Abstract
The Escherichia coli protease inhibitor ecotin is believed to be implicated in the evasion of host defenses during infection. The protein has also attracted attention as a scaffold for the design of novel, specific protease inhibitors. Ecotin interacts with its targets through two sites. Key hydrophobic residues in both sites (Leu-64, Trp-67, Tyr-69, Met-84, and Met-85) were mutated to alanine and the effects on the inhibition of trypsin, chymotrypsin, and elastase were assessed. Each of these mutant ecotin proteins tested in kinetic assays with these enzymes exerted less inhibitory potency compared to wild-type ecotin. However, these effects were relatively small and not additive.
| Abbreviations: | ||
| IC50, | = | concentration of inhibitor required to produce half-maximal inhibition |
| Ki, | = | inhibition constant |
| Pkal, | = | plasma kallikrein |
| uPA, | = | urokinase-type plasminogen activator |
| V0, | = | rate at zero inhibitor concentration |
| V∞, | = | rate at infinite inhibitor concentration |
Introduction
Proteases are involved in a range of cellular processes. Given that many of the cell’s key molecules are proteins, the potentially destructive power of proteases must be regulated. In part this is achieved through proteinaceous protease inhibitors, many of which have high affinity and specificity for their targetsCitation1. One such inhibitor is the Escherichia coli protein ecotin (E. coli trypsin inhibitor)Citation2. Unusually it has a wide specificity, being able to inhibit a range of serine proteases with high affinityCitation2,Citation3. It is a periplasmic protein, and is believed to be secreted by invading bacteria during infection in order to inhibit host proteases of the innate immune response such as neutrophil elastaseCitation4.
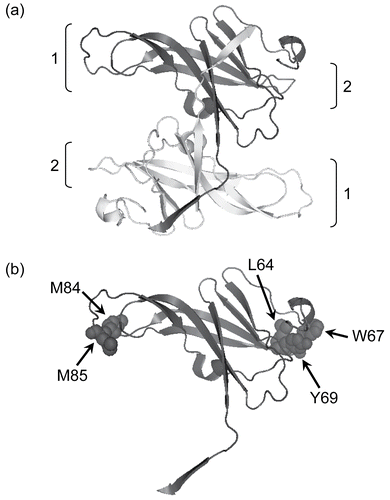
Ecotin exists in solution as a non-covalent homodimer in which two elongated, 18 kDa monomers associate in a head to tail fashion, mediated by hydrogen bond interactions between residues 125 and 142 in the C-terminal protruding tails of the monomers ()Citation5,Citation6. This gives the dimer two essentially identical ends, both of which can interact with proteases. The physiologically relevant complex is believed to consist of two ecotin monomers and two serine protease moleculesCitation5. These interactions involve residues from both monomers of the dimerCitation5. Consequently, each monomer has residues (referred to as site 1 and site 2 respectively) which contribute to binding and inhibition of both protease molecules. Of these, site 1 is inserted into the active site of the serine protease. The main contribution of site 2 is to anchor the inhibitor to the protease and thus contribute additional free energy of interactionCitation5.
Figure 1. The structure of ecotin (PDB ID: 1ECZCitation17). In (a) a ribbon diagram of the antiparallel homodimer is shown with protease interaction sites 1 and 2 indicated. In (b) a single monomer is shown with the residues mutated in this paper illustrated in space-filling format. The diagram was made using PyMol (www.pymol.com).
The broad specificity of ecotin has attracted attention in the development of protease inhibitors with modified specificities. Some success has been achieved. For example, the quadruple mutant, M84R/D70R/N110R/K112Q, resulted in a 24,000-fold higher affinity (compared to wild-type ecotin) for the urokinase-type plasminogen activator (uPA) proteaseCitation7. A mutant incorporating eight mutations in three of its binding loops resulted in a six-fold decrease in Ki for plasma kallikrein (Pkal)Citation3. However, less work has been done to understand the contribution that individual amino acids make to specificity and affinity. Here we report the consequences of substituting key hydrophobic residues in both site 1 and site 2 with alanine.
Materials and methods
Expression and purification of wild-type and mutant ecotin
DNA encoding ecotin was amplified by polymerase chain reaction (PCR) from E. coli XL1Blue genomic DNA using primers which incorporated NcoI and XhoI restriction sites in the forward and reverse primers respectively. The PCR product was purified, cut with these restriction enzymes, and ligated into pET21d (Merck Biosciences, Nottingham, UK), which had been previously cut with the same enzymes. Recombinant plasmids were identified by restriction digestion and by DNA sequencing (Fusion Antibodies, Dunmurry, Northern Ireland). Site-directed mutagenesis was carried out using the QuikChange protocolCitation8, and these mutations were verified by DNA sequencing.
