Abstract
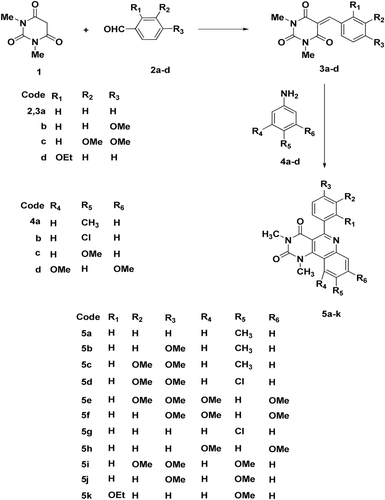
A series of pyrimido[5,4-c]quinoline-2,4-dione derivatives 5a–k were synthesized in moderate yields via a thermolysis reaction of equimolar ratio of 5-arylidine-1,3-dimethylbarbituric acid derivatives 3a–d with aniline derivatives 4a–d at 150–180 °C for 1–2 h. Eight of the synthesized compounds were chosen for a primary in vitro one-dose anticancer assay performed using the full NCI 60 cell panel. Only compound 5b showed moderate GI% at the used dose (10 μM) against four of the tested cell lines corresponding to leukemia SR (GI%: 51), non small-cell lung cancer HOP-92 (GI%: 63), melanoma UACC-62 (GI%: 53) and renal cancer UO-31 (GI%: 69). On the other hand, antimicrobial screening of the whole set of the synthesized compounds was performed against three Gram +ve and two Gram −ve bacterial strains. Results of the antimicrobial screening showed that compounds 5d, 5e, 5f, 5h and 5k have broad-spectrum antibacterial efficacy being moderately active against all the tested Gram +ve and two Gram −ve bacteria. Also, compound 5a showed interesting results being only active against Streptococcus faecalis and both tested Gram −ve strains viz. E. coli and P. aeruginosa. In order to compare the binding mode of the most active compounds 5e and 5f along with the inactive compound 5c we docked these compounds into the empty binding site of topoisomerase II DNA gyrase (PDB ID: 1KZN), and results were compared with the bound inhibitor Clorobiocin.
Introduction
Pyrimidines and their annelated derivatives are of particular importance being widely spread in nature chiefly in the nucleobases found in nucleic acids viz the three pyrimidines cytosine, thymine, and uracil as well as the purines adenine and guanineCitation1. Consequently, derivatives thereof have been extensively studied and many have been found useful in the field of chemotherapy. Among the pyrimidine-containing chemotherapeutics currently in use are the anticancer 5-fluorouracil, the antiviral zidovudine and the antifungal flucytosine in addition to many other antimicrobial agentsCitation2–7.
On the other hand, quinolines play a fairly important role as chemotherapeutics, many derivatives of which being of significant importance as synthetic antimalarials, a disease that claims 1 to 3 million lives annually, for example chloroquineCitation8,Citation9. Also, many antibiotics and UT antiinfectives are based on the quinoline scaffold, like norfloxacin, levofloxacin and othersCitation10,Citation11.
In DNA replication, one group of enzymes has proved to be effective target for therapeutic agents, which is topoisomerase enzyme. DNA gyrase is a type II topoisomerase found in all bacteria and controls the topological state of DNACitation12. DNA gyrase consists of two subunits GyrA (875 amino acids) and GyrB (804 amino acids) with the active species being a heterotetramer A2B2Citation13. Mechanistic studies have revealed the steps involved in the gyrase supercoiling reactionCitation14. This process involves the wrapping of DNA around the A2B2 complex, cleavage of this DNA on both strands, and the passage of a segment of DNA through the double strand break. Relegation of the break results in the introduction of two negative supercoils. These processes require the binding and hydrolysis of ATPCitation15. Inhibition of DNA gyrase blocks relaxation of supercoiled DNA, relaxation being a requirement for transcription and replication. DNA gyrase is a selective target for antibacterial agents, such as the most studied quinolone and coumarin antibiotics. Quinolone drugs (e.g. ciprofloxacin) affect the protein subunit GyrA and coumarins (1-benzopyran-2-ones) (e.g. novobiocin, clorobiocin) act on GyrBCitation16. Computer docking technique plays an important role in the drug design and discovery, as well as in the mechanistic study by placing a molecule into the binding site of the target macromolecule in a non-covalent fashionCitation17,Citation18, and predicting the binding geometry and hydrogen bonds formed with the surrounding amino acids as well as the mode of the ligand to the receptor, which all contribute to the docking score. MOE as a flexible docking program facilitates the prediction of favorable protein–ligand complex structures with reasonable accuracy and speed (MOE 2008.10 of Chemical Computing Group. Inc.).
Given the promising properties of pyrimidines and quinolines and in continuation of our interest in preparing biologically active heterocycles, we decided to study the effect of combining the pyrimidine and quinoline scaffolds in one molecule on the biological activity of such a ring system. Therefore, this study covers the synthesis and molecular modeling study of a series of substituted 5-aryl-pyrimido[5,4-c]quinoline-2,4-diones and evaluation of their anticancer and antimicrobial activities.
