Abstract
Twenty-three phenolic compounds were isolated from Dioscorea opposita by bioactivity-guided method and their inhibitory effect against pancreatic lipase was evaluated. A total of 15 isolates reduced lipase activity at IC50 values of less than 50 µM and 3,3′,5-trihydroxy-2′-methoxybibenzyl showed the highest inhibition with an IC50 value of 8.8 µM. This study is a first to reveal the pancreatic lipase inhibitory activity by both D. opposita and its isolated compounds.
Introduction
Lipid digestion is initiated in the stomach and completed in the small intestine by the action of diverse gastrointestinal lipasesCitation1. The human lipases include the pre-duodenal lingual and gastric lipase and the extra-duodenal pancreatic, hepatic, lipoprotein and endothelial lipaseCitation2,Citation3. Of these several lipases, pancreatic lipase is a predominant lipolytic enzyme in humans responsible for the hydrolysis and absorption of 50–70% of total dietary fats in the intestinal lumenCitation2,Citation4. Therefore, pancreatic lipase plays a key role in preventing obesity, and the pancreatic lipase inhibition is a well-known mechanism for the determination of natural products with anti-obesity propertyCitation5.
Phenolic compounds are secondary aromatic plant metabolites possessing one or more hydroxyl substituentsCitation6. Naturally occurring polyphenols have beneficial health-related properties, which are based on their strong antioxidant activitiesCitation6,Citation7. According to recent studies, many polyphenol-rich extracts are effective inhibitors of pancreatic lipase in vitroCitation5,Citation8. Furthermore, phenolics from plant extracts, such as flavonols and tannins, also reduced obesity in vivo together with lipase inhibitionCitation8,Citation9. These results suggest that phenolic materials may be useful to decrease dietary fat absorption and accumulation.
Dioscorea opposita has been cultivated in China, Japan and Korea as a food and used for digestive problems with poor absorption for a long timeCitation10. In the preliminary experiment, the extract of D. opposita Thunb. (Dioscoreaceae) effectively suppressed lipase activity. Phytochemical investigations of D. opposita have revealed many chemical components such as allantoin, batatasin, diosgenin and dioscinCitation11,Citation12. It has also been reported to possess diverse pharmacological properties such as antioxidant, anti-inflammation and anti-diabetic effectsCitation13,Citation14. However, the lipase inhibitory effects of D. opposita and its phenolic components have not been reported previously. This study attempts to isolate and identify the active compounds from D. opposita through bioactivity-guided fractionation and investigate their inhibitory activities against porcine pancreatic lipase in vitro.
Materials and methods
General methods
1H- and 13C-NMR, COSY, HMQC, HMBC and NOESY spectral data were run on Bruker AVANCE 400 and 500 spectrometer (Bruker, Karlsruhe, Germany). EIMS and HREIMS were recorded on JEOL JMS 700 spectrometer (Tokyo, Japan). UV spectra were measured on a Perkin Elmer Lambda 25 UV/Vis spectrophotometer (California). IR spectral data were taken on a JASCO FT/IR-300E spectrophotometer (Tokyo, Japan). CPC was carried using an LLB-M high-performance centrifugal partition chromatography (Sanki Engineering, Kyoto, Japan) with a four-way switching valve operating in either descending or ascending mode together with binary Gilson 321 pump, a Gilson UV/Vis-151 detector, a Gilson FC 203B fraction collector and Rheodyne valve with a 5 mL sample loop for manual injection (Cotati, Sonoma County, CA). A Gilson HPLC system (Gilson, Fairfield, NJ) was used to isolate compounds, and was equipped with two 321 pumps, a UV/Vis-151 detector, an autosampler 234 and a fraction collector 204. Silica gel (230–400 mesh, Merck, Darmstadt, Germany) and Sephadex LH-20 gel (25–100 µm mesh, Pharmacia, Stockholm, Sweden) were used in column chromatography (CC).
Plant material
The rhizomes of D. opposita Thunb. (Dioscoreaceae) were provided by Tong Yang Moolsan Co., Ltd (Nonsan, South Korea) and identified by Prof. Gwang Jin Chang of the Korea National College of Agricultural and Fisheries. A voucher specimen (SNUPH-0822) has been deposited in the Medicinal Herb Garden, Seoul National University.
