Abstract
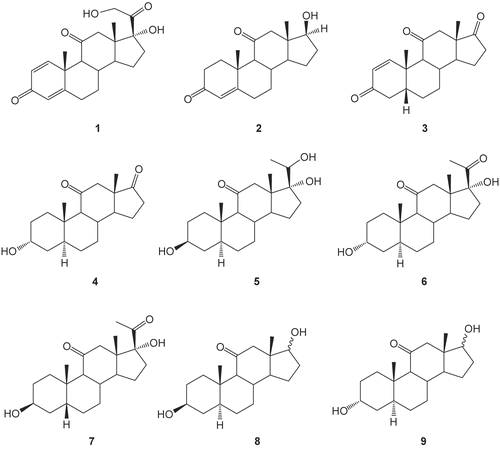
Anaerobic incubation of prednisone 1 with human intestinal bacteria (HIB) afforded nine metabolites: 5β-androst-1-ene-3,11,17-trione 3, 3α-hydroxy-5α-androstane-11,17-dione 4, 3β,17α,20-trihydroxy-5α-pregnan-11-one 5, 3α,17α-dihydroxy-5α-pregnane-11,20-dione 6, 3α,17α-dihydroxy-5β-pregnane-11,20-dione 7, 3β,17β-dihydroxy-5α-androstan-11-one 8β, 3β,17α-dihydroxy-5α-androstan-11-one 8α, 3α,17β-dihydroxy-5α-androstan-11-one 9β, and 3α,17α-dihydroxy-5α-androstan-11-one 9α. The structures of these metabolites (3–9) were elucidated using several spectroscopic techniques. Computer-aided prediction of potential biological activities of the isolated prednisone metabolites (3–9) revealed potential inhibition of prostaglandin E2 9-ketoreductase (PGE2 9-KR). Docking studies applied to PGE2 9-KR allowed recommendation of the metabolites 4, 8β, and 8α for further pharmacological study as PGE2 9-KR inhibitors.
Introduction
Microbial transformation studies have been conducted successfully as model systems to predict metabolic pathways in humans or to increase the efficiency of drugs by metabolic activationCitation1,Citation2. However, it was not until 1955 that a systematic study of steroid transformations by microorganisms belonging to the same genus was reported. In 1958, Lizuka et al. investigated the ability of 473 members of the genus Aspergillus to transform progesteroneCitation3,Citation4. A detailed description of the microbial transformation of steroids has been reviewed by Lizuka and NatioCitation5. In many studies, feces have served as a sample of the gastrointestinal floraCitation6–8. The vast majority (99.9%) of the estimated 400 different bacterial species in the human colon are strict anaerobes. The intestinal microflora is composed of numerous bacterial species and strains, which produce numerous kinds of enzymes that perform many chemical reactionsCitation9–11. The major biotransformation reactions such as dehydroxylation of the 21-hydroxyl groupCitation12, reduction of ring ACitation13,Citation14, epimerization of the 3-hydroxyl group15–18, introduction of a double bond conjugated to a keto groupCitation19, side chain cleavageCitation20, reduction of 20-ketone to an alcoholCitation14,Citation17,Citation20, and 16α-dehydroxylationCitation20 are reported in 21-C steroidal transformations. Heretofore, metabolic studies of 1 involving anaerobic human intestinal flora have not been reported. In the present article, we report the isolation and structural determination of nine metabolites obtained after anaerobic incubation of 1 with a fecal bacterial mixture from a human subject. Furthermore, the potential inhibitory activity of the isolated metabolites (3–9) as substrates for prostaglandin E2 9-ketoreductase (PGE2 9-KR) has been studied using the computerized system PASS (Prediction of Activity Spectra for Substance). The docking of prednisone 1 and its respective metabolites (3–9) has also been investigated since these structures are of most relevance for PGE2 9-KR enzyme inhibition.
Materials and methods
Human intestinal bacterial biotransformation of prednisone 1
Melting points were determined on an electrothermal melting point apparatus (Stuart Scientific Co.) in capillary tubes and were uncorrected. Citation1H and Citation13C nuclear magnetic resonance (NMR) spectra were recorded using either a Jeol JNM ECA-500 spectrometer at the Laboratory Center, Faculty of Science, Alexandria University, or a Varian Jeol JNM EX-400 (Citation1H, 500 MHz; Citation13C, 100 MHz) spectrometer at the NMR Laboratory Center, Assiut University, Egypt. Chemical shifts are given in δ (ppm) relative to the internal standard tetramethylsilane (TMS). Heteronuclear multiple quantum coherence (HMQC) experiments were performed with the usual pulse sequence, and data processing was obtained with standard Varian software. Electron impact (EI) mass spectra were recorded with a Jeol JMS-600 spectrometer at an ionization voltage of 70 eV (Jeol, Tokyo, Japan) at the Central Laboratory, Assiut University, and at the Microanalytical Center, Faculty of Science, Cairo University, Egypt. An anaerobic Shel-lab CO2 incubator (model 2200; Sheldon Manufacturing Inc., Cornelius, OR, USA) was used.