An E. coli strain, JW2197-1, which is deleted for ecotin, was obtained from the Keio collectionCitation9, supplied by the E. coli Genetic Resource Center, Yale University, Connecticut. This strain was modified with the DE3 lysogen to enable expression of proteins under the control of the T7 promoter, using a kit purchased from Merck and following the manufacturer’s instructions. Expression plasmids for the wild-type and mutant ecotins were transformed into competent JW2197-1(DE3) cells and plated onto Luria–Bertani (LB) plates supplemented with 100 μg/mL ampicillin and 30 μg/mL kanamycin. Single colonies were picked, grown overnight in 5 mL of LB (supplemented with 100 μg/mL ampicillin and 30 μg/mL kanamycin) at 37°C, then transferred into 1 L of LB (supplemented with 100 μg/mL ampicillin and 30 μg/mL kanamycin). This culture was grown until A600 (optical density) reached between 0.6 and 1.0 (typically 5 h), induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and grown for a further 3 h. The cultures were harvested by centrifugation (10 min at 4200g) and the periplasmic fraction was isolated using a method based on that previously described for the purification of α-synucleinCitation10. The cell pellets were resuspended in a total volume of 50 mL osmotic shock buffer (30 mM Tris-HCl, 2 mM EDTA (ethylenediaminetetraacetic acid), 40% (w/v) sucrose, pH 7.2) and incubated at room temperature for 10 min. This suspension was centrifuged at 27,000g for 15 min at 4°C and the supernatant was discarded. The pellet was resuspended in 9 mL of cold sterile water and 3.8 μL of saturated magnesium chloride added. This suspension was kept on ice for 5 min and centrifuged at 27,000g and 4°C for 15 min. The periplasmic proteins were collected in the supernatant and stored frozen at −80°C. Further purification was achieved by column chromatography. The periplasmic fraction (5 mL) was applied to a ResourceQ column (Pharmacia) (total volume 6 mL, flow rate 6 mL/min), which had been previously equilibrated in buffer A (20 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid)-OH, pH 7.5, 10% (v/v) glycerol). The column was then washed with three column volumes of buffer A, and a linear gradient from 0 to 2 mM NaCl in buffer A was applied over five column volumes. Fractions containing purified ecotin were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protease inhibition assays
Wild-type ecotin and the mutants were assayed for their ability to inhibit elastase (prepared as described previouslyCitation11), trypsin (Sigma), and chymotrypsin (Sigma) using the chromogenic substrates MeOSuc-Ala-Ala-Pro-Val-pNA, Bz-Phe-Val-Arg-pNa, and Suc-Ala-Ala-Pro-Phe-pNA (Bachem) respectively. In each case, the Michaelis constant (Km) for the substrate was determined under the conditions of the experiment, i.e. at 22°C in assay buffer (50 mM HEPES-OH, pH 7.8; 150 mM NaCl; 10% (v/v) glycerol). These Km values were: elastase, 75 μM; trypsin, 45 μM; and chymotrypsin, 45 μM. Inhibition assays were carried out in triplicate at 22°C over a 5 min period using the substrate concentrations equal to the experimentally determined Km and enzyme concentrations of 50 nM in a total reaction volume of 250 μL. Assay buffer volumes were added to the reaction mixes first, followed by substrate, and then inhibitor. Enzyme was the final reactant added and started the reaction. Then, steady state rates were determined in triplicate over a wide range of ecotin (or mutant) concentrations (between a maximum of 15 μM and a minimum of 3 pM). These data were fitted to the equation v = V∞ + (V0 – V∞)/(1 + 10(log[Ecotin] − logIC50)) using non-linear curve-fittingCitation12 as implemented by the program GraphPad Prism (GraphPad software), where v is the measured rate, V∞ is the rate at infinite inhibitor concentration, V0 is the rate at zero inhibitor concentration, and IC50 is the concentration of inhibitor required to produce half-maximal inhibition. In all cases the fitted value for V∞ was close to zero, indicating that there was little background hydrolysis of the peptide. Results were plotted as the logarithm to the base 10 of the IC50 for each mutant compared to the wild-type.