Extraction and isolation
Fresh rhizomes of D. opposita (19 kg) were sliced into small pieces and freeze-dried prior to extraction. The dried samples were extracted with 95% EtOH (20 L × 3) and evaporated under reduced pressure. The 95% EtOH extract (380 g) was suspended in distilled water (600 mL) and the suspension was partitioned with CHCl3 (1 L), EtOAc (1 L) and n-BuOH (1 L), sequentially.
The active n-BuOH soluble fraction (50 g) was separated by silica gel CC (CH2Cl2:n-hexane (1:1) → CH2Cl2:MeOH (25:1) →CH2Cl2:MeOH (12.5:1) → CH2Cl2:MeOH (6.25:1) → CH2Cl2:MeOH (3:1) → CH2Cl2:MeOH (1:1)) to yield six fractions. Fraction 1 (9.3 g) was subjected to CPC with two-phase solvent system composed of the upper phase of n-hexane:MeOH:water (10:9:1, v/v) as a stationary phase (solvent A) and the lower phase as a mobile phase (solvent B), eluted with the mixture of solvent B and water (0–100% solvent B), to yield 7 subfractions (Frs 1-1–1-7). Fraction 1-2 (587.2 mg) was subjected to Sephadex LH-20 CC eluted with the same solvent of MeOH and 6 subfractions (Frs 1-2-1–1-2-6) were collected.
Fraction 1-2-2 was submitted to RP HPLC (YMC-pack ODS-A C18, S-5 µm, 250 × 10 mm, Kyoto, Japan; eluent, MeCN:H2O (60:40); detection, UV at 254 nm; flow rate, 4 mL/min) to yield compounds 8 (3.5 mg, tR 16.8 min), 10 (4.1 mg, tR 20.5 min) and 6 (5.3 mg, tR 27.1 min). Compounds 15 (2.4 mg, tR 28.5 min), 5 (11.0 mg, tR 40.2 min), 9 (6.9 mg, tR 43.7 min), 13 (1.9 mg, tR 52.5 min) and 20 (2.1 mg, tR 66.0 min) were isolated from fraction 1-2-4 using Phe-Hex HPLC (YMC-pack Ph, S-5 µm, 250 × 10 mm; eluent, MeCN:H2O (50:50); detection, UV at 280 nm; flow rate, 2 mL/min). Fraction 1-2-6 was subjected to Phe-Hex HPLC (YMC-pack Ph, S-5 µm, 250 × 10 mm; eluent, MeCN–H2O (50:50); detection, UV at 280 nm; flow rate, 2 mL/min) to obtain compounds 14 (6.5 mg, tR 20.2 min), 4 (10.6 mg, tR 23.0 min), 11 (2.7 mg, tR 27.5 min) and 16 (1.4 mg, tR 31.0 min).
Fraction 1-4 (420 mg) was subjected to Sephadex LH-20 CC eluted with an isocratic elution of MeOH to yield six subfractions (Frs 1-4-1–1-4-6). Fraction 1-4-2 was subjected to RP HPLC (YMC-pack ODS-H80, S-4 µm, 250 × 20 mm; eluent, MeCN–H2O (80:20); detection, UV at 254 nm; flow rate, 4 mL/min) to yield compounds 12 (11.0 mg, tR 12.1 min) and 7 (3.2 mg, tR 18.1 min). Fraction 1-6 (165 mg) was applied to a Sephadex LH-20 CC, using MeOH, 5 subfractions (Frs 1-6-1–1-6-5) were collected and fraction 1-6-2 yielded compound 19 (14.5 mg, tR 19.8 min) by RP HPLC (Zorbax column Eclipse XDB-C18, S-5 µm, 250 × 9.4 mm, Agilent, CA; eluent, MeCN:H2O (50:50); detection, UV at 210 nm; flow rate, 4 mL/min). Fraction 1-7-2 from the first Sephadex chromatography for fraction 1-7 (230 mg) was purified by RP HPLC (Zorbax column Eclipse XDB-C18, S-5 µm, 250 × 9.4 mm; detection, UV at 210 nm; flow rate, 4 mL/min) eluted with MeCN–H2O (55:45), affording compounds 23 (2.7 mg, tR 20.4 min) and 21 (2.6 mg, tR 24.9 min).