Thin layer chromatography (TLC) was carried out on pre-coated silica gel 60 GF254 plates (0. 25 mm thickness; Merck, Darmstadt, Germany) and the spots were detected under UV light at 254 nm or after spraying with H2SO4 reagent (20% H2SO4 in methanol) followed by heating. Column chromatography was carried out using silica gel 60 (0.063–0. 200 mm; Merck). The high performance liquid chromatography (HPLC) system consisted of a Knauer model 64, solvent delivery module (Knauer, Germany), Knauer variable wavelength UV detector, 20-μL sample loop, and a Shimadzu CR-6A Chromatopac integrator (Shimadzu, Tokyo, Japan), and the retention time (Rt) was recorded in minutes. The column used was a Phenomenex Luna C18 model ( 150 mm × 4. 6 mm i.d., 5 μm). The effluent was monitored at 240 or 206 nm at a flow rate of 1 mL/min. The mobile phase consisted of acetonitrile/water (2:3).
Prednisone 1 was obtained by extraction from Hostacortin® tablets as described by USP XXIICitation21. Two hundred tablets, equivalent to about 1 g of prednisone, were pulverized and extracted with 1 L of solvent hexane, with frequent agitation and slight warming, for 15 min. The supernatant liquid was decanted and discarded. Then the residue was extracted with 400 mL of chloroform for 15 min, and the mixture was filtered. Methanol (400 mL) was added to the filtrate, mixed, and evaporated to dryness on a steam bath with the aid of a current of air. The residue was dried at 105°C for 30 min and used without further purification as working reference material. All other chemicals and solvents were of analytical grade.
Anaerobic incubation of prednisone with human intestinal bacteria (HIB)
HIB suspension:
A fresh fecal sample (150 g) was homogenized with 1500 mL potassium phosphate buffer at 37°C (50 mmol, pH 7.3) in a sterilized 2 L flask and the supernatant was decantated and used immediately. To 975 mL of the decant, 27 g thioglycolate medium was added and mixed well under CO2.
Prednisone solution:
One gram of prednisone was dissolved in 25 mL dioxane/methanol mixture (3:1).
Test suspension:
This was prepared by adding prednisone solution (25 mL) to HIB suspension (975 mL) to give a final concentration of 1 mg/mL and then mixing well under CO2 using a magnetic stirrer.
Substrate control:
An aliquot of prednisone solution was added to potassium phosphate buffer (50 mmol, pH 7.3) and thioglycolate medium to give a final dilution of 1 mg/mL and used as the control solution.
HIB culture control:
An aliquot of HIB suspension was used in the control experiments.
Procedure The test suspension, the substrate control solution, and the HIB culture control were anaerobically incubated at 37°C for 4 days. At 24 h intervals the reaction mixture was reconstituted by a bubbling stream of CO2 for 1 min, and 2 mL samples were withdrawn from the solution and suspensions and transferred to separate test tubes. Then, to each tube, 2 mL ethyl acetate was added and shaken by vortex three times each for 1 min. Ethyl acetate extract was applied on a TLC plate and then developed using hexane/ethyl acetate (1:1), and spots were visualized by spraying with sulfuric acid reagent followed by heating over a hotplate.
At the end of the incubation time, the test experiment was extracted with butanol (500 mL × 4) and the combined extract was dried over anhydrous sodium sulfate. The residue after evaporation of butanol in vacuo was chromatographed over a silica gel column ( 25 cm length × 1. 5 cm i.d.) and eluted with hexane/ethyl acetate using the gradient elution method, starting with 100% hexane and ending with 70% hexane in ethyl acetate. Five fractions, A–E, were discerned on monitoring the hexane/ethyl acetate eluates by TLC. The scheme of handling the fractions is illustrated in .
Figure 1. Separation of prednisone metabolites (3–9) after anaerobic incubation with human intestinal bacteria (HIB).
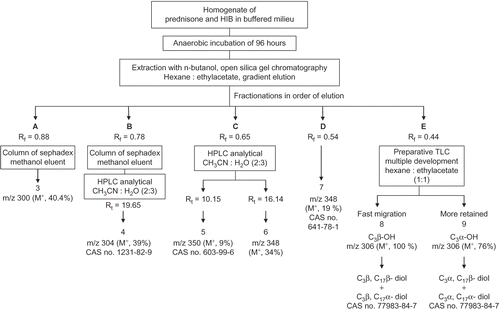
Fraction A (Rf = 0.88) was separated then passed over a column of sephadex ( 25 cm × 1 cm i.d.) eluted with methanol, evaporated in vacuo, and dried using a vaccum desiccator to yield metabolite 3 (4 mg).
5β-Androst-1-ene-3,11,17-trione 3: Colorless crystalline solid; UV (MeOH) λmax 240 nm; Rt = 20.5 min (CH3–CN:H2O, 2:3); Citation1H-NMR (CDCl3, 500 MHz), δ 7.50 (1H, m, H-1), δ 5.90 (1H, m, H-2); Citation13C-NMR (CDCl3, 125 MHz), ; EI-MS m/z (rel. int. %) 300 [M]+ (40.4), 285 (100).
Table 1. 13C-NMR assignments of prednisone 1 and its anaerobic metabolites 3–9.
Fraction B (Rf = 0.78) was separated then passed over a column of sephadex ( 25 cm × 1 cm i.d.) eluted with methanol. The methanolic eluent was evaporated in vacuo and dried using a vaccum desiccator to yield crude metabolite 4 (12 mg). Metabolites in the eluent were resolved by multiple injections on analytical HPLC eluted with CH3CN:H2O (2:3), and the major component eluted at Rt = 19.65 was collected. The combined eluent was evaporated in vacuo to obtain 10 mg of metabolite 4.
