Abstract
Spectroscopic, enzyme-inhibition, and free-radical scavenging properties of a series of hydrazide ligands and their vanadium(IV) complexes have been investigated. Analytical and spectral data indicate the presence of a dimeric unit with two oxovanadium(IV) ions (VO2+) coordinated with two hydrazide ligands along with two water molecules. All complexes are stable in the solid state, but exhibit varying degrees of stability in solution. Binding of the coordinating solvent such as DMSO is indicated at the 6th position of vanadium in the dimeric unit followed by conversion to a monomeric intermediate species, [VOL(DMSO)3]1+ (L = hydrazide ligand). The free hydrazide ligands are inactive against snake venom phosphodiesterase I (SVPD), whereas oxovanadium(IV) complexes of these ligands show varying degrees of inhibition and are found to be non-competitive inhibitors. The superoxide and nitric oxide radical scavenging properties have been determined. Hydrazide ligands are inactive against these free radicals, whereas their V(IV) complexes show varying degrees of inhibition. Structure–activity relationship studies indicate that the electronic and/or steric factors that change the geometry of the complexes play an important role in their inhibitory potential against SVPD and free radicals.
Introduction
Vanadium has the exceptional ability to interact with biomolecules in both cationic and anionic forms and in its several oxidation statesCitation1–3. Among its biological roles, many important therapeutic effects have been reported, including hormonal, cardiovascular, anticarcinogenic, and insulin mimetic activitiesCitation4,Citation5. It is found at the active site of haloperoxidases and nitrogenasesCitation4,Citation6–9. As an analog of phosphate, vanadium inhibits or stimulates various phosphate metabolizing enzymesCitation10, and thus possibly attains a general role in most living organisms. This role includes its insulin-like properties, for which extensive research is under way to identify a potent antidiabetic agent.
Phosphodiesterases (PDs) form a group of catalytic enzymes that play an important role in many cellular processes, including carbohydrate metabolismCitation11,Citation12. They catalyze the hydrolysis of diesters of phosphoric acidCitation13. Many biological compounds, such as cyclic nuc1eotides, nucleic acids, and phospholipids, contain phosphodiester bonds that are hydrolyzed in vivoCitation14–16. This enzyme family has been involved in diverse biological activities, including osteoarthritis, insulin resistance in type II diabetes, and tumor cell motilityCitation17. It has been proposed that the insulin mimetic properties of vanadium may be attributed to the inhibition of PD, although controversy existsCitation11,Citation12.
Free radicals have been implicated in several diseases such as liver cirrhosis, cancer, and diabetes. Increased oxidative stress is a widely accepted cause in the development and progression of diabetes and its complicationsCitation18. Diabetes is usually associated with increased production of reactive oxygen species (ROS), including hydroxyl (OH•)Citation19,Citation20 and nitric oxide (NO•) free radicalsCitation21. Free radicals are formed disproportionately in diabetes by glucose oxidation, glycosylation of proteins, activation of transcription factors, and subsequent oxidation of glycated proteinCitation22. Compounds that can scavenge free radicals have great potential in treating these diseasesCitation23. Also, the antioxidant affect of vanadate has been reported in streptozotocin-induced diabetic ratsCitation24. It shows the importance of enzyme-inhibition studies of these complexes and their actions against free radicals, which are addressed in this article.
The majority of enzyme-inhibition studies of vanadium complexes contains oxygen-coordinating atoms, which are known to show high hydrolytic but low redox stabilityCitation25,Citation26. However, vanadium complexes with mixed oxygen and nitrogen atoms are known to have high hydrolytic and redox stabilityCitation27. Recognizing this fact, we chose hydrazides as bidentate ligands with N and O coordinating atoms. Herein, we report the synthesis and characterization of V(IV) complexes, as well as their PD inhibition and radical scavenging properties.
Experimental
Materials
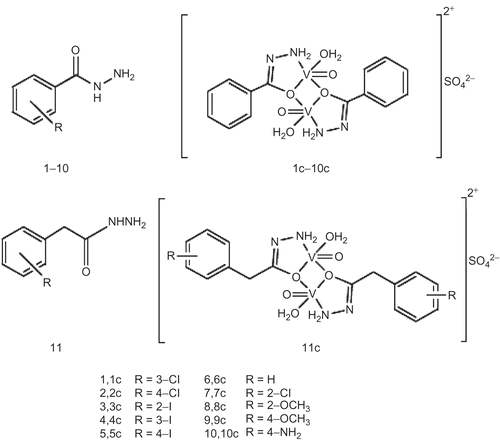
The analytical data of 2-chlorobenzohydrazide (7), 2-methoxybenzohydrazide (8), 4-methoxybenzohydrazide (9), and 4-aminobenzohydrazide (10) were reported previouslyCitation28. All reagent-grade chemicals and snake venom phosphodiesterase I (EC 3.1.4.1) were obtained from Aldrich and Sigma chemical companies and were used without further purification. All the chemicals used for enzyme-inhibition studies were of biochemical grade. Structures of hydrazide ligands and vanadium complexes are given in .