Experimental section
Biology
Anticancer screening of pyrimido[5,4-c]quinoline-2,4-dione derivatives
Experimental design
The chosen compounds were subjected to the in vitro disease-oriented NCI 60 human cells screening panel assay to be evaluated for their in vitro anticancer activity. In this protocol, all compounds submitted to the screen are tested initially at a single high dose (10 μM) in the full NCI 60 cell panel including leukemia, non-small cell lung, colon, CNS, melanoma, ovarian, renal, prostate and breast cancer cell lines. A 48 h continuous drug exposure protocol was used, and a sulphorhodamine B (SRB) protein assay was employed to estimate cell viability or growthCitation19–21. The data obtained are a mean graph of the percent growth of treated cells, and presented as percentage growth inhibition (GI %) caused by the tested compounds.
Experimental procedure
Cells were plated in 96-multiwell microtiter plate (104 cells/well) for 24 h before treatment with the test compound to allow attachment of cells to the wall of the plate. Tested compounds were dissolved in DMSO and diluted with saline to the appropriate volume. The compounds under test were added to the cell monolayer. Monolayer cells were incubated with the compound(s) for 48 h at 37 °C and in atmosphere of 5% CO2. After 48 h, cells were fixed, washed, and stained for 30 min with 0.4% (wt/vol) SRB dissolved in 1% acetic acid. Excess unbound dye was removed by four washes with 1% acetic acid and attached stain was recovered with Tris–EDTA buffer. Color intensity was measured in an ELISA reader at a wave length of 570 nm.
Antimicrobial screening of pyrimido[5,4-c]quinoline-2,4-dione derivatives
Experimental design
An extensive assessment of the antimicrobial activity of the 11 newly synthesized pyrimido[5,4-c]quinoline derivatives 5a–k was performed. The screening was carried out against 5 different microbial strains; three organisms representing Gram +ve bacteria viz. Bacillus subtilis, Staphylococcus Aureus and Streptomyces faecalis; two Gram −ve bacterial strains Escherichia coli and Pseudomonas aeruginosa. The activity of the tested compounds was evaluated using disc diffusion method comparing the diameter of the inhibition zone produced by the new derivatives with those of the standard antibacterial agent Tetracycline.
Experimental procedure
Antimicrobial activity of the tested samples was determined using a modified Kirby-Bauer disc diffusion methodCitation22–25.
Blank paper discs with a diameter of 8.0 mm were impregnated with 10 µL of the stock solutions of the tested compounds and placed on agar where the chemical diffused from the disc into the agar only around the disc. Organisms susceptible to the tested compound do not grow in the area around the disc in what is known as a “Zone of inhibition” whose diameters were measured (in mm) with slipping calipers.
Standard discs of Tetracycline (Antibacterial agent) served as positive controls for antimicrobial activity while filter discs impregnated with 10 µL of the solvent (DMSO) were used as a negative control.
Docking methodology
Docking studies were performed using MOE software Version 2008.10. The X-ray crystal structure of the antimicrobial agent Clorobiocin bound to topoisomerase II DNA gyrase was obtained from the RCSB Protein Data Bank (PDB ID: 1KZN). The enzyme was prepared for docking as follows: (a) Clorobiocin interactions were determined to reveal the different types of interaction and a validation for the docking was performed (). (b) Clorobiocin and solvent molecules were removed from the binding site. (c) The hydrogens were added with their standard geometry. (d) The newly synthesized compounds were constructed and were energy minimized for 1000 iterations reaching a convergence of 0.01 kcal/molA. (e) Docking of the energy minimized ligand molecules was carried out using the default MOE-dock parameters. The docking aims at searching for favorable binding conformations. The quality of the docked structures was evaluated by measuring the intermolecular energy of the ligand-DNA gyrase-B assembly dG calculated in kcal/molA (S value in MOE)Citation26,Citation27.
Figure 1. Left panel: 2D sketch of the binding mode of Clorobiocin into its binding site of DNA gyrase-B, showing three hydrogen bonds as green dotted lines (with residues Arg136, Asp73 and Asn46) and one Pi-cationic interaction with Arg76; right panel: Docking validation of Clorobiocin with DNA gyrase-B: crystal structure ligand (red) docked ligand (yellow); RMSD 0.41 Å.
Chemistry
Melting points were recorded using a Gallenkamp melting-point apparatus and are uncorrected. Thin-layer chromatography (TLC) analysis was carried out on silica gel 60 F254 precoated aluminum sheets and detected under ultraviolet (UV) light. Infrared (IR) spectra were recorded using KBr wafer technique on a Shimadzu 5800 Fourier transform FT-IR Spectrometer at College of Science, King Faisal University. 1H NMR spectra were recorded employing a Varian Unity Plus 300 spectrometer and chemical shifts (δ) are in parts per million (ppm) relative to tetramethylsilane (TMS) as internal standard. Mass spectra were recorded on a GC-MS (Schimadzu QP-1000 EX) spectrometer. Elemental analyses were performed on Perkin-Elmer 2400 analyser at the microanalytical laboratories of the Faculty of Science, Cairo University and are within ±0.4 of the theoretical values. All chemicals and solvents were purchased from Aldrich Chemical Co., Fisher Scientific. All solvents were reagent grade. 5-Benzylidene-1,3-dimethylpyrimidine-2,4,6-trione (3a) was prepared according to the reported literatureCitation28.
Preparation of 5-arylidine-1,3-dimethylbarbituric acids 3b–d
5-(4-Methoxybenzylidene)-1,3-dimethylpyrimidine-2,4,6-trione (3b): To a solution of 1,3-dimethylbarbituric acid (15.6 g, 100 mmol) and 4-methoxybenzaldehyde (13.62 g, 100 mmol) in methanol (150 mL) was added few drops of conc. HCl. The reaction mixture was heated at reflux for 3 h and the precipitate was filtered off, washed with methanol and recrystallized from ethanol/EtOAc to afford compound 3b in 79% yield, mp 154–155 °C; Lit.Citation29 mp 149–151 °C.