3α-Hydroxy-5α-androstane-11,17-dione 4: Colorless crystals; UV (MeOH) λmax 206; Rt = 19.65 min (CH3–CN:H2O, 2:3); Citation13C-NMR (CDCl3, 125 MHz), ; EI-MS m/z (rel. int. %) 304 [M]+ (39), 286, 271 (21), 243 (14), 147 (51), 55 (100); CAS 1231-82-9.
Fraction C (Rf = 0.65) analyzed by HPLC eluted with acetonitrile/water (2:3) revealed two signals at Rt = 10.15 and 16.14. The two compounds were separated and collected using an analytical column and multiple injections. The eluents at Rt = 10.15 and 16.14 were separately combined and evaporated in vacuo to obtain metabolites 5 (20 mg) and 6 (25 mg) respectively.
3β,17α,20-Trihydroxy-5α-pregnan-11-one 5: Colorless crystals; m.p. 265°C (decomp.); UV λmax 206 nm; Rt = 10.15 min (CH3–CN:H2O, 2:3); Citation13C-NMR (CDCl3, 125 MHz), ; EI-MS m/z (rel. int. %) 350 [M]+ (9), 348 (58), 330 (20), 305 (27), 261 (100); CAS 603-99-6.
3α,17α-Dihydroxy-5α-pregnane-11,20-dione 6: Colorless crystals; m.p. 226–228°C; UV λmax 206 nm; Rt = 16.14 min (CH3–CN:H2O, 2:3); Citation13C-NMR (CDCl3, 125 MHz), ; EI-MS m/z (rel. int. %) 348 [M]+ (34), 330 (19), 305 (23), 287 (22), 243 (34), 261 (84), 147 (45), 55 (100).
Fraction D (Rf = 0.54) was separated and evaporated in vacuo to yield metabolite 7 (18 mg).
3α,17α-Dihydroxy-5β-pregnane-11,20-dione 7: Colorless crystals; m.p. 204–206°C; UV λmax 206 nm; Rt = 15.36 min (CH3–CN:H2O, 2:3); Citation13C-NMR (CDCl3, 125 MHz), ; EI-MS m/z (rel. int. %) 348 [M]+ (19), 330 (19), 287 (39), 243 (90), 261 (16), 147 (26), 55 (100); CAS 641-78-1.
Fraction E (Rf = 0.44) was separated and evaporated in vacuo to yield a fraction of 5 mg. This fraction was rechromatographed using a preparative multiple development TLC technique and eluted with hexane/ethylacetate (1:1), where two bands were resolved. Each band was scratched and extracted with hot methanol. The fast migrating band yielded the mixture of metabolite 8 while the slow migrating one yielded the mixture 9.
3β,17α/β-Dihydroxy-5α-androstan-11-one 8: Colorless crystals; m.p. 228–230°C; UV λmax 206 nm; Rt = ND; Citation1H-NMR (CDCl3, 500 MHz), δ 3.84 (1H, m, OH-17), 3.56 (1H, m, OH-3), and 3.48 (1H, m, OH-17); Citation13C-NMR (CDCl3, 125 MHz), ; EI-MS m/z (rel. int. %) 306 [M]+ (100), 193 (80), 149 (52), 147 (60), 57 (79); CAS 77983-84-7.
3α,17α/β-Dihydroxy-5α-androstan-11-one 9: Colorless crystals; UV λmax 206 nm; Rt = ND; Citation1H-NMR (CDCl3, 500 MHz), δ 4.03 (1H, s, OH-3), 3.84 (1H, d, J = 6.85, OH-17), and 3.49 (1H, d, J = 6.85, OH-17); Citation13C-NMR (CDCl3, 125 MHz), ; EI-MS m/z (rel. int. %) 306 [M]+ (76), 303 (100), 193 (89), 149 (40), 147 (78), 57 (54); CAS 77983-84-7.
Prediction of biological activity spectra for prednisone metabolites (3–9)
The computerized system PASS (Prediction of Activity Spectra for Substance) was used for simultaneous suggestion of the biological effects of testosterone, prednisone 1, and the isolated steroidal metabolites (3–9). Only the structural formula is necessary to estimate which kinds of activity are more probable for the metabolite. The PASS 4.20 training set, including about 10,000 biologically active substances, was used to predict the probabilities of presence/absence for 114 biological actions simultaneously. Testosterone, prednisone 1, and the isolated steroidal metabolites (3–9) were screened for prediction of the possible biological activities at levels of more than 75% probability. The result of prediction is presented as a list of activities with appropriate Pa (probability as active) and Pi (probability as inactive), sorted in descending order.
Docking studies
Docking simulations were conducted using Molecular Operating Environment (MOE) version 2005.06 (Chemical Computing Group Inc., Montreal, Canada). The program was operated under the Windows XP system installed on an Intel Pentium IV PC with a 2.8 MHz processor and 512 RAM. Prednisone 1, testosterone 2, and HIB metabolites (3–9) were constructed using the builder interface of the MOE program and subjected to energy minimization using the included MOPAC 7.0 tool. The model produced was then subjected to systematic conformational search, where all items were set as default with a root mean square (RMS) gradient of 0.01 kcal/mol and RMS distance (RMSD) of 0.1 å.