Physical measurements
Ultraviolet (UV)-visible spectra were recorded on a Shimadzu 1601 UV-visible spectrometer with UVPC v3.9 software, from 200 to 900 nm. Solutions were made in air and the spectra were collected at room temperature. Solutions of the complexes (2.5 × 10−4 M) were freshly prepared. Spectra were recorded immediately after complete dissolution. Infrared (IR) spectra were recorded in KBr on a Shimadzu 460 IR spectrometer. Magnetic measurements were made on powders by employing a Sherwood Magway MSB Mk1 magnetic susceptibility balance using sealed-off MnCl2 solution as the calibrant. Metal contents were determined by iodometric titrationCitation29 and also confirmed by 3100 PerkinElmer atomic absorption spectrophotometer. 1H nuclear magnetic resonance (NMR) spectroscopic analysis of ligands was done on a Bruker spectrometer at 400 and 500 MHz. Electron ionization mass spectroscopic (EI-MS) analysis of ligands was done on a Finnigan MAT 311-A apparatus. Elemental (C, H, N) analysis was performed on a PerkinElmer 2400 CHN elemental analyzer. Sulfate was determined by the precipitation methodCitation30. Conductivity measurements were made using a Hanna (HI-8633) conductivity meter (Romania).
Syntheses
Synthesis of hydrazide ligands
Hydrazide ligands were synthesized using a published procedure, and they exhibited similar structures to those reported elsewhere for other hydrazide ligandsCitation28,Citation31. The spectral and analytical data of ligands 1–6 are given in , whereas those of 7–11 are reported elsewhereCitation28,Citation31.
Table 1. Physical, analytical, and spectral data of ligands 1–6.
General procedure for the synthesis of oxovanadium(IV)–hydrazide complexes
To a solution of VOSO4·5H2O (5 mmol) in methanol (10 mL), a solution of the appropriate hydrazide (5 mmol) in methanol (5 mL) was added under stirring. The mixture was heated at reflux for 2 h, during which time the complex precipitated out. The hot mixture was filtered, and the resulting green solid was washed with methanol to remove unreacted vanadyl salt and hydrazide ligand. The product was dried in air. Physical and analytical data of oxovanadium complexes 1c–6c are given in , whereas those of 7c–11c are reported elsewhereCitation28,Citation31.
Table 2. Physical and analytical data of complexes 1c–6c.
Phosphodiesterase inhibition
Activity against snake venom phosphodiesterase I (Sigma P 4631; EC 3.1.4.1) was determined using the reported methodCitation32 with some modifications.
Enzyme inhibition was measured at pH 8.8, at body temperature (37°C), in a solution containing 30 mM magnesium acetate with bis-(p-nitrophenyl)phosphate as a substrate and 0.000742 U/well enzyme. Cysteine and ethylenediaminetetraacetic acid (EDTA) were used as positive controls (IC50 = 748 ± 0.15 μM, 274 ± 0.07 μM, respectively)Citation13,Citation33–35. After 30 min incubation, the enzyme activity was monitored spectrophotometrically at 37°C by following the release of p-nitrophenol from p-nitrophenyl phosphate at 410 nm. The increment in absorption was continuously monitored. All kinetics experiments were performed in 96-well microtiter plates using a SpectraMax 340 reader. IC50 values, the inhibitor concentrations inhibiting 50% enzyme activity, were determined by monitoring the effect of various concentrations of the inhibitors in the assays on the inhibition values. The IC50 values were calculated using the EZ-Fit Enzyme Kinetics program (Perella Scientific, Amherst, USA).
Assays were conducted in triplicate, and graphs were plotted using the GraFit programCitation36. Values of the correlation coefficient, slope, and intercept and their standard errors were obtained by linear regression analysis using the same software. The correlation coefficient for all lines on all graphs was found to be > 0.99, each point in the constructed graphs representing the means of three experiments.