Results and discussion
Expression and purification of recombinant ecotin
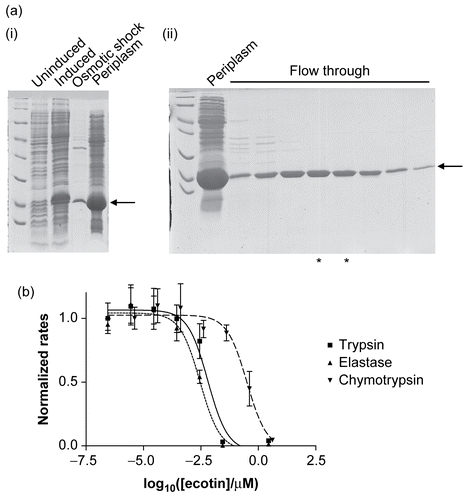
Recombinant wild-type ecotin, and its mutants, were purified by extracting the periplasmic fractions and then subjecting them to further purification by ion exchange chromatography. The method used here is different to previously published onesCitation2,Citation13,Citation14 in that the protein fails to interact with the column, whereas the majority of the impurities bind. Thus the material flowing through the column was purified ecotin (as judged by SDS-PAGE, ) in low ionic strength buffer. Consequently it could be used in subsequent experiments without further purification, buffer exchange, or dialysis. During the process of making the site-directed mutations, an additional mutant was produced (presumably as a consequence of an error in PCR). This mutant (M84A/M85A/W67C) was included in all subsequent experiments. The recombinant ecotin was active as a serine protease inhibitor () and is approximately 100 times less effective against chymotrypsin compared to trypsin and elastase.
Figure 2. (a) The expression and purification of recombinant ecotin. The recombinant protein is indicated by an arrow and runs close to the 18 kDa marker, as expected. (i) Extraction of a periplasmic extract. Uninduced and induced refer to cell extracts immediately before and 3 h after induction of the cells with IPTG. Osmotic shock refers to a sample of the bacterial suspension following treatment with osmotic shock buffer, and the last lane is the periplasmic extract. (ii) Purification by ion-exchange chromatography. The periplasmic extract was loaded onto a ResourceQ column (see “Materials and methods”). Ecotin washed through the column under the conditions of the experiment whereas most of the impurities remained bound. Periplasm refers to a sample of the periplasmic proteins prior to application to the ion-exchange column. Flow through refers to samples of fractions from the column. The fractions marked with asterisks were used without further purification. (b) Recombinant ecotin inhibits trypsin (▪ and solid line), elastase (▴ and dotted line), and chymotrypsin (▾ and dashed line). To enable the data to be shown on a single graph, the rates were normalized such that the rate in the absence of ecotin was equal to 1.0. Each point represents the mean of three separate determinations, the error bars the standard error of these means, and the lines the non-linear curve fit obtained as described in “Materials and methods.”
Effects of mutations on the ability of ecotin to inhibit serine proteases
All the mutations affected the ability of ecotin to inhibit the three serine proteases (; ). In general, the magnitudes of the effects were similar for the various alanine mutations. Furthermore, these effects were not additive. For example, while both the single mutations M84A and M85A resulted in reduced inhibition, the double mutant M84A/M85A gave similar results to both the single mutants with all three proteases. A similar pattern was seen with the mutations in site 2. As noted previouslyCitation15, deletion of two side chains does not, necessarily, result in an additive effect on the free energy of interaction. Although it has been shown previously that more radical changes at Met-84 and Met-85 (e.g. M84R) can result in alterations in ecotin’s specificityCitation3,Citation15, the changes reported here do not alter the specificity of the inhibitor for the proteases tested.
Figure 3. The effects of alanine mutations on the interaction between ecotin and (a) trypsin, (b) chymotrypsin, and (c) elastase. In each case, the negative logarithm of the IC50 is plotted as a bar which represents the mean of three separate determinations; the error bars represent the standard deviations of these means.
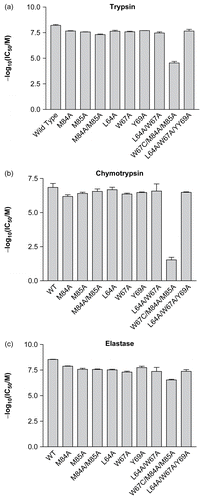
Table 1. Experimentally determined IC50 values (nM) for wild-type and mutant ecotin with trypsin, chymotrypsin, and elastase.