The X-ray crystallographic structure of prostaglandin E2 9-ketoreductase (PGE2 9-KR) complexed with nicotinamide adenine dinucleotide (NAD), SO4 ion, and testosterone was obtained from the Protein Data Bank. The enzyme was prepared for docking studies where: (i) the ligand molecule with any existing solvent molecules was removed from the enzyme active site; (ii) hydrogen atoms were added to the structure with their standard geometry; and (iii) MOE Alpha Site Finder was used for the active sites search in the enzyme structure, and dummy atoms were created from the obtained alpha spheres. The obtained ligand–enzyme complex model was then employed in calculating the energy parameters, using the MMFF94x force field energy calculation, and predicting the ligand–enzyme interactions at the active site.
Results and discussion
HIB biotransformation of prednisone 1
Anaerobic incubation of prednisone 1 with HIB was carried out parallel with two control experiments, one for the substrate prednisone 1 and the other for the HIB mixture. During the 96 h incubation time, control and test experiments were monitored by TLC at 24 h intervals. Control experiments did not show spots on TLC other than those located at the incubation zero time, indicating that the spots revealed by the time of incubation of prednisone 1 with HIB can be attributed to the enzymatic bacterial machinery in the HIB mixture. The n-butanol extract of the test was concentrated to a few milliliters, and then chromatographed on the silica gel column; the eluent was then monitored by TLC. Five fractions, A, B, C, D, and E, were collected and further processed separately and subjected to elucidation of structure. The structures of the metabolites (3–9) are shown in .
Fraction A (Rf = 0.88) contains a single component showing m/z 300 (M+, 40.4%), i.e. 58 m/z less than the substrate, assigned as metabolite 3. The Citation13C-NMR spectrum of this metabolite 3 () revealed 19 carbon signals, devoid of the two signals assigned to the α-ketol group in prednisone 1, indicating an androstane skeleton. The downfield shifted signal at δ 217.03 ppm assigned to C17 in metabolite 3 illustrates that the cleavage of the α-ketol group has been followed by oxidation of the C17-αOH group to the corresponding ketone. This was confirmed by the vanished signal of proton singlet at δ 5.58 ppm assigned to C17-OH in prednisone 1. The Citation1H-NMR spectrum of 3 showed two downfield olefinic proton signals at δ 7.50 and 5.90, integrated by one proton each. The HMQC spectrum showed coupling between these two protons and olefinic carbon at δ 160.46 and 126.95, assigned to C1 and C2 respectively, indicating a C1–C2 double bond in ring ACitation22,Citation23. The DEPT (distortionless enhancement polarization techniques) spectrum of 3 at 45° revealed the signals of 14 carbons, indicating the presence of five quaternary carbonsCitation22. Two of the quaternary carbons have δ values matching the carbonyls at C3 and C11 in prednisone 1 and a third already assigned to that of C17 at δ 217.03, while the last two are evidently assigned to C10 and C13 (). DEPT at 90° showed six methine carbons assigned to C1, C2, C5, C8, C9, and C14. Finally, DEPT at 135° revealed two methyl groups, C18 and C19, in addition to the six methine carbons, and on the other side six signals assigned to methylene carbons: C4, C6, C7, C12, C15, and C16.
Assignment of metabolite 3 to the β-androstane series was realized through matching the observed δ value of C19 at δ 20.73 with reported values of quite similar compounds. 3α-Hydroxy-5β-androstan-17-one showed C19 at δ 23.3, while in the 5α series the C19 signal was located upfield at δ 11.2 ppmCitation24. Consequently, by analogy, metabolite 3 was related to the 5β-androstane series. The accumulated spectral data strongly supported the suggested structure of metabolite 3 as 5β-androst-1-ene-3,11,17-trione.
Fraction B (Rf = 0.78) was chromatographed over a sephadex column and eluted with methanol. After evaporation of methanol the crude product obtained was fractionated by multiple injection on analytical HPLC, and eluted with acetonitrile/water (2:3). The major component eluted at Rt = 19.65 min was collected and the solvent evaporated to yield metabolite 4. The mass spectrum of 4 displayed m/z 304 (M+, 39%), i.e. 54 m/z less than that of prednisone 1. Preliminary investigation of structure 4 was interpreted as in metabolite 3. According to Citation13C-NMR of 4 () that revealed 19 carbons, devoid of C20 and C21 signals, in the spectrum of prednisone 1, and the emerged downfield signal at δ 217.93 assigned to C17 carbonyl, we designated 4 as an androstan-17-one derivative. Matching Citation13C-NMR and DEPT spectra at 45° enabled the assignment of two carbonyl carbons at C11 and C17 in addition to two other quaternary carbons. DEPT at 90° showed five methine carbons and at 135° showed eight methylene carbons in addition to two methyls. This indicated the absence of spCitation2 carbons in 4 other than those of the two carbonyls at C11 and C17. It seems that the carbonyl signal of C3 in prednisone was reduced to the C3-αOH group, revealing a δ value at 66.27 lower than the value assigned for the C3-βOH signal (>70 ppm) ()Citation24. Assignment of 4 as either the 5α- or 5β-androstane series was undertaken by comparing the pattern of δ values of both C9 and C19 in 3α-hydroxy-5α-androstan-17-one and that of the 5β-isomerCitation23,Citation24. In the α-series the C9 and C19 are assigned at 54.5 and 11.2 respectively, while in the 5β-series the δ values are 40.8 and 23.3Citation23. Metabolite 4 showed the pattern that compares with that of the 5α-series since it displayed signals at δ 64.95 and 14.74 assigned for C9 and C19 respectively. The interpreted spectra of metabolite 4 displayed in led us to designate this metabolite as 3α-hydroxy-5α-adronstan-11,17-dione.