Radical scavenging
Superoxide anion scavenging assay
Superoxide scavenging activities of compounds were determined using the modified method described by Gaulejac et al.Citation37. The reaction was performed in triplicate in a 96-well plate and the absorbance measured on a multiplate reader (SpectraMax 340). The reaction mixture contained 40 μL of the reduced form of nicotinamide adenine dinucleotide (NADH), 40 μL nitroblue tetrazolium (NBT), 90 μL 0.1 M phosphate buffer, pH 7.5, and 10 μL of the test compound. The reaction was initiated by the addition of 20 μL of phenazine methosulfate (PMS) at 28°C and monitored at 560 nm. The control contained 10 μL of dimethylsulfoxide (DMSO) instead of the test compound. The final concentrations of NADH, NBT, and PMS, in a total reaction mixture volume of 200 μL, were 280, 80, and 8 μM, respectively, while the concentration of tested compounds was kept at 1000 μM. The solutions of NBT, NADH, and PMS were prepared in phosphate buffer, while the test compounds were dissolved in DMSO. The radical scavenging activity (RSA; in %) was calculated as: RSA = [100 – (AS/AC) × 100], where AS is the absorbance of the superoxide radical and formazan dye in the presence of the test sample, and AC is the corresponding absorbance without the sample (control).
Nitric oxide radical scavenging assay
Sodium nitroprusside in aqueous solution at physiological pH spontaneously generates nitric oxide to produce nitrite ion, which can be estimated using the Griess Illosvoy reaction. In the present investigation, Griess Illosvoy reagent was modified by the use of naphthylethylenediamine dihydrochloride (0.1% w/v) instead of 1-naphthylamine (5%) scavenger of nitric oxideCitation38. The reaction mixture, contained 10 μL sample (0.25 mM), 20 μL potassium phosphate buffer (0.1 mM, pH 7.4), and 70 μL sodium nitroprusside (10 mM). This was incubated at 25°C for 90–100 min followed by the addition of 50 μL naphthylethylenediamine dihydrochloride (0.1%) and sulfanilic acid (50 μL, 0.33% in 20% glacial acetic acid), and the absorbance at 540 nm was noted against the blank solution in a microtiter plate using an enzyme-linked immunosorbent assay (ELISA) reader.
IC50 determination
Concentrations of compounds at which 50% of the radicals were scavenged or inhibited by the test compound, IC50 values, were determined by monitoring the effect of different concentrations of test compound at 1–1000 μM. The IC50 values of the compounds were calculated using the EZ-Fit Enzyme Kinetics program.
Results and discussion
Synthesis and physicochemical properties
Ligands 1–6 () required for synthesis of the vanadium complexes were prepared in yields 52–84% by refluxing hydrazine hydrate, (NH2)2·H2O, with the appropriate ester in EtOH. The structures of these ligands were determined spectroscopically and by elemental analysis as detailed in . Mass spectral data showed the parent peaks corresponding to the appropriate m/z with different fragmentation patterns. The oxovanadium(IV)–hydrazide (1:1) complexes 1c–6c were synthesized by heating a mixture (1:1) of the appropriate ligand and vanadyl sulfate in an equimolar ratio. Analytical data of complexes is given in and IR and UV-visible spectral data of complexes are given in and , respectively.
Table 3. IR spectroscopic data of ligands 1–6 and their complexes 1c–6c.
The V(IV)–hydrazide complexes were characterized by elemental analysis (C, H, N, V), gravimetric analysis, IR spectroscopy conductivity and magnetic measurements. The complexes exhibited molar conductivities (ΛM) in the range 42.5–62.4 ω−1 cm2 mol−1, which suggested a 1:1 ionic ratio, pointing towards non-coordination of the sulfate ion with the dimeric unit of the V(IV)–hydrazide cation, supporting the formation of outer-sphere complexesCitation39. For monomeric or oligomeric units, the ionic ratio would be different. Gravimetric analysis of sulfate further supported that this counter-ion does not coordinate to the V(IV) center.
The above studies indicated dimeric structures of 1c–6c, with each V(IV) center exhibiting square-pyramidal geometry, coordinated with a bidentate hydrazide ligand and an H2O molecule at the equatorial position, and an oxo (=O) atom coordinated at the apex position (). The hydrazides are, thus, acting as bridging ligands, forming five- and four-membered rings. The magnetic properties supported the dimeric nature of the complexes. The magnetic moments (μeff) were in the range 1.04–1.48 μB (Bohr magnetons; BM), which is lower than that exhibited by an unpaired electron in V(IV) complexes. This may be attributed to a lowered magnetic moment due to antiferromagnetic couplingCitation40,Citation41. Several reports exist in the literature of oxovanadium(IV) binuclear complexes that exhibit magnetic moments in the range reported hereCitation42,Citation43.