5-(3,4-Dimethoxybenzylidene)-1,3-dimethylpyrimidine-2,4,6-trione (3c): Adopting the same methodology used for preparation of 3b, starting with 3,4-dimethoxybenzaldehyde instead of 4-methoxybenzaldehyde. Yield 89%, mp 229–231 °C; Lit.Citation29 mp 226–228 °C; Lit.Citation30 mp 208–210 °C. IR (KBr) ν’ 3122, 3004 (aromatic CH), 2947, 2906, 2839 (CH3), 1720, 1651 (CO), 1598, 1556, 1502 (C=C) cm−1. 1H NMR (CDCl3); δ 3.39, 3.40 (2s, 6H; 2 × CH3), 3.97, 3.98 (2s, 6H; 2 × OCH3), 6.94 (d, J = 8.7 Hz, 1H), 7.78 (dd, J = 8.7 Hz, 2.1 Hz, 1H), 8.38 (d, J = 2.1 Hz, 1H), 8.48 (s, 1H).
5-(2-Ethoxybenzylidene)-1,3-dimethylpyrimidine-2,4,6-trione (3d): Adopting the same methodology used for preparation of 3b, starting with 2-ethoxybenzaldehyde instead of 4-methoxybenzaldehyde. Yield 85%, mp 165–166 °C; Lit.Citation31 melting point not reported. IR (KBr) ν’ 3114, 3095, 2987, 2950, 2900 (CH), 1730, 1670 (CO), 1577, 1541 (C=C) cm−1. 1H NMR (CDCl3); δ 1.37 (t, J = 7.2 Hz, 3H), 3.28 (s, 3H), 3.35 (s, 3H), 4.07 (q, J = 7.2 Hz, 2H), 6.84 (d, J = 8.4 Hz, 1H), 6.92 (t, J = 7.2 Hz, 1H), 7.39 (t, J = 7.2 Hz, 1H), 7.98 (d, J = 8.4 Hz, 1H), 8.85 (s, 1H). MS (EI) m/e (rel.int.); 288 (M+, 24), 243 (100). Anal. Calc. for C15H16N2O4 (288.30): C, 62.49; H, 5.59; N, 9.72. Found. C, 62.21; H, 5.73; N, 9.48.
General Procedure for preparation of pyrimido[5,4-c]quinoline-2,4-diones 5a–k
A mixture of 5-arylidene-1,3-dimethylbarbituric acid derivatives 3a–d (10 mmol) and aniline derivatives 4a–d (10 mmol) was fused in an oil bath at 150–180 °C for 1–2 h. The reaction mixture was cooled and triturated with ethanol, the precipitated products were filtered off and purified by either recrystallization from EtOH/DMF or column chromatography on silica gel using petroleum ether (35–60 °C)/EtOAc (7:3) as an eluent.
1,3,9-Trimethyl-5-phenylpyrimido[5,4-c]quinoline-2,4-dione (5a): adopting the reported methodologyCitation32, mp >300 °C.
5-(4-Methoxyphenyl)-1,3,9-trimethylpyrimido [5,4-c]quinoline-2,4-dione (5b): adopting the reported methodologyCitation32, mp 251–252.5 °C.
5-(3,4-Dimethoxyphenyl)-1,3,9-trimethylpyrimido [5,4-c]quinoline-2,4-dione (5c): Yield 53 %, mp 276–277 °C (EtOH/DMF). IR (KBr) ν’ 2991, 2935, 2837 (CH), 1708, 1668 (CO), 1635, 1606, 1569 (C=N, C=C) cm−1. 1H NMR (DMSO-d6); δ 2.35 (s, 3H), 3.18 (s, 3H), 3.69 (s, 3H), 3.70 (s, 3H), 3.87 (s, 3H), 6.72 (dd, J = 8.1 Hz, 2.1 Hz, 1H), 6.82 (d, J = 2.1 Hz, 1H), 7.07 (d, J = 8.1 Hz, 1H), 7.16 (d, 2.1 Hz, 1H), 7.68 (dd, J = 8.4 Hz, 2.1 Hz, 1H), 7.86 (d, J = 8.4 Hz, 1H). MS (EI) m/e (rel.int.); 391 (M+, 100), 392 (M+ + 1, 20), 376 (10), 363 (10). Anal. Calc. for C22H21N3O4 (391.42): C, 67.51; H, 5.41; N, 10.74. Found. C, 67.16; H, 5.62; N, 10.89.
9-Chloro-5-(3,4-dimethoxyphenyl)-1,3-dimethyl pyrimido[5,4-c]quinoline-2,4-dione (5d): Yield 47 %, mp 276–278 °C (EtOH/DMF). IR (KBr) ν’ 3080, 3008, 2958, 2927, 2837 (CH), 1712, 1672 (CO), 1602, 1568, 1517 (C=N, C=C) cm−1. 1H NMR (DMSO-d6); δ 3.17 (s, 3H), 3.67 (s, 3H), 3.70 (s, 3H), 3.87 (s, 3H), 6.75 (dd, J = 8.1 Hz, 1.8 Hz, 1H), 6.85 (d, J = 1.8 Hz, 1H), 7.11 (d, J = 8.4 Hz, 1H), 7.29 (d, J = 2.4 Hz, 1H), 7.83 (dd, J = 9.0 Hz, 2.4 Hz, 1H), 7.95 (d, J = 9.0 Hz, 1H). MS (EI) m/e (rel.int.); 411 (M+, 100), 412 (M+ + 1, 12), 396 (49). Anal. Calc. for C21H18ClN3O4 (411.84): C, 61.24; H, 4.41; N, 10.20. Found. C, 61.19; H, 4.49; N, 10.13.