Fraction 2 (15.0 g) was subjected to CPC, eluted with the same solvent mixture as referred to above, to yield seven subfractions (Frs 2-1–2-7). Fraction 2-3 was re-fractionated by CC on Sephadex LH-20 eluted with MeOH to give five fractions as Frs 2-3-1–2-3-5. Fraction 2-3-5 was successively purified by Phe-Hex HPLC (YMC-pack Ph, S-5 µm, 250 × 10 mm; eluent, MeCN:H2O (20:80); detection, UV at 254 nm; flow rate, 2 mL/min) to afford compounds 1 (5.1 mg, tR 25.6 min) and 3 (8.8 mg, tR 30.4 min), respectively. Fraction 2-4 was re-chromatographed on a Sephadex LH-20 as described above. A total of five fractions (Frs 2-4-1–2-4-5) were collected and fraction 2-4-2 was submitted to RP HPLC (YMC J’sphere ODS-H80, S-4 µm, 250 × 20 mm; eluent, MeOH:H2O (90:10); detection, UV at 203 nm; flow rate, 4 mL/min) to provide compound 18 (2.2 mg, tR 15.0 min).
Fraction 2-6 was chromatographed on a Sephadex LH-20 as an eluent of MeOH to give five fraction groups (Frs 2-6-1–2-6-5) in order of elution. Fraction 2-6-4 gave compound 17 (2.2 mg, tR 12.0 min) using RP HPLC (Phenomenex Gemini C18 110A, S-5 µm, 250 × 21.20 mm, Phenomenex, CA; eluent, MeCN–H2O (85:15); detection, UV at 210 nm; flow rate, 5 mL/min). Fraction 2-7 was subjected to Sephadex LH-20 CC (MeOH) to yield six subfractions (Frs 2-7-1–2-7-6). Compounds 2 (4.2 mg, tR 18.3 min) and 22 (6.9 mg, tR 42.0 min) were isolated from fraction 2-7-5 by RP HPLC (YMC-pack Pro C18 RS, S-5 µm, 250 × 20 mm; eluent, MeCN:H2O (30:70); detection, UV at 254 nm; flow rate, 4 mL/min). The purities of isolated compounds were evaluated as over 98% by HPLC method, and they were used as test samples in this study.
In vitro pancreatic lipase assay
Pancreatic lipase (type II, from porcine pancreas, Sigma-Aldrich, St. Louis, MO) activity was determined using 4-methylumbelliferyl oleate (4-MU oleate, Sigma-Aldrich) as a substrateCitation15,Citation16. Various concentrations of test compound (5, 10, 25, 50 and 100 µM) were dissolved in DMSO (final culture concentration 0.1%) and 4-MU oleate and lipase were diluted in 0.1 M McIlvane buffer containing 0.1 M citric acid–Na2HPO4 (pH 7.4). The mixed solution that composed of 100 µL of 0.1 mM 4-MU oleate, 40 µL of the above-mentioned buffer and 10 µL of test samples was prepared before adding lipase. The enzymatic reaction was started by adding 0.05 mL of 1 U/mL pancreatic lipase and incubation was carried out for 20 min at 37 °C. The amount of 4-methylumbelliferon released by the enzyme was monitored at an emission of 450 nm and excitation wavelength of 320 nm using a fluorescence spectrophotometer. The inhibition activity was calculated using the following formula:
where A0 is the absorbance of the control without inhibitor, A1 the absorbance in the presence of the test sample and A2 the absorbance sample blank (without porcine pancreatic lipase). IC50 was determined to be the sample concentration decreasing the initial enzymatic activity by 50% of under the experimental conditions given. The value was obtained to the linear portion of the log concentration–response curves by the least square regression method.
The Ki value was obtained from the Cheng–Prusoff equationCitation17:
where Ki is the dissociation constant, Km the Michaelis constant and [S] the concentration of the substrate. The Km constant was determined using the Lineweaver–Burk plots from the relevant Michaelis–Menten equations.
Statistical analysis
Data are expressed as the mean ± SD. The values were expressed as percent changes from the mean value of the control experiment. Statistical analyses were performed by a one-way analysis of variance using statistical package. P values less than 0.05 were considered statistically significant.