Spectroscopy
Infrared spectroscopy
The IR spectroscopic data of ligands 1–6 and their complexes 1c–6c are given in . All hydrazide ligands exhibited a pair of fairly sharp stretching bands in the range 3311– 3060 cm−1, which were tentatively assigned to H-bonded NH groups. The corresponding V(IV) complexes showed broad absorption bands centered at ~ 3250 cm−1 due to a combination of NH and OH stretching vibrations originating from hydrazide ligands and coordinated H2O molecules, respectively, which appear in the same region.
All the ligands exhibited strong C=O stretching absorptions at 1648 ± 18 cm−1, which is within the reported range (1670– 1640 cm−1)Citation44. Upon V(IV) complexation, the intense amide C=O band was shifted to a lower frequency, indicating coordination through the amide O-atom. All complexes exhibit strong V=O symmetric stretching absorptions around 981 ± 25 cm−1, which are close to the V=O stretching frequency reported for other related oxovanadium(IV) complexesCitation31,Citation45–48. These peaks are absent in parent ligands. The appearance of C=O stretching frequencies at lower energy supports the suggested dimeric structure of the complexes, in which the double-bond character of the C=O moiety is expected to be lower than in the parent ligand due to resonance effects. This is supported by an increase in the C–N frequency from 1338 ± 12 cm−1 in the free ligand to ~1352 ± 12 cm−1 in the complexes, indicating a certain double-bond character of the C–N bond in the V(IV) complexes.
UV-visible spectroscopy
The observed electronic transitions exhibited by complexes 1c–6c and free ligands 1–6 recorded in DMSO are collected in . All ligands showed absorption in the UV region assigned to π–π* transitions originating from the π bonds of the hydrazide ligands. These transitions were shifted to higher energy upon coordination with vanadium, which indicates lowering of the energy of the π orbital. However, the V(IV) complexes exhibited another transition in the UV region, and at lower energy than the free ligands. This second peak was tentatively assigned to the π–π* transition of the C=N bond, which is present in the complex, but absent in the free ligand. This electron transition supports the proposed structure of the vanadium complexes as presented in . Oxovanadium complexes 1c–6c exhibited another low intensity transition above 700 nm that is assigned to d–d transitionCitation49.
Table 4. UV-visible spectroscopic data of ligands 1–6 and their complexes 1c–6c.
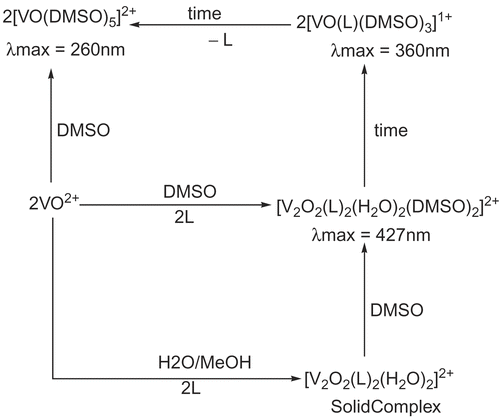
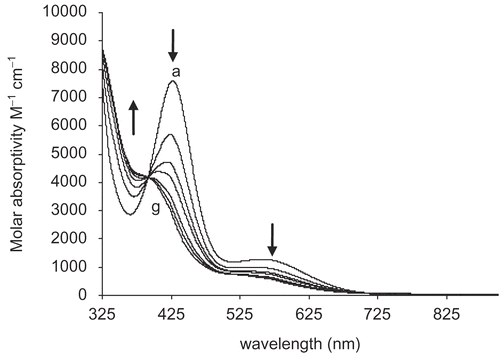
Time-dependent solution stability studies of oxovanadium(IV) complexes in DMSO revealed interesting behavior. shows the absorption pattern of 3c in which an increase in absorbance at 360 nm along with a decrease in absorption at 427 and 559 nm with accompanying shift of peaks is observed. All complexes first showed an increase in absorbance for all transitions with time, before the absorbance at maximum wavelength decreased. The observed initial increase in absorbance was tentatively attributed to the formation of a new dimeric species in which the solvent (DMSO) is coordinated at the sixth position while keeping the original binuclear unit intact (). This assignment was supported by the observation that freshly prepared solutions of the complexes did not show any peaks above 700 nm just after dissolution. These peaks only appeared over time, indicating slow coordination of DMSO.