5-(3,4-Dimethoxyphenyl)-8,10-dimethoxy-1,3-dimethylpyrimido[5,4-c]quinoline-2,4-dione (5e): Yield 40 %, mp 302–303.5 °C (EtOH/DMF). IR (KBr) ν’ 2997, 2943, 2837 (CH), 1708, 1668 (CO), 1618, 1585, 1552 (C=N, C=C) cm−1. 1H NMR (DMSO-d6); δ 3.11 (s, 3H), 3.38 (s, 3H), 3.63 (s, 3H), 3.68 (s, 3H), 3.81 (s, 3H), 3.91 (s, 3H), 6.41 (d, J = 2.1 Hz, 1H), 6.57 (dd, J = 8.4 Hz, 2.1 Hz, 1H), 6.74 (d, J = 2.1 Hz, 1H), 6.88–6.91 (m, 2H). MS (EI) m/e (rel.int.); 437 (M+, 100), 438 (M+ + 1, 32), 422 (26), 407 (11). Anal. Calc. for C23H23N3O6 (437.45): C, 63.15; H, 5.30; N, 9.61. Found. C, 63.23; H, 5.46; N, 9.72.
8,10-Dimethoxy-5-(4-methoxyphenyl)-1,3-dimethylpyrimido[5,4-c]quinoline-2,4-dione (5f): Yield 38 %, mp >300 °C (EtOH/DMF). IR (KBr) ν’ 2954, 2912, 1835 (CH), 1708, 1668 (CO), 1635, 1616, 1560 (C=N, C=C) cm−1. 1H NMR (DMSO-d6); δ 3.13 (s, 3H), 3.33 (s, 3H), 3.68 (s, 3H), 3.83 (s, 3H), 3.94 (s, 3H), 6.44 (s, 1H), 6.89 (d, J = 8.4 Hz, 2H), 6.94 (s, 1H), 6.99 (d, J = 8.4 Hz, 2H). MS (EI) m/e (rel.int.); 407 (M+, 100), 408 (M+ + 1, 20), 379 (20). Anal. Calc. for C22H21N3O5 (407.42): C, 64.86; H, 5.20; N, 10.31. Found. C, 64.49; H, 5.41; N, 10.32.
9-Chloro-1,3-dimethyl-5-phenylpyrimido[5,4-c]quinoline-2,4-dione (5g): Yield 45 %, mp 263-265 °C (EtOH/DMF). IR (KBr) ν’ 3078, 3026, 2956, 2866 (CH), 1716, 1676 (CO), 1652, 1602, 1566 (C=N, C=C) cm−1. 1H NMR (DMSO-d6); δ 3.18 (s, 3H), 3.71 (s, 3H), 7.14 (d, J = 2.4 Hz, 1H), 7.22–7.26 (m, 2H), 7.52–7.55 (m, 3H), 7.88 (dd, J = 9.0 Hz, 2.4 Hz, 1H), 8.01 (d, J = 9.0 Hz, 1H). MS (EI) m/e (rel.int.); 351, 353 (M+, 100, 36: chlorine isotopes), 316 (17). Anal. Calc. for C19H14ClN3O2 (351.79): C, 64.87; H, 4.01; N, 11.94. Found. C, 64.55; H, 4.06; N, 12.03.
8,10-Dimethoxy-1,3-dimethyl-5-phenylpyrimido[5,4-c]quinoline-2,4-dione (5h): Yield 40 %, mp >300 °C (EtOH/DMF). IR (KBr) ν’ 3067, 3006, 2952 (CH), 1708, 1664 (CO), 1618, 1585, 1556 (C=N, C=C) cm−1. 1H NMR (DMSO-d6); δ 3.12 (s, 3H), 3.16 (s, 3H), 3.69 (s, 3H), 3.95 (s, 3H), 6.43 (s, 1H), 6.95–7.09 (m, 4H), 7.31 (s, 2H). MS (EI) m/e (rel.int.); 377 (M+, 100), 378 (M+ + 1, 71), 349 (45), 234 (40). Anal. Calc. for C21H19N3O4 (377.39): C, 66.83; H, 5.07; N, 11.13. Found. C, 66.59; H, 5.27; N, 11.39.
5-(3,4-Dimethoxyphenyl)-9-methoxy-1,3-dimethylpyrimido[5,4-c]quinoline-2,4-dione (5i): Yield 43 %, mp 253–255 °C(EtOH/DMF). IR (KBr) ν’ 3010, 2950 (CH), 1709, 1668 (CO), 1652, 1622, 1569, 1558, 1506 (C=N, C=C) cm−1. 1H NMR (CDCl3); δ 3.40 (s, 3H), 3.69 (s, 3H), 3.87 (s, 3H), 3.93 (s, 3H), 4.00 (s, 3H), 6.71–6.81 (m, 3H), 7.06 (d, J = 8.1 Hz, 1H), 7.48 (dd, J = 9.0 Hz, 2.7 Hz, 1H), 8.05 (d, J = 9.0 Hz, 1H). MS (EI) m/e (rel.int.); 407 (M+, 100), 392 (7). Anal. Calc. for C22H21N3O5 (407.42): C, 64.86; H, 5.20; N, 10.31. Found. C, 64.73; H, 5.18; N, 10.25.
