longitudinal septa. For the genus Aspergillus, the conidiophores, vesicles, and phialides with chains of conidia were observed. In the case of the genus Trichoderma, branched conidiophores slightly wider than the characteristic form for the genus were seen to be present. For Cladosporium, erect conidiophores were observed, bearing catenulate conidia on each branch while for Epicoccum pale yellow septated hyphae and small conidiophores in clusters were observed.
Of the isolated strains, 32% were from the genus Aspergillus, 23% were from the genus Fusarium, 15% from genus Acremonium, 6% from the genus Cladosporium, 6% from the genus Trichoderma, and 3% from each of the genus Kuskia, Alternaria, Monilia, Curvularia, Sarocladium, and Epicoccum.
Molecular identification
In this study, species identity was verified for the four active marine endophytic fungal isolates by using a phylogenetic analysis based on the sequences of the ITS region. The isolates LAEE 01, LAEE 03, LAEE 07, and LAEE 17 were identified morphologically as belonging to the genera Cladosporium, Curvularia, and Fusarium for the last two strains accordingly.
Isolate LAEE 01 demonstrated 99% similarity with Cladosporium cladosporioides Fresen G.A. de Vries (Cladosporiaceae), placed in C. cladosporioides clade on the dendogram, separated from other species of Cladosporium. This strain was isolated from the marine sponge Amphimedon compressa as an endophyte fungus.
Isolate LAEE 03 shared a sequence similarity of 99.64% with Curvularia trifolii (Kauffman) Boedijn (Pleosporaceae) and is placed in C. trifolii clade on a dendrogram that is clearly separated from the C. geniculata group (). Strains of these closely related species which sequences deposited in GeneBank are known as plant pathogens (Crous et al., Citation2011). The strain LAEE 03 was isolated from a sponge Amphimedon compressa in marine environment and this finding can be considered as a first record of the species C. trifolii as the marine endophyte.
Figure 1. Maximum-likelihood phylogenetic tree based on ITS sequences of the marine endophytic isolates of this study and reference strains. Accession numbers in GeneBank are given in parentheses. Numbers at the nodes are bootstrap support values for 1000 replicates. The scale bar indicates the number of substitutions per site.
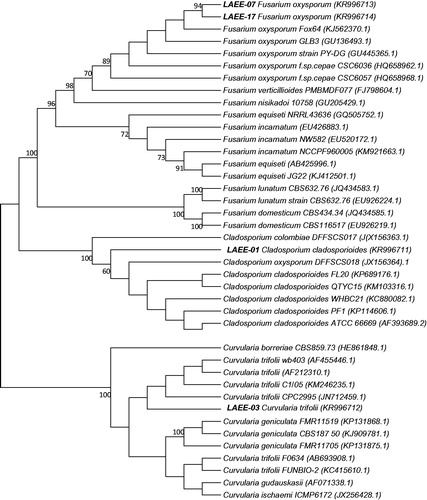
Isolate LAEE 17 demonstrated 99.24% similarity with Fusarium oxysporum E.F. Sm. & Swingle (Nectriaceae) ITS sequences deposited in GeneBank. On the dendrogram (), F. oxysporum species complex is clearly differentiated from F. incarnatum-F. equiseti (Desm.) Sacc. (Nectriaceae) species complex and the other Fusarium species. Thus it can be concluded that the strain LAEE17 isolated from the marine basin (from a sponge Diploria strigosa) belongs to F. oxysporum species complex. Isolate LAEE 07 as well was identified as F. oxysporum with a similarity of 99%, but the isolation was carried out from a different organism, the marine sponge Aplysina sp. Thus we concluded that the strain LAEE 07 also belong to the F. oxysporum species complex.
Antiproliferative activity
As a model to study, the antiproliferative activity of the obtained extracts, we used the representative panel of human solid tumor cell lines: HBL-100 (breast), HeLa (cervix), SW1573 (lung), T-47D (breast), and WiDr (colon). The extracts were assayed following the protocol and recommendations of the National Cancer Institute (Grever et al., Citation1992). Therefore, all extracts were tested at a single concentration of 250 μg/mL regardless of solubility, i.e., particulate matter may be present (Monks et al., Citation1991). The effect is given as percentage growth (PG) and the criterion for determination of activity was adopted as follows. Test sample showing PG <50% at 250 μg/mL is considered to be active. Both the broth and the biomass of all 34 isolated strains were analyzed for their antiproliferative activity. shows the PG data of active extracts.
Table 2. Class of extracts showing which fungal strain presented a PG % of under 50 and can be considered as bioactive.
The organisms collected from the coral reef showed a total of 34 endophyte fungal strains, from which 68 extracts were obtained and assayed on cancer cell lines, six of the extracts exhibited a different degree of activity against the growth of one or more of the cancer cell lines employed. The active strains were identified as C. trifolii, F. oxysporum, and C. cladosporioides. The remaining samples resulted inactive. Active extracts were obtained from four different strains: two strains of Fusarium, a strain of Cladosporium and a strain of Curvularia.