Figure 1. Time-dependent change in the UV-visible spectrum of 3c in DMSO solution. A decrease in absorbance at 427 and 559 nm and an increase at 360 nm was observed. Spectrum “a” was recorded after 4 days; each subsequent spectrum was recorded with an interval of at least 1 day; spectrum “g” was recorded after 11 days.
Over time, the absorption at 427 nm decreased, with a concomitant increase in a transition around 360 nm. The new species is tentatively assigned to a monomeric complex [VOL(DMSO)3]1+. This complex is not observed in the absence of the ligand and/or DMSO, indicating that both ligand and DMSO are coordinated with the metal. The final product displays peaks at 260 nm and 810 nm, that are assigned to the [VO(DMSO)5]2+ species. This conclusion was verified by dissolving vanadyl sulfate in DMSO, which gives species that show the same transitions. The signal at 260 nm is assigned to π–π* transition, whereas the 810 nm peak is assigned to d–d transition. The presence of solution species could not be verified by any other technique. Therefore, some uncertainty in the nature and exact composition of these species remains.
Enzyme inhibition studies
The hydrazide ligands 1–11 and their oxovanadium(IV) complexes 1c–11c were investigated for their phosphodiesterase inhibitory activities. All free hydrazide ligands were found to be inactive. Complexes 1c–11c were active, exhibiting pure non-competitive-type inhibition, based upon kinetic parameters, which will be reported elsewhere. The IC50 values for phosphodiesterase enzyme with V(IV) complexes are presented in . IC50 values of V(IV)–hydrazide complexes (7–15 μM) compared to vanadium salt VOSO4·5H2O (29 μM) indicate the role that hydrazide ligands play in inhibiting this enzyme. Complexes with chloro-substituted ligands exhibited higher inhibition compared to iodo-substituted ligands, indicating that electronic factors may play an important role in their inhibitory potential. Similar IC50 values for iodo-substituted complexes at ortho, meta, and para positions suggest little effect of steric hindrance.
Table 5. IC50 values for hydrazidesa and their oxovanadium complexes against phosphodiesterase.
The observation that free hydrazide ligands did not show any inhibition, in contrast to the corresponding V(IV) complexes, demonstrates the essential role of the metal center in terms of inhibitory potential. These complexes contain binuclear centers in which each vanadium ion is five-coordinated, leaving the 6th coordination site vacant or weakly coordinated with solvent molecules. Protein side chains are known to bind at this 6th position to inhibit or activate enzymesCitation50. This type of interaction may, thus, be crucial for the mechanism of inhibition of phosphodiesterase by these complexes. Hydrophobic or H-bonding interactions of enzyme side chains with coordinated hydrazide ligands may also contribute. Detailed kinetic studies to understand the mechanism of inhibition are currently under way and will be reported elsewhere.
Superoxide dismutase activity
Free ligands 1–11 exhibited no superoxide dismutase (SOD) activity, whereas their oxovanadium(IV) complexes 1c–11c exhibited moderate to weak activity (IC50 = 90–350 μM). The IC50 values are collected in . Complex 6c with unsubstituted ligand exhibited an IC50 of 141 μM. Substitution of the chloro group at ortho, 7c, and meta, 1c, decreased this value to 113 μM and 90 μM, respectively, whereas it increased to 170 μM for the para-substituted chloro ligand, 2c. This suggests that steric hindrance may increase SOD activity, whereas electronic effects may not play a significant role. Patel and co-workersCitation51 reported IC50 values of oxalato-bridged Cu/Zn (205 μM) and Cu/Ni (216 μM) dimeric complexes. All dimeric V(IV) complexes reported in this study except 10c exhibit lower IC50 values than the reported values for Cu/Zn and Cu/Ni dimers. This may suggest the important role that vanadium may play in SOD activity. Complexes with amino-substituted ligands showed the lowest SOD activity, which may be attributed to the H-bonding interactions of protein side chains with the vanadium center.
Table 6. IC50 values of superoxide scavenging activities of oxovanadium–hydrazide complexesa.
It is interesting to note that the radical scavenging potential of vanadium salt, VOSO4·H2O, is very low (IC50 = 433 μM) compared to V(IV)–hydrazide complexes. Except for 10c, all other complexes exhibit a significantly higher radical scavenging potential (IC50 = 90–184 μM). This suggests the important role of hydrazide ligands coordinated with vanadium centers in SOD activity.