9-Methoxy-5-(4-methoxyphenyl)-1,3-dimethylpyrimido[5,4-c]quinoline-2,4-dione (5j): purified by column chromatography on silica gel using petroleum ether (35–60 °C)/EtOAc (7:3) as an eluent, Rf = 0.40. Yield 55 %, mp 268–269.5 °C. IR (KBr) ν’ 3001, 2962, 2941 (CH), 1709, 1664 (CO), 1635, 1622, 1569 (C=N, C=C) cm−1. 1H NMR (CDCl3); δ 3.40 (s, 3H), 3.69 (s, 3H), 3.90 (s, 3H), 3.93 (s, 3H), 6.71 (d, J = 2.7 Hz, 1H), 7.10 (d, J = 8.7 Hz, 2H), 7.17 (d, J = 8.7 Hz, 2H), 7.47 (dd, J = 9.0 Hz, 2.7 Hz, 1H), 8.03 (d, J = 9.0 Hz, 1H). MS (EI) m/e (rel.int.); 377 (M+, 100), 362 (6). Anal. Calc. for C21H19N3O4 (377.39): C, 66.83; H, 5.07; N, 11.13. Found. C, 66.95; H, 5.12; N, 10.88.
5-(2-Ethoxyphenyl)-9-methoxy-1,3-dimethylpyrimido[5,4-c]quinoline-2,4-dione (5k): purified by column chromatography on silica gel using petroleum ether (35-60 °C)/EtOAc (7:3) as an eluent, Rf = 0.46. Yield 46 %, mp 255–257 °C. IR (KBr) ν’ 3060, 2979, 2954 (CH), 1710, 1660 (CO), 1622, 1568 (C=N, C=C) cm−1. 1H NMR (CDCl3); δ 1.05 (t, J = 7.2 Hz, 3H), 3.40 (s, 3H), 3.68 (s, 3H), 3.88 (s, 3H), 4.00 (q, J = 7.2 Hz, 2H), 6.70 (d, J = 2.7 Hz, 1H), 7.07–7.14 (m, 3H), 7.43–7.50 (m, 2H), 7.94 (d, J = 9.0 Hz, 1H). MS (EI) m/e (rel.int.); 391 (M+, 58), 346 (100). Anal. Calc. for C22H21N3O4 (391.42): C, 67.51; H, 5.41; N, 10.74. Found. C, 67.39; H, 5.54; N, 10.46.
Results and discussion
Chemistry
In view of the significant therapeutic value of quinolines, it was considered worthwhile to incorporate quinoline moieties into the C5–C6 position of the oxypyrimidine nucleus. Thus, a series of the pyrimido[5,4-c]quinoline-2,4-dione derivatives 5a–k were synthesized in moderate yields via a thermolysis reaction of equimolar ratio of benzalbarbituric acid derivatives 3a–d with the respective aniline derivatives 4a–d at 150–180 °C for 1–2 h. The structures of pyrimido[5,4-c]quinolines 5a–k were confirmed from their elemental analyses as well as their spectral data. Thus, 1H NMR spectrum of 5c does not show the signals corresponding to AA’-BB’ system for a p-substituted benzene ring, however, it showed a new ABX system attributed for 1,2,4-trisubstituted benzene. Previously, we reported a suggested mechanism for the formation of 5aCitation32. In general, the suggested mechanism for the formation of pyrimido[5,4-c]quinolines 5a–k, involves addition reaction of aniline derivative 4a–d to the 5-arylidine derivative 3a–d, followed by cyclization to dihydropyrimido[5,4-c]quinoline intermediate which was subsequently autooxidized (). 5-Arylidine-1,3-dimethylbarbituric acids 3a–d were prepared via a direct acid-catalyzed condensation reaction between equimolar ratios of 1,3-dimethylbarbituric acid and the respective benzaldehyde derivatives 2a–d in methanol as a solvent. The structures of compounds 3c and 3d were confirmed from their spectral data. Thus, 1H NMR spectrum of 3c showed ABX system attributed for 1,2,4-trisubstituted benzene moiety indicated by the presence of three protons signals attributed to H-2; H-5 and H-6; and singlet signal at δ 8.48 attributed for a methine proton, in addition to four singlet signals attributed for protons of four methyl groups.
Biology
Anticancer screening of pyrimido[5,4-c]quinoline-2,4-dione derivatives
Eight of the synthesized compounds 5a–h were chosen by the National Cancer Institute (NCI, Betheseda, MD, USA) for the in vitro disease-oriented human cells screening panel assay to be evaluated for their in vitro cytotoxic activity. A primary in vitro one-dose anticancer assay was performed using the full NCI 60 cell panel in accordance with the current protocol of the Drug Evaluation Branch, NCI, Bethesda, MD, USA. These cell lines were incubated with one concentration (10 µM) for each tested compound. A 48 h continuous drug exposure protocol was used, and a sulphorhodamine B (SRB) protein assay was employed to estimate cell viability or growth.