Fraction C, located as a single spot on TLC (Rf = 0.65), was fractionated by analytical HPLC to give two components, 5 and 6, at Rt = 10.15 and 16.14 min respectively. The mass spectrum of 5 showed m/z 350 (M+, 9.2%) and a base peak at m/z 261 that has been already recognized among the fragmentation pattern of the C17-OH pregnane nucleusCitation15. The mass, in addition to 21 carbons displayed by the Citation13C-NMR, spectrum supported the postulation of a pregnane nucleus to metabolite 5. Assignment of the C-atoms to their respective signals in the Citation13C-NMR spectrum has been achieved as previously discussed on the basis of DEPT measurements and in comparison to the chemical shifts of the corresponding C-atoms of the parent compound, prednisone 1. The absence of the signal at δ 185.5 ppm originating the C3 carbonyl in 1 and the presence of a new signal at δ 71.04 ppm indicate the reduction of the carbonyl group of the parent compound to the corresponding alcohol. This value of δ 71.04 ppm is characteristic of the 3β-OH configurationCitation23,Citation24. DEPT at 135° revealed three methyl carbons, two of which have δ values matching with those of prednisone 1. The third methyl, relatively downfield shifted at δ 27.9, was assigned to C21 that most probably emerged from the biotransformation of C21H2OH located at δ 66.6 in prednisone 1. The 5α-pregnane configuration was assigned to 5 by analogy to metabolite 4 where the chemical shifts of C9 and C19 are consistent with the reported δ values of the 5α-pregnane seriesCitation25. Assignment of the α/β configuration of C20HOH was interpreted in light of available data designating the high melting point (m.p.) 253–257°C to the α-isomer and the low m.p. 133–135°C to the β-isomerCitation26,Citation27. The found m.p. of 5, 265°C, is closer to the reported value of C20Hα-OH. Consequently, metabolite 5 was assigned the structure of 3β,17α,20α-trihydroxy-5α-pregnan-11-one.
The pregnane nucleus with saturated ring A was assigned to metabolite 6 on the basis of mass spectral data: m/z 348 (M+, 34%), m/z 261 (84.6%), and 21 carbon signals revealed by Citation13C-NMR. DEPT of metabolite 6 showed three methyl and eight methylene carbons with δ values equal to those in metabolite 5, in addition to five methines and five quaternary carbons. The chemical shifts of carbons of the same nature in 6 are practically matching with those in metabolite 5 and with that in prednisone, and were assigned to the respective sited carbons (). Peak by peak comparison of 5 and 6 spectra revealed some distinctive signal differences between them. Metabolite 6 showed a carbon signal assigned to C20 carbonyl with a δ value equal to that in prednisone. A second difference observed is the downfield shift of C3HOH at 66.38 in 6 relative to the respective signal in 5 at 71.04, indicating the α-configuration of OH at this carbon. Assignment of the pregnane structure to the 5α-series in 6 was interpreted as in 5, considering the δ values of C9 and C19. Accordingly, the structure of metabolite 6 was assigned as 3α,17α-dihydroxy-5α-pregnane-11,20-dione.
Fraction D, identified at Rf = 0.54 by TLC and at Rt = 15.36 min by HPLC, showed m/z 348 (M+, 19%) and m/z 261 (16.4%) and was labeled as metabolite 7. The Citation13C-NMR and DEPT spectra of 7 are identical to those of metabolite 6 except for the δ values revealed by C3, C9, C18, and C19 (). The downfield shift of the C3HOH signal relative to that in 6 was taken as an indication of the β-oriented OH group. On the other hand, the pattern of chemical shifts at C9 and C19 was related to the 5β-pregnane nucleus. Based on the available data listed in , metabolite 7 was specified as a 3-epimer of 6: 3β,17α-dihydroxy-5β-pregnane-11,20-dione.