Nitric oxide radical scavenging activity
Eleven free hydrazide ligands, 1–11, and three vanadium complexes did not exhibit nitric oxide scavenging (NOS) activity. Six oxovanadium complexes showed varying degrees of activity, with IC50 values ranging from 94 to 443 μM, as collected in . Only one V(IV) complex, 2c, with a para-substituted chloro group, showed similar inhibitory potential (IC50 = 94 μM) to standard 7,8-dihydroxy flavonel (DHF) (IC50 = 74 μM). All other complexes exhibited low inhibitory potential (IC50 > 237 μM).
Table 7. IC50 values of nitric oxide scavenging activities of oxovanadium–hydrazide complexesa.
Five complexes showed IC50 values of more than 200 μM, whereas only one compound, 2c, showed similar inhibitory potential (IC50 = 94 μM) to standard DHF (IC50 = 74 μM). Complex 2c contains a chloro group at the para position that may not exhibit steric hindrance; however, it will decrease the electron density on the vanadium center by an inductive effect. Reduced electron density on the metal center may help in binding nitric oxide radical with the vanadium center to transfer electrons. Complex 7c with an ortho-substituted chloro group is expected to exhibit similar electronic effects; however, its greater steric hindrance may be responsible for its low inhibition potential. The inactivity of three complexes (1c, 8c, and 11c) may be attributed to decomposition of the complexes under the nitric oxide assay conditions. Similar behavior is exhibited by methoxy-substituted complexes in which the ortho-substituted methoxy complex is inactive and the para-substituted compound exhibits moderate inhibition potential (IC50 = 282 μM). This supports the above conclusion that higher steric hindrance decreases the radical scavenging potential. It indicates that more studies are needed to establish a clear structure–activity relationship.
Conclusions
Several V(IV)–hydrazide complexes have been synthesized and characterized in solid and solution states. Oxovanadium(IV) salt and free ligands are either inactive or exhibit low enzyme inhibition and radical scavenging activity compared to V(IV)–hydrazide complexes. Steric hindrance does not seem to play a role in phosphodiesterase inhibition activity, whereas it increases SOD activity and decreases NOS activity. Electron-withdrawing substituents on the hydrazide ligands decrease their enzyme inhibition activity, whereas no effect is observed for their SOD activity. These studies suggest that fine-tuning is needed for the ligands to exhibit appropriate radical scavenging and enzyme inhibition potential.
Acknowledgments
The authors are thankful to the Higher Education Commission (HEC), Pakistan, for financial support under “National Research Grants Program for Universities”, Grant No. 20-224/2nd Phase/R&D/03/442, and the Ministry of Labour, Manpower and Overseas Pakistanis National Talent Pool for the visit of M.M.T.
Declaration of interest: The authors report no conflicts of interest.
References
- Nekola H, Wang D, Gruning C, Gatjens J, Behrens A, Rehder D. Thiofunctional vanadium complexes. Inorg Chem 2002;41:2379.
- Buglyo P, Nagy EM, Sovago I. Vanadium(III) binding strengths of small biomolecules. Pure Appl Chem 2005;77:1583.
- Crans DC, Smee JJ, Gaidamauskas E, Yang L. Bioinorganic vanadium chemistry. Chem Rev 2004;104:849.
- Sheela A, Roopan SM, Vijayaraghavan R. New diketone based vanadium complexes as insulin mimetics. Eur J Med Chem 2008;43:2206.
- Soares SS, Martins H, Gutiérrez-Merino C, Aureliano M. Vanadium and cadmium in vivo effects in teleost cardiac muscle: metal accumulation and oxidative stress markers. Comp Biochem Physiol C Toxicol Pharmacol 2008;147:168.
- Niu Y, Liu W, Tian C, Xie M, Gao L, Chen Z, et al. Effects of bis(α-furancarboxylato)oxovanadium(IV) on glucose metabolism in fat-fed/streptozotocin-diabetic rats. Eur J Pharmacol 2007;572:213.
- Trends in the use of V. A report of the National Materials Advisory Board, National Academy of Science, 1970, Publication NMAB-267. Springfield, VA: Clearinghouse for Federal Scientific and Technical Information, 1970:46.
- Stern A, Yin X, Tsang SS, Davison A, Moon J. Vanadium as a modulator of cellular regulatory cascades and oncogene expression. Biochem Cell Biol 1993;71:103.
- Fukui K, Ueki T, Ohya H, Michibata H. Vanadium-binding protein in a vanadium-rich ascidian Ascidia sydneiensissamea: CW and pulsed EPR studies. J Am Chem Soc 2003;125:6352.
- Gresser MJ, Tracey AS, Stankiewicz PJ. The interaction of vanadate with tyrosine kinases. J Adv Protein Phosphatases 1987;4:35.