Data obtained from results of the anticancer screening of the eight tested quinoline derivatives can be summarized as follows, while seven of the tested compounds showed no growth inhibitory activity against all the 60 tested cancer cell lines at the used dose (10 μM), only compound 5b possessed some cytotoxic activity. It showed moderate GI% against four of the tested cell lines corresponding to leukemia SR (GI%: 51), non small-cell lung cancer HOP-92 (GI%:63), melanoma UACC-62 (GI%: 53) and renal cancer UO-31 (GI%: 69). It also exhibited poor activity against seven cell lines comprising three breast cancer cell lines (MCF, MDA-MB-231/ATCC and HS 578T) with GI% 31, 38 and 33, respectively; two non-small cell lung cancer cell lines HOP-62 (GI%: 30) and NCI-H522 (GI%: 37); one renal cancer cell line (A498, GI%: 32) and one leukemia cell line (RPMI-8226, GI%: 30).
Antimicrobial screening of pyrimido[5,4-c]quinoline-2,4-dione derivatives
The newly synthesized compounds 5a-k were tested against a panel of susceptible and resistant Gram +ve bacteria (Bacillus subtilis, Staphylococcus Aureus and Streptomyces faecalis) and Gram −ve (Escherichia coli and Pseudomonas aeruginosa) bacteria. Results of the antimicrobial screening () showed that compounds 5d, 5e, 5f, 5h and 5k have broad-spectrum antibacterial efficacy being moderately active against all the tested Gram +ve and Gram −ve bacteria. Compound 5i was active against four of the tested bacterial strains. Interestingly, compound 5b showed selectivity only towards Streptococcus faecalis which is known to be a relatively resistant strain of Gram +ve bacteria. Also, compound 5a showed interesting results being only active against Streptococcus faecalis and both tested Gram −ve strains viz. E. coli and P. aeruginosa. Finally, Compound 5j was only slightly active against Staph. aureus and E.coli.
Table 1. In vitro antimicrobial activity of pyrimido[5,4-c]quinoline-2,4-diones 5a–k.
In conclusion, results of the antibacterial screening showed that pyrimido[5,4-c]quinoline-2,4-diones 5d, 5e, 5f, 5h and 5k have broad-spectrum antibacterial efficacy being moderately active against all the tested Gram +ve and two Gram −ve bacteria as compared to the standard drug Tetracycline. The following few lines display the antibacterial activity of some pyrimidine and quinoline derivatives either in the market or a few of those recently reported for the sake of comparison. Whereas, pyrimidine is pharmacologically inactive, its synthetic derivatives possess an important role in modern medicineCitation33. Amongst the 2,4-diaminopyrimidine drugs, trimethoprim, an antibacterial drug is also a selective inhibitor and selectively inhibits bacterial enzyme dihydrofolate reductase (DHFR)Citation34. Brodimoprim, is also found to be an effective antibacterial compoundCitation35. Recently, some novel reported pyrazolopyrmidine-diones showed highest antimicrobial activity against Pseudomonas aeruginosa, Enterococcus faecalis and Bacillus pumilus as compared to the standard drug StreptomycinCitation36.
On the other hand, quinolones represent a well-known class of synthetic broad-spectrum antibacterial agents including the market antibiotics levofloxacin and ciprofloxacinCitation37,Citation38. These drugs are active against a diverse assembly of Gram −ve and Gram +ve bacteria. It is well stated that the pharmacophore of this group of drugs is the quinolone ring system which is responsible for the biological activity of these drugsCitation39. Furthermore, Holla et al.Citation40 described that their newly synthesized quinoline substituted pyrazolo[3,4-d]pyrimidin-4-ones showed maximum antibacterial activity against staphylococcus aureus in comparison to the standard drug in that study Streptomycin. Recently, a series of novel tetrahydroquinoline annulated heterocycles exhibited good antibacterial activity against microorganisms of which one of them was found to be as active as the antibiotic ciprofloxacinCitation41.
Finally, while our newly synthesized compounds are less potent than the pyrimidines or quinolines in the market they compare well with some reported ones.
Docking studies
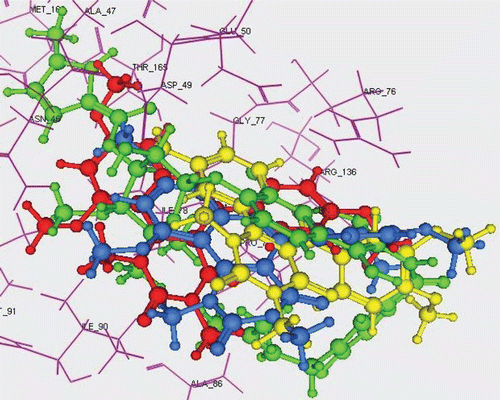
In an attempt to explain the observed antimicrobial data on a structural basis, automated docking studies were carried out using MOE 10.2008 program installed on 2.0G Core 2 Duo. The protein–ligand complex based on the X-ray crystal structure of the topoisomerase II DNA gyrase-B enzyme with its bound inhibitor Clorobiocin available through the RCSB Protein Data Bank (PDB entry 1KZN) was employed for the docking study of the new compoundsCitation42. The active site of the enzyme was defined to include residues within a 10.0Å radius to any of the inhibitor atoms. The binding poses as well as the binding interaction modes of the docked compounds with the surrounding amino acids in the active pocket of DNA gyrase-B were used to try to understand and rationalize the observed antimicrobial activity of the most active compounds 5e and 5f along with the inactive compound 5c. The binding mode of topoisomerase II DNA gyrase-B (1KZN) with its bound inhibitor Clorobiocin () showed that the binding complex involves 3 hydrogen bonds with residues Arg136, Asp73 and Asn46 and one Pi-cationic interaction with Arg76. Docking validation was performed with an obtained RMSD value of 0.41Å ensuring precision and reproducibility of the docking process (). The energy minimized structures of the most active compounds 5e and 5f as well as of the inactive compound 5c with were docked in the active site of topoisomerase II DNA gyrase-B. The 2D ligand interactions of these 3 derivatives with DNA gyrase-B displayed in shows that the inactive compound 5c possesses only a pi–cationic interaction with Arg76 which is also conserved with the other two compounds 5e and 5f. Moreover, the active compounds 5e and 5f demonstrated the presence of extra H-bond interactions (one with Arg136 for 5e; two with Arg136 and Asn46 for 5f) which can be regarded as the reason behind their improved activity (). For further understanding, 3D overlay of Clorobiocin, 5c, 5e and 5f in DNA-gyrase active pocket was performed which revealed that the orientations of 5e and 5f are similar with a slight displacement however 5c possessed a totally different orientation with the pyrimido[5,4-c]quinolone fused ring system slightly protruding out of the active pocket thus preventing the formation of essential binding interactions ().