Fraction E, identified as a single spot on TLC at Rf = 0.44, was resolved into two well separated bands by the preparative multiple development TLC technique. The two bands were labeled as E1, the fast migrating, and as E2, the most retained one. After extraction from silica, product 8 isolated from E1 and product 9 isolated from E2 were subjected to spectral elucidation of structure. Metabolites 8 and 9 revealed common features of the mass and Citation13C-NMR spectra. The molecular ion peak m/z 306 and a common fragmentation pattern at m/z 288, 193, 180, 149, 147, 109, 105, 91, 81, and 79 with practically close abundance percentages were observed. Furthermore, Citation13C-NMR of either 8 or 9 revealed 21 carbon signals, i.e. two carbon signals more than expected from the M+ value. DEPT spectra disclosed three quaternary carbons assigned to C10, C11, and C13, seven methines, nine methylenes, and two methyl carbon signals (). There were no signals perceived in the olefinic range of δ 119–172, indicating an absence of olefinic C-atomsCitation28. The chemical shifts of carbons in 8 and 9 were very close, except one signal at the CHOH scale, that afforded a clue to differentiate between metabolites 8 and 9. Metabolite 8 revealed a carbon signal at δ 71.1, a value assigned to C3Hβ-OH by analogy to 3β-hydroxy-5α-androstan-17-oneCitation24, while metabolite 9 displayed a signal at δ 66.4, already assigned to C3Hα-OHCitation24. Therefore, 8 and 9 can be regarded as C3 epimers. Metabolite 8 with C3Hα-OH configuration was the most retained by TLC, while the less polar 9 with C3Hβ-OH configuration was the fast migrating one. Our observation of the difference in migration rate between 8 and 9 associated with the difference in polarity confined to the configuration of OH at C3 is in accord with that reported by Bokkenheuser et al.Citation15. In addition, there are two downfield bands in 9 in the same region of the CHOH scale at δ 80.2 and 77.7 ppm that were assigned to C17β-OH and C17α-OH, respectively, by analogy to reported values for 17-hydroxyandrostanesCitation24. Consequently, metabolites 8 and 9 are regarded as a mixture of C17 epimers (C17β-OH and C17α-OH isomers). The perceived mixture of C17 epimers might be expected to affect appreciably the vicinal C16 methylene protons with quasi axial/equatorial orientations. The observed signals at δ 32.2 and 30.94 ppm matched with values reported for C16 methylene vicinal to epimeric C17HOH. It was assumed that the δ value of C16 at δ 32.2 ppm can be assigned to the C17Hβ-OH epimer at δ 80.2 ppm, while the δ value of C16 at 30.94 ppm can be assigned to the vicinal C17Hα-OH epimer at δ 77.7 ppm. Accordingly, the two signals revealed by Citation13C-NMR spectra of 8 and 9 more than that assumed for the androstane nucleus can be rationalized by assignment of two signals for C16 and two signals for C17 instead of one for each.
Docking studies
In this work we exploit the benefit of PASS predictive potentials of the isolated steroids obtained by incubation of prednisone 1 with an HIB mixtureCitation29. The isolated steroidal metabolites (3–9) were screened for prediction of the possible biological activities at levels of more than 75% probability. The results afforded a valuable guide for further study of the undesirable side effects and/or favorable activities of the metabolites. The most common biological activity pointed out by PASS for all our prednisone metabolites (3–9) was the potential inhibition of prostaglandin E2 9-ketoreductase (PGE2 9-KR) enzyme (). The occurrence of PGE2 9-KR has been reported in a variety of animal species and tissues. The activity of the enzyme is dependent on NADPH (reduced nicotinamide adenine dinucleotide phosphate) in tissuesCitation30,Citation31 and consequently the tissue levels of E and F prostaglandinsCitation32, which play an important role in cardiovascular diseasesCitation33,Citation34. The dock tool of MOE was applied. This might enable prediction of the orientation of these metabolites (3–9) in the PGE2 9-KR active site. The structure of Protein Data Bank (PDB) entry 1Q13 was used for the docking simulations of PGE2 9-ketoreductase. The 1Q13 structure contains one molecule of PGE2 9-KR complexed with an NAD, SO4 ion, and testosterone molecule. To generate a docking target with a steroid accessible binding cavity, steroid ligand was removed from crystal structures together with any existing solvent molecules. The enzymeNADP+ complex (E-NADP+) targets were used for the initial validation run where testosterone 2 was docked into PGE2 9-KR. The RMSD value between the docked conformation and the conformation in the crystal structure was evaluated by applying superimposition of the docked and crystallographic structures, and was found to be 0.8 å. The enzyme–NAD target was used for docking of prednisone metabolites (3–9), since these structures are of most relevance for enzyme inhibition or ketosteroid reduction. The output of docking simulation are the scoring functions, which reflects the binding free energy dG in kcal/mol (S), value proportional to the sum of Gaussian R1R2exp(–0.5d2) where R1 and R2 are the radii of the atoms in angstroms (å) and d is the distance between the pair in å (ASE), and is a linear combination of S, ASE, and Econf where Econf is an estimated self-energy of the ligand in kcal/mol (E). The saved pose for the ligand–enzyme complex of each molecule was subjected to detailed 3D analysis for its interactions at the enzyme active site. In the PGE2 9-KR ternary complex model, testosterone seems mainly stabilized at one end by interactions with residues His 117 and Tyr 55, which are involved in a hydrogen bond network with the C3-ketone group of the steroid. Testosterone is oriented with its C3-ketone group pointing toward the hydroxyl group of the side-chain of Tyr 55 (2.7 å), the general acid/base donor or acceptor in catalysis (). Metabolites 4 and 8β are orientated with their C3-hydroxyl group pointing toward the imino group of His 117 (3.06 å), the general acid/base donor or acceptor in catalysis. Hydrophobic interactions with residues Phe 118, Trp 86, Phe 310, and Phe 54 are observed for metabolite 4, while hydrophobic interactions of 8β were found between C1, C5, and C6 and Val 306, Phe 118, Phe 54, and Trp 86 respectively. Metabolite 8α is oriented with its C3-hydroxyl group pointing toward the hydroxyl group of Tyr 55 (2.9 å) and imino group of His 117 (2.9 å), the general acid/base donor or acceptor in catalysis. Another hydrogen bond interaction of the 17α-OH group with Glu 224 (3.23 å) and Tyr 305 (2.74 å) was also observed for metabolite 8α. Hydrophobic interactions were found between the C6, C7, and C19 of the 8α and the hydrophobic pocket formed by the benzene ring of Phe 54, Phe 118, and Val 306 respectively. From the above findings, it is evident that Tyr 55 and His 117 are the most essential amino acids involved in hydrogen bond interactions with the metabolites, testosterone, and prednisone. Most of the ligands revealed at least two hydrogen bond interactions like the most active ligand, testosterone. On the other hand, Phe 54, Phe 118, and Phe 310 in addition to Val 306 and Trp 310 are the amino acids involved mainly in hydrophobic bonding with the ligands. The metabolites docked on PGE2 9-KR revealed dG ≈ dGtestosterone ± 1 kcal/mol as shown in . 3α-Hydroxy-5α-androstane-11,17-dione 4, 3β,17β-dihydroxy-5α-androstan-11-one 8β, and 3β,17α-dihydroxy-5α-androstan-11-one 8α are prednisone metabolites showing the highest dG values of −6.96, −7.06, and −7.25 kcal/mol, respectively. show the orientation of these prednisone metabolites (4, 8β, 8α, respectively) in the PGE2 9-KR enzyme active site.