- Sakurai H, Kojima Y, Yoshikawa Y, Kawabe K, Yasui H. Antidiabetic vanadium(IV) and zinc(II) complexes. Coord Chem Rev 2002;226:187.
- Kawabe K, Yoshikawa Y, Adachi Y, Sakurai H. Possible mode of action for insulinomimetic activity of vanadyl(IV) compound in adipocytes. Life Sci 2006;78:2860.
- Razzell WE. Phosphodiesterases. Methods Enzymol 1963;6:236.
- Deutscher MP. The metabolic role of Rnases. Trends Biochem Sci 1988;13:136.
- Deutscher MP. Ribonuclease multiplicity, diversity, and complexity. J Biol Chem 1993;268:13011.
- Sprinkle TJ. 29,39-cyclic nucleotide 39-phosphodiesterase, an oligodendrocyte-Schwann cell and myelin-associated enzyme of the nervous system. Crit Rev Neurobiol 1989;4:235.
- Frank R, Sucheta V, Kristen J, Ira G, Betty M, Petra S, et al. PC-1 nucleoside triphosphate pyrophosphohydrolase deficiency in idiopathic infantile arterial calcification. Am J Pathol 2001;158:543.
- Ceriello A. Oxidative stress and glycemic regulation. Metabolism 2000;49:27.
- Takasu N, Komiya I, Asawa T, Nagasawa Y, Yamada T. Streptozocin- and alloxan-induced H2O2 generation and DNA fragmentation in pancreatic islets. H2O2 as mediator for DNA fragmentation. Diabetes 1991;40:1141.
- Crouch RK, Gandy SE, Kimsey G, Galbraith RA, Galbraith GM, Buse MG. The inhibition of islet superoxide dismutase by diabetogenic drugs. Diabetes 1981;30:235.
- Kaneto H, Fujii J, Seo HG, Suzuki K, Matsuoka T, Nakamura M, et al. Apoptotic cell death triggered by nitric oxide in pancreatic beta-cells. Diabetes 1995;44:733.
- Wolff SP, Dean RT. Glucose autoxidation and protein modification. The potential role of “autoxidative glycosylation” in diabetes. Biochem J 1987;245:243.
- Wilson RL. Free radicals and tissue damage, mechanistic evidence from radiation studies. In: Biochemical Mechanisms of Liver Injury. New York: Academic Press, 1998:123.
- Sekar N, Kanthasamy A, William S, Balasubramaniyan N, Govindasamy S. Antioxidant effect of vanadate on experimental diabetic rats. Acta Diabetol Lat 1990;27:285.
- Sun Y, James BR, Retting SJ, Orvig C. Oxidation kinetics of the potent insulin mimetic agent bis(maltolato)oxovanadium(IV) (BMOV) in water and in methanol. Inorg Chem 1996;35:1667.
- Elvingson K, Baro AG, Pettersson L. Speciation in vanadium bioinorganic systems. 2. An NMR, ESR, and potentiometric study of the aqueous H(+)-vanadate-maltol system. Inorg Chem 1996;35:3388.
- Melchior M, Thompson KH, Jong JM, Rettig SJ, Shuter E, Yuen VG, et al. Vanadium complexes as insulin mimetic: coordination chemistry and in vivo studies of oxovanadium(IV) and dioxovanadate(V) complexes formed from naturally occurring chelating oxazolinate, thiazolinate, or picolinate units. Inorg Chem 1999;38:2288.
- Maqsood ZT, Khan KM, Ashiq U, Jamal RA, Chohan ZH, Mahroof-Tahir M, et al. Oxovanadium(IV) complexes of hydrazides: potential antifungal agents. J Enzyme Inhib Med Chem 2006;21:37.
- Jeffery GH, Bassett J, Denney RC. Titrimetric analysis. In: Vogel’s Textbook of Quantitative Chemical Analysis, 5th ed. Harlow, UK: Longman, 1989:404.
- Jeffery GH, Bassett J, Denney RC. Gravimetry. In: Vogel’s Textbook of Quantitative Chemical Analysis, 5th ed. Harlow, UK: Longman, 1989:490.
- Ara R, Ashiq U, Mahroof-Tahir M, Maqsood ZT, Khan KM, Lodhi MA, et al. Chemistry, urease inhibition, and phytotoxic studies of binuclear vanadium(IV) complexes. Chem Biodivers 2007;4:58.
- Mamillapalli R, Haimovitz R, Ohad M, Shinitzky M Enhancement and inhibition of snake venom phosphodiesterase activity by lysophospholipids. FEBS Lett 1998;436:256.