When considering the source of the active samples, four samples correspond to biomass extracts, while two derived from broth extracts. Interestingly, both broth and biomass extracts from F. oxysporum showed activity against all cell lines and in similar potential. However, the most active strain was C. trifolii (quantitative data).
There are reports on the isolation of active compounds from marine derived fungi of the aforementioned genus. Representative examples are mangicols isolated from a marine Fusarium which have shown a moderated cytotoxicity (Renner et al., Citation2000) or Cladosin C isolated from deep sea-derived Cladosporium which showed a moderate antiviral activity (Wu et al., Citation2014).
Nevertheless, the most active strain against the tested cell lines was C. trifolii, and there are no previous reports on isolation of this particular fungus from marine environment. Thus it could suggest that C. trifolii marine strain LAEE03 produces novel bioactive metabolites. The results obtained in this study indicate that the extract from a marine derived C. trifolii have an antiproliferative effect, suggesting that these organisms are good candidates for further analysis of their metabolites.
Furthermore this work shows the importance of SAV as a good source of microorganisms from which it is possible to obtain bioactive compounds as shown by the determination of the inhibition of bacterial quorum sensing using aquatic fungi from this coral reef (Martín-Rodríguez et al., Citation2014). Our preliminary results strongly suggest the importance of a deeper study related to this kind of organisms natives from these reef endangered by human activities.
Disclosure statement
Special thanks to CONACyT for the financial support throughout the project 181820 and for the scholarship number 308291 to the Alan Couttolenc, Virgilio Arenas Fuentes and Miguel Ángel Lozano Aburto from the “Instituto de Ciencias Marinas y Ecología” Universidad Veracruzana, Mexico. J. M. P. thanks the EU Research Potential (FP7-REGPOT-2012-CT2012-31637-IMBRAIN), the European Regional Development Fund (FEDER), and the Spanish Instituto de Salud Carlos III (PI11/00840) for financial support. G.B.P. thanks Fundación CajaCanarias for a postgraduate grant.
References
- Ake-Castillo JA. (2011). Temporal dynamics of Trichodesmium erythraeum (Cyanophyta) in the National Park “Sistema Arrecifal Veracruzano” in the Gulf of Mexico. J Environ Biol 32:395–9.
- Barnett HL, Hunter BB. (1998). Illustrated Genera of Imperfect Fungi. St. Paul, MN: American Phytopathological Society Press.
- Bungi T, Ireland C. (2004). Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat Prod Rep 21:143–63.
- Crous PW, Summerel BA, Swart L, et al. (2011). Fungal pathogens of Proteaceae. Persoonia 27:20–45.
- Gareth EB. (2011). Fifty years of marine mycology. Fungal Divers 50:73–112.
- Golubic S, Radtke G, Le Campion-Alsumard T. (2005). Endolithic fungi in marine ecosystems. Trends Microbiol 13:229–35.
- Grever MR, Schepartz SA, Chabner BA. (1992). The National Cancer Institute: Cancer drug discovery and development program. Semin Oncol 19:622–38.
- Kupka J, Anke T, Steglich W, Zechlin L. (1981). Antibiotics form basidiomycetes. The biological activity of siccayne, isolated form marine fungus Halocyphina villosa. J Antibiot 34:298–304.
- Lima A, Furtado M. (2007). Espécies do género Curvularia (fungos anamórficos: Hyphomycetes) na ilha de Santiago, Cabo Verde. Portugaliae Acta Biol 22:145–56.
- Liu D, Coloe S, Baird R, Pedersen J. (2000). Rapid mini-preparation of fungal DNA for PCR. J Clin Microbiol 38:471–5.
- Martín-Rodríguez AJ, Reyes F, Martín J, et al. (2014). Inhibition of bacterial quorum sensing by aquatic fungi: First report from marine endophytes. Mar Drugs 12:5503–26.
- Monks A, Scudiero D, Skehan P, et al. (1991). Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 83:757–66.
- Renner M, Jansen P, Fenical W. (2000). Mangicols: Structures and biosynthesis of a new class of sesterpenes polyols from a marine fungus of the genus Fusarium. J Org Chem 65:4843–52.
- Saleem M, Shaiq-Ali M, Hussain S, et al. (2007). Marine natural products of fungal origin. Nat Prod Rep 24:1142–52.
- Shigemori H, Kasai Y, Komatsu K, et al. (2004). Sporolides A and B, new cytotoxic twelve-membered macrolides form a marine-derived fungus Cladosporium species. Mar Drugs 2:164–9.
- Skehan P, Storeng R, Scudiero D, et al. (1990). New colorimetric cytotoxicity assay for anticancer drug screening. J Natl Cancer Inst 82:1107–12.
- Tamura K, Stecher G, Peterson D, et al. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–9.
- Wu G, Sun X, Yu G, et al. (2014). Cladosins A–E, hybrid polyketides from a deep-sea-derived fungus, Cladosporium sphaerospermum. J Nat Prod 77:270–5.
- Zhang Y, Mu J, Feng Y, et al. (2009). Broad-spectrum antimicrobial epiphytic and endophityc fungi from marine organism: Isolation, bioassay and taxonomy. Mar Drugs 7:97–112.