Figure 3. The steroid-binding site of PGE2 9-ketoreductase. Structures of PGE2 9-KR (taken from PGE2 9-KR, NAD,·testosterone complex, light gray). Residues that contact the steroid ligands such as Phe 54, Tyr 55, His 117, Phe 118, Ile 129, Val 306, and Phe 310 are shown in stick representation.
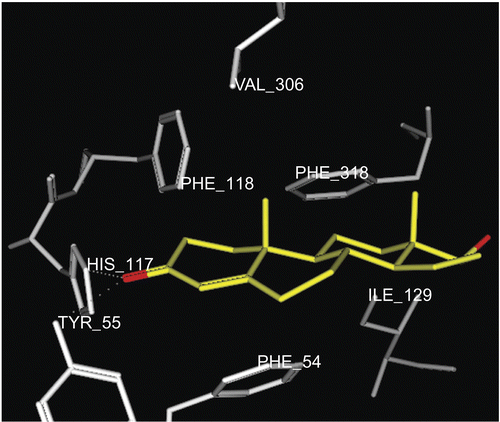
Figure 4. Docking of metabolite 4 (yellow stick) in the active site of PGE2 9-KR. Hydrogen atoms of the amino acid residues have been removed to improve clarity.
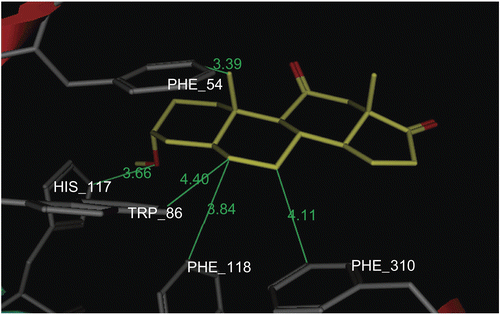
Figure 5. Docking of metabolite 8β (yellow stick) in the active site of PGE2 9-KR. Hydrogen atoms of the amino acid residues have been removed to improve clarity.
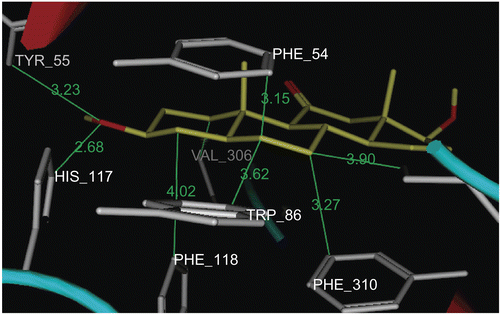
Figure 6. Docking of metabolite 8α (yellow stick) in the active site of PGE2 9-KR. Hydrogen atoms of the amino acid residues have been removed to improve clarity
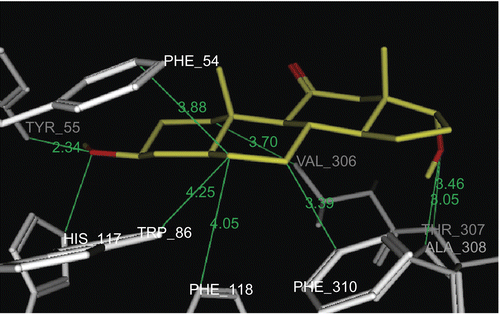
Table 2. Results of PASS predicted potential and molecular modeling of prednisone metabolites (3–9).
Conclusions
Human intestinal bacterial anaerobic incubation of prednisone 1 afforded nine metabolites (3–9). All the isolated prednisone metabolites (3–9) exhibited biological activity as PGE2 9-KR inhibitors with probabilities ranging from 78 to 91%. Docking simulation revealed that the isolated prednisone metabolites (3–9) inhibit the PGE2 9-KR enzyme by binding in a quite similar manner to the same active site as the already known inhibitor, testosterone 2. Exploration of the PGE2 9-KR enzyme inhibition potential of these metabolites (4, 8β, and 8α) will be the subject of further investigations.
Declaration of interest: The authors report no conflicts of interest.
References
- Rosazza JP. Microbial Transformation of Bioactive Compounds, 1st edn. New York: CRC, 1982: vol I; and references therein.
- Azerad R. Advances in Biochemical Engineering/Biotechnology. Berlin: Springer-Verlag, 1999.
- Sardinas JL, Pisano MA. Steroid transformations by species of Cephalosporium and other fungi. Appl Microbiol 1967;15:277–84; and references therein.
- Madyastha KM, Joseph T. Transformation of dehydroepiandrosterone and pregnenolone by Mucor piriformis. Appl Microbiol Biotechnol 1995;44:339–43; and references therein.