- Razzell WE, Khorana HG. Studies on polynucleotides. III. Enzymic degradation; substrate specificity and properties of snake venom phosphodiesterase. J Biol Chem 1959;234:2105.
- Nakabayashi T, Matsuoka Y, Ikezawa H, Kimura Y. Alkaline phosphodiesterase I release from eucaryotic plasma membranes by phosphatidylinositol-specific phospholipase C–IV. The release from Cacia porcellus organs. Int J Biochem 1994;26:171.
- Gomez JL, Costas MJ, Ribeiro JM, Fernandez A, Romero A, Avalos M, et al. Glycine-enhanced inhibition of rat liver nucleotide pyrophosphatase/phosphodiesterase-I by EDTA: a full account of the reported inhibition by commercial preparations of acidic fibroblast growth factor (FGF-1). FEBS Lett 1998;421:77.
- Leatherbarrow RJ. GraFit, 4.09 ed. Staines, UK: Erithacus Software Ltd, 1999.
- Gaulejac NSC, Glories Y, Vivas N. Free radical scavenging effect of anthocyanins in red wines. Food Res Int 1999;32:327.
- Badami S, Gupta MK, Suresh B. Antioxidant activity of the ethanolic extract of Striga orobanchioides. J Ethnopharmacol 2003;85:227.
- Geary W. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord Chem Rev 1971;7:81.
- Boas LV, Pesos JC. In: Wilkinson G, Gillard RD, McCleverty JA, eds. Comprehensive Coordination Chemistry. Oxford: Pergamon Press, 1987;3:487.
- Rao SN, Mishra DD, Maurya RC, Rao NN. Oxovanadium binuclear (IV) Schiff base complexes derived from aroyl hydrazones having subnormal magnetic moments. Polyhedron 1997;16:1825.
- Salib KAR, Stefan SL, Abu El-Wafa SM, El-Shafiy HF. Metal complexes of novel symmetrical Schiff base ligands. Syn React Inorg Met Org Chem 2001;31:895.
- Dutta SK, Tiekink ERT, Chaudhury M. Mono- and dinuclear oxovanadium (IV) compounds containing VO(ONS) basic core: synthesis, structure and spectroscopic properties. Polyhedron 1997;16:1863.
- Linert W, Herlinger E, Margl P, Boca R. Spectroscopic, electrochemical and quantum mechanical investigations of vanadyl(IV)-acetylacetonate in non-aqueous solutions. J Coord Chem 1993;28:1.
- Ghosh T, Bhattacharya S, Das A, Mukherjee G, Drew MGB. Synthesis, structure and solution chemistry of mixed-ligand oxovanadium(IV) and oxovanadium(V) complexes incorporating tridentate ONO donor hydrazone ligands. Inorg Chim Acta 2005;358:989.
- Mahroof-Tahir M, Brezina D, Fatima N, Choudhary MI, Rahman AU. Synthesis and characterization of mononuclear oxovanadium(IV) complexes and their enzyme inhibition studies with a carbohydrate metabolic enzyme, phosphodiesterase I. J Inorg Biochem 2005;99:589.
- Yue H, Zhang D, Shi Z, Feng S. Dinuclear oxovanadium(IV) compounds from designed amino acid derivatives. Inorg Chim Acta 2007;360:2681.
- Xing Y-H, Zhang B-L, Zhang Y-H, Yuan H-Q, Sun Z, Ge M-F. Insulin-like model complexes: synthesis, spectra characterization and crystal structure of two novel oxovanadium complexes with poly(pyrazolyl)borate ligands, VO(HB(pz)3)(H2B(pz)2) and VO(B(pz)4)2. Polyhedron 2007;26:3037.
- Remya PN, Suresh CH, Reddy MLP. Rapid reduction and complexation of vanadium by 1-phenyl-3-methyl-4-toluoyl-5-pyrazolone: spectroscopic characterization and structure modeling. Polyhedron 2007;26:5016.
- Cornman CR, Zovinka EP, Meixner MH. Vanadium(IV) complexes of an active-site peptide of a protein tyrosine phosphatase. Inorg Chem 1995;34:5099.
- Patel RN, Singh N, Shukla KK, Chauhan UK, Chakraborty S, Niclos-Gutierrez J, et al. X-ray, spectral and biological (antimicrobial and superoxide dismutase) studies of oxalato bridged CuII–NiII and CuII–ZnII complexes with pentamethyldiethylenetriamine as capping ligand. J Inorg Biochem 2004;98:231.