Figure 3. 2D Ligand interaction of the 5-aryl-pyrimido[5,4-c]quinoline-2,4-diones with DNA gyrase-B; panel (1) compound 5c showing only pi-cationic interaction with Arg76; panel (2) compound 5e showing pi-cationic interaction with Arg76 and H-bond with Arg136; panel (3) compound 5f showing pi-cationic interaction with Arg76 and two H-bonds with Arg136 and Asn46.
![Figure 3. 2D Ligand interaction of the 5-aryl-pyrimido[5,4-c]quinoline-2,4-diones with DNA gyrase-B; panel (1) compound 5c showing only pi-cationic interaction with Arg76; panel (2) compound 5e showing pi-cationic interaction with Arg76 and H-bond with Arg136; panel (3) compound 5f showing pi-cationic interaction with Arg76 and two H-bonds with Arg136 and Asn46.](/cms/asset/75a2483f-6a01-4217-88a4-648f66e13f16/ienz_a_654113_f0003_b.gif)
Figure 4. 3D overlay of Clorobiocin (green), 5c (yellow), 5e (blue) and 5f (red) in DNA-gyrase active pocket.
After studying the docking poses and binding modes of the docked compounds, the necessity of hydrogen bond formation for enhancing the activity of this class of compounds can be highly advocated.
Acknowledgement
This work was supported by Deanship of Scientific Research, King Faisal University, Project No. 110010. The in vitro anticancer screening was done by the National Cancer Institute (NCI, Betheseda, MD, USA).
Declaration of interest
The authors declare no conflict of interest.
References
- Brown DJ. The Pyrimidines. Interscience, New York, NY. 1994.
- Macllwain C. Activists now urge caution on approval of new AIDS drug. Nature 1993;365:378.
- Wamberg M, Pedersen EB, El-Brollosy NR, Nielsen C. Synthesis of 6-arylvinyl analogues of the HIV drugs SJ-3366 and Emivirine. Bioorg Med Chem 2004;12:1141–1149.
- Jones AS, Sayers JR, Walker RT, De Clercq E. Synthesis and antiviral properties of (E)-5-(2-bromovinyl)-2′-deoxycytidine-related compounds. J Med Chem 1988;31:268–271.
- De Clercq E. Potential of bromovinyldeoxyuridine in anticancer chemotherapy. Anticancer Res 1986;6:549–556.
- Miyasaka T, Tanaka H, Baba M, Hayakawa H, Walker RT, Balzarini J et al. A novel lead for specific anti-HIV-1 agents: 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine. J Med Chem 1989;32:2507–2509.
- Zoorob HH, Abou Elzahab M, Abdel-Mogib M, Ismail MA, Abdel-Hamid M. 1,3-Dimethylpyrimidoheterocycles as antibacterial agents. Arzneimittelforschung 1997;47:958–962.
- Foley M, Tilley L. Quinoline antimalarials: mechanisms of action and resistance and prospects for new agents. Pharmacol Ther 1998;79:55–87.
- Savarino A, Gennero L, Chen HC, Serrano D, Malavasi F, Boelaert JR et al. Anti-HIV effects of chloroquine: mechanisms of inhibition and spectrum of activity. AIDS 2001;15:2221–2229.
- Mittmann NN, Jivarj FF, Wong AA and Yoon AA. Oral fluoroquinolones in the treatment of pneumonia, bronchitis and sinusitis. Can J Infect Dis 2002;5:293–300.
- Childs SJ. Safety of the Fluoroquinolone antibiotics: focus on molecular structure. Infect Urol 2000;13:3–10.
- Menzel R, Gellert M. The biochemistry and biology of DNA gyrase. Adv Pharmacol 1994;29A:201–225.
- Maxwell A, Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem 1986;38:69–107.
- Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci USA 1999;96:8414–8419.
- Brino L, Urzhumtsev A, Mousli M, Bronner C, Mitschler A, Oudet P et al. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J Biol Chem 2000;275:9468–9475.
- Flatman RH, Eustaquio A, Li SM, Heide L, Maxwell A. Structure-activity relationships of aminocoumarin-type gyrase and topoisomerase IV inhibitors obtained by combinatorial biosynthesis. Antimicrob Agents Chemother 2006;50:1136–1142.