The greatest effects were seen with the mutant produced serendipitously (M84A/M85A/W67C). This mutation resulted in substantial changes in IC50—particularly with trypsin and chymotrypsin. It might be thought that the effects of this triple mutation result from the dramatic change (tryptophan to cysteine) causing gross structural changes to the overall fold of the protein. However, given that this mutation has markedly less effect on ecotin’s ability to inhibit elastase (compared to trypsin and chymotrypsin), this is unlikely to be the case. Furthermore, since it has been previously shown that alterations to the secondary site have very little effect on the conformation of the primary siteCitation16, it seems likely that the large changes observed with this mutation are largely due to the alteration in the secondary site. In the absence of a crystal structure of ecotin bound to elastase, it is difficult to make detailed predictions about why this change in the inhibitor is better tolerated by this enzyme. Indeed, modeling suggests that ecotin’s secondary site interactions with trypsin and elastase will be similarCitation5. Nevertheless, this mutation highlights the importance of the secondary site in contributing toward the stability of the complex with all three proteases.
Conclusions
Overall, the results demonstrate that ecotin is robust in its ability to tolerate the substitution of individual side chains in its two interaction sites. Each of the hydrophobic residues studied here contributes to the interaction, but removal of any one of them does not result in loss of binding. Taken together, the results reported here reinforce the idea that ecotin is an ideal scaffold for building protease inhibitors with novel specificities.
Acknowledgments
MTCM acknowledges a studentship from the Department of Employment and Learning (Northern Ireland).
Declaration of interest: The authors report no conflicts of interest.
References
- Joanitti GA, Freitas SM, Silva LP. Proteinaceous protease inhibitors: structural features and multiple functional faces. Curr Enzyme Inhib 2006;2:199–217.
- Chung CH, Ives HE, Almeda S, Goldberg AL. Purification from Escherichia coli of a periplasmic protein that is a potent inhibitor of pancreatic proteases. J Biol Chem 1983;258:11032–8.
- Stoop AA, Craik CS. Engineering of a macromolecular scaffold to develop specific protease inhibitors. Nat Biotechnol 2003;21:1063–8.
- Eggers CT, Murray IA, Delmar VA, Day AG, Craik CS. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem J 2004;379:107–18.
- McGrath ME, Erpel T, Bystroff C, Fletterick RJ. Macromolecular chelation as an improved mechanism of protease inhibition: structure of the ecotin-trypsin complex. EMBO J 1994;13:1502–7.
- McGrath ME, Gillmor SA, Fletterick RJ. Ecotin: lessons on survival in a protease-filled world. Protein Sci 1995;4:141–8.
- Laboissiere MC, Young MM, Pinho RG, Todd S, Fletterick RJ, Kuntz I, Craik CS. Computer-assisted mutagenesis of ecotin to engineer its secondary binding site for urokinase inhibition. J Biol Chem 2002;277:26623–31.
- Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. BioTechniques 1999;26:680–2.
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006;2:2006.0008.
- Huang C, Ren G, Zhou H, Wang CC. A new method for purification of recombinant human α-synuclein in Escherichia coli. Protein Expr Purif 2005;42:173–7.
- McCrudden MT, Dafforn TR, Houston DF, Turkington PT, Timson DJ. Functional domains of the human epididymal protease inhibitor, eppin. FEBS J 2008;275:1742–50.
- Marquardt D. An algorithm for least squares estimation of nonlinear parameters. SIAM J Appl Math 1963;11:431–41.
- Pal G, Sprengel G, Patthy A, Graf L. Alteration of the specificity of ecotin, an E. coli serine proteinase inhibitor, by site directed mutagenesis. FEBS Lett 1994;342:57–60.
- Palmer SM, St John AC. Characterization of a membrane-associated serine protease in Escherichia coli. J Bacteriol. 1987;169:1474–9.
- Yang SQ, Wang CI, Gillmor SA, Fletterick RJ, Craik CS. Ecotin: a serine protease inhibitor with two distinct and interacting binding sites. J Mol Biol 1998;279:945–57.
- Gillmor SA, Takeuchi T, Yang SQ, Craik CS, Fletterick RJ. Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases. J Mol Biol 2000;299:993–1003.
- Shin DH, Song HK, Seong IS, Lee CS, Chung CH, Suh SW. Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability. Protein Sci 1996;5:2236–47.