- Briggs MH, Christie GA. Steroid biochemistry and Pharmacology. New York: Academic Press, 1972; and references therein.
- Rosazza JP. Microbial Transformation of Bioactive Compounds, 1st edn. New York: CRC, 1982: vol II; and references therein.
- Basit AW, Newton JM, Lacey LF. Susceptibility of the H2-receptor antagonists cimetidine, famotidine and nizatidine, to metabolism by the gastrointestinal microflora. Int J Pharm 2002;237:23–33.
- Basit AW, Lacey LF. Colonic metabolism of ranitidine: implications for its delivery and absorption. Int J Pharm 2001;227:157–65.
- Scheline RR. Metabolism of foreign compounds by gastrointestinal microorganisms. Pharmacol Rev 1973;25:451–523.
- Wasowska-Krolikowska K. Composition and significance of gastrointestinal tract bacterial flora and antibiotic therapy. Case Rep Clin Pract Rev 2001;2:163–7.
- Lenoir-Wijnkoop I. The Intestinal Microflora. Understanding the Symbiosis. New York: John Libbey Eurotext Press, 2003.
- Bokkenheuser VD, Winter J, Dehazya P, Kelly WG. Isolation and characterization of human fecal bacteria capable of 21-dehydroxylating corticoids. Appl Environ Microbiol 1977;34:571–5; and references therein.
- Bokkenheuser VD, Winter J, Cohen BI, O’Rourke S, Mosbach EH. Inactivation of contraceptive steroid hormones by human intestinal Clostridia. J Clin Microbiol 1983;18:500–4.
- Martin F, Peltonen J, Laatikainen T, Pulkkinen M, Adelercreutz H.Excretion of progesterone metabolites and estriol in faeces from pregnant women during ampicillin administration. J Steroid Biochem 1975;6:1339–46.
- Winter J, Bokkenheuser VD, Ponticortivo L. Bacterial metabolism of corticoids with particular reference to the 21-dehydroxylation. J Biol Chem 1979;254:2626–9.
- Winter J, Bokkenheuser VD. 21-Dehydroxylation of corticoids by anaerobic bacteria isolated from human fecal flora. J Steroid Biochem 1978;9:379–84.
- Bokkenheuser VD, Winter J. Biotransformation of steroid hormones by gut bacteria. Am J Clin Nutr 1980;33:2502–6.
- Gustafsson JA. Steroids in germfree and conventional rats; identification of C19 and C21 steroids in faeces from conventional rats. Eur J Biochem 1968;6:248–55.
- Joseph J, Smith LL. 16α-Hydroxy steroids; effect of medium composition on isomerization of 9α-fluoro-16α-hydroxyhydrocortisone and 9α-fluoro-16α-hydroxyprednisolone (triamcinolone) during microbiological fermentation. Appl Environ Microbiol 1960;8:363–6.
- Macdonald IA, Bokkenheuser VD, Winter J, McLernon AM, Mosbach EH. Degradation of steroids in the human gut. J Lipid Res 1983;24:675–700; and references therein.
- USP Pharmacopeia XXII. Rockville, MD: United States Pharmacopeia Convention, 1999.
- Atta-Ur-Rahman. One and Two Dimensional NMR Spectroscopy. New York: Elsevier Science, 1989.
- Breitmaier E, Voelter W. Carbon-13 NMR Spectroscopy, 3rd edn. Weinheim: VCH Verlag, 1987.
- Blunt JW, Stothers JB. 13C-NMR spectra of steroids: a survey and commentary. Org Magn Reson 1977;9:439–64.
- Choudhary MI, Siddiqui ZA, Musharraf SG, Nawaz SA, Atta-Ur-Rahman. Microbial transformation of prednisone. Nat Prod Res 2005;19:311–17.
- Faramarzi MA, Yazdi MT, Shafiee A, Zarrini G. Microbial transformation of hydrocortisone by Acremonium Strictum PTCC 5282. Steroids 2002;67:869–72.
- Yazdi MT, Arabi H, Faramarzi MA, Ghasemi Y, Amini M, Shokravi S, Mohseni FA. Biotransformation of hydrocortisone by a natural isolate of Nostoc muscorum. Phytochemistry 2004;65:2205–9.
- Laan RFJM, Van Riel PLCM, Van Rning LJTO, Lemmens JAM, Ruijs SHJ, Van De Putte LBA. Vertebral osteoporosis in rheumatoid arthritis patients: effect of low dose prednisone therapy. Br J Rheumatol 1992;31:91–6.
- http://ibmc.p450.ru/PASS/Abstract/art002.htm#4.
- Watanabe K. Prostaglandin F. synthase. Prost Other Lipid Med 2002;68:401–7.
- Goff AK. Steroid hormone modulation of prostaglandin secretion in the ruminant endometrium during the estrous cycle. Biol Reprod 2004;71:11–16.
- Ziboh VA, Lord JT, Penneys NS. Alterations of prostaglandin E2 9-ketoreductase activity in proliferating skin. J Lipid Res 1977;18:37–43.
- Arosh JA, Banu SK, Chapdelaine P, Madore E, Sirois J, Fortier MA. Prostaglandin biosynthesis, transport, and signaling in corpus luteum: a basis for autoregulation of luteal function. Endocrinology 2004;145:2551–60.
- Chan PS, Cervoni P. Prostaglandins, prostacyclin, and thromboxane in cardiovascular diseases. Drug Dev Res 2004;7:341–59.