- Kontoyianni M, McClellan LM, Sokol GS. Evaluation of docking performance: comparative data on docking algorithms. J Med Chem 2004;47:558–565.
- Wang R, Lu Y, Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J Med Chem 2003;46:2287–2303.
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990; 82:1107–1112.
- McCaffrey TA, Agarwal LA, Weksler BB. A rapid fluorometric DNA assay for the measurement of cell density and proliferation in vitro. In Vitro Cell Dev Biol 1988;24:247–252.
- Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 1991;83:757–766.
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 1966;45:493–496.
- Pfaller MA, Burmeister L, Bartlett MS, Rinaldi MG. Multicenter evaluation of four methods of yeast inoculum preparation. J Clin Microbiol 1988;26:1437–1441.
- National Committee for Clinical Laboratory Standards. 1993. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. National Committee for Clinical Laboratory Standards, Villanova, PA.
- National Committee for Clinical Laboratory Standards. 1993. Performance VOL. 41, 1997 antimicrobial susceptibility of Flavobacteria
- Ali HI, Ashida N, Nagamatsu T. Antitumor studies. Part 4: Design, synthesis, antitumor activity, and molecular docking study of novel 2-substituted 2-deoxoflavin-5-oxides, 2-deoxoalloxazine-5-oxides, and their 5-deaza analogs. Bioorg Med Chem 2008;16:922–940.
- Abdel-Aziz AA, ElTahir KE, Asiri YA. Synthesis, anti-inflammatory activity and COX-1/COX-2 inhibition of novel substituted cyclic imides. Part 1: Molecular docking study. Eur J Med Chem 2011;46:1648–1655.
- Naya S, Miyagawa M, Nitta M. Novel photo-induced oxidative cyclization of 1,3-dimethyl-5-(1-arylmethylidene)pyrimidine-2,4,6(1,3,5H)-triones: Synthesis and properties of areno[5,6]pyrano[2,3-d]pyrimidine-2,4(1,3 H)-dionylium ions and their photo-induced autorecycling oxidizing reaction toward some amines. Tetrahedron 2005;61:4919–4930.
- Shi D, Shi J, Rong S. A facile and clean synthesis of pyrimidine derivatives via three-component reaction in aqueous media. Chin J Chem 2010;28:791–796.
- Shul’ts EE, Andreev GN, Shakirov MM, Komarova NI, Bagryanskaya IY, Gatilov YV, Tolstikov GA. Diels-Alder reactions with cyclic sulfones: VIII. Organic catalysis in the synthesis of spiro[1-benzothiophene-4,5′-pyrimidine]-2′,4′,6′-trione 1,1-dioxides and 2′-thioxo-spiro[1-benzothiophene-4,5′-pyrimidine]-4′,6′-dione 1,1-dioxides. Russian Journal of Organic Chemistry 2009; 45:87–101.
- Krasnov KA, Kartsev VG, Gorovoi AS. Chemical modification of plant alkaloids. I. Aminomethylation of barbituric acid derivatives by cytisine. Chemistry of Natural Compounds 2000;36: 192–197.
- Zoorob HH, Abou-Elzahab M, Abdel-Mogib M, Ismail MA. Peculiar reaction behaviour of barbituric acid derivatives towards aromatic amines. Tetrahedron 1996; 52(30):10147–10158.
- Mishra R, Tomar I. Pyrimidine: The Molecule of diverse biological and medicinal importance. International Journal of Pharmaceutical Sciences and Research 2011;2:758–771.
- Cheng CC, Roth B. Recent Progress in the Medicinal Chemistry of 2,4-Diaminopyrimidines. Prog Med Chem 1982;19:269–331.
- Kompis I, Wick A. The Synthesis of 4-Halogen-substituted Analogs of Trimethoprim. Helv Chim Acta 1977;60:3025–3034.
- Bazgir A, Khanaposhtani MM, Soorki AA. One-pot synthesis and antibacterial activities of pyrazolo[4′,3′:5,6]pyrido[2,3-d]pyrimidine-dione derivatives. Bioorg Med Chem Lett 2008;18: 5800–5803.
- Nelson JM, Chiller TM, Powers JH, Angulo FJ. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin Infect Dis 2007;44:977–980.
- Ivanov DV. [Antibiotic susceptibility and molecular mechanisms of cephalosporins resistance in Klebsiella isolates from patients with hospital-acquired infections]. Antibiot Khimioter 2006;51:29–36.
- Schaumann R., Rodloff AC. Activities of quinolones against obligately anaerobic bacteria. Anti-Infective Agents in Medicinal Chemistry 2007;6:49–56.
- Holla BS, Mahalinga M, Karthikeyan MS, Akberali PM, Shetty NS. Synthesis of some novel pyrazolo[3,4-d]pyrimidine derivatives as potential antimicrobial agents. Bioorg Med Chem 2006;14:2040–2047.
- Ramesh E, Manian RD, Raghunathan R, Sainath S, Raghunathan M. Synthesis and antibacterial property of quinolines with potent DNA gyrase activity. Bioorg Med Chem 2009;17:660–666.
- Lafitte D, Lamour V, Tsvetkov PO, Makarov AA, Klich M, Deprez P et al. DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5′-methyl group of the noviose. Biochemistry 2002;41:7217–7223.
![Figure 2. Synthesis of pyrimido[5,4-c]quinoline-2,4-dione derivatives 5a–k.](/cms/asset/654390cd-59a3-45f9-beac-bc1b6703ddac/ienz_a_654113_f0002_b.gif)