Results and discussion
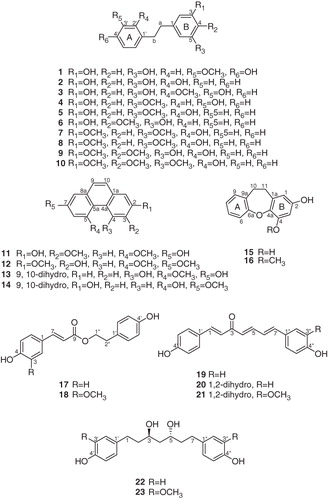
The n-BuOH fraction from 95% EtOH extract of D. opposita appeared the lipase inhibition (57% at final concentration of 10 µg/mL) and was further partitioned on the basis of polarity to find the active fractions. Of the six subfractions, fractions 1 and 2 were most active against pancreatic lipase, implying that they might have active components with lipase inhibitory activity ().
Figure 1. Effect of D. opposita on pancreatic lipase in the presence or absence of samples at the final concentration of 10 µg/mL. Inhibition of lipase by 95% EtOH extract and fractions of D. opposita (A) and the subfractions of D. opposita n-BuOH fraction (B). Each values are expressed as the % control means ± SD of triplicate experiments; *p < 0.05, **p < 0.01 and ***p < 0.001 compared with the control.
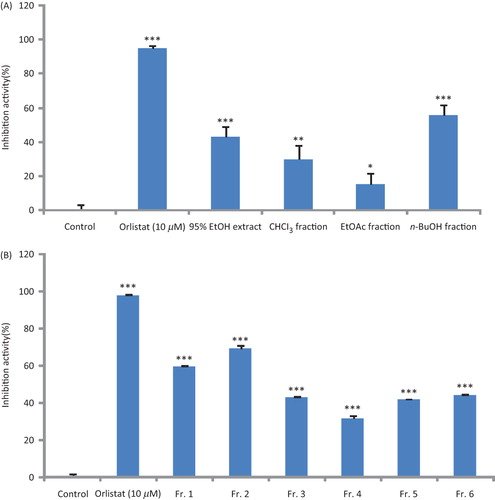
Bioassay-guided fractionation of D. opposita extract led to the isolation and identification of 23 phenolic compounds including 10 dihydrostilbenes, tristinCitation18 (1), 2′,3,5-trihydroxybibenzylCitation19 (2), 3,3′,5-trihydroxy-2′-methoxybibenzylCitation14 (3), batatasin IIICitation20 (4), batatasin IVCitation19 (5), 2′,4-dihydroxy-3,5-dihydroxy-4-methoxybibenzylCitation14 (6), 3,5-dimethoxy-2′-hydroxybibenzylCitation21 (7), 3,5-dimethoxybibenzylCitation22 (8), 3,4-dimethoxy-2′,5-dihydroxybibenzylCitation23 (9), batatasin VCitation24 (10), four phenanthrenes, 3,5-dimethoxy-2,7-phenanthrenediolCitation25 (11), batatasin ICitation26 (12), hircinolCitation26 (13) and 2,5-dihydroxy-7-methoxy-9,10-dihydrophenanthreneCitation27 (14), two dibenzoxepins, 9,10-dihydro-dibenzoxepin-2,4-diolCitation14 (15) and 9,10-dihydro-4-methoxy-dibenzoxepin-2-olCitation14 (16), five diarylheptanoids, p-hydroxyphenylethyl p-coumarateCitation28 (17), p-hydroxyphenethyl trans-ferulateCitation29 (18), (1E,4E,6E)-1,7-bis(4-hydroxyphenyl)-1,4,6-heptatrien-3-oneCitation30 (19), (4E,6E)-1,7-bis(4-hydroxyphenyl)-4,6-heptadien-3-oneCitation31 (20) and (4E,6E)-7-(4-hydroxy-3-methoxyphenyl)-1-(4-hydroxyphenyl)-4,6-heptadien-3-oneCitation32 (21), two p-hydroxyphenylethyl-p-hydroxyphenyl propenoic acids, (3R,5R)-3,5-dihydroxy-1,7-bis(4-hydroxyphenyl)-3,5-heptanediolCitation33 (22) and (3R,5R)-1,7-bis(4-hydroxy-3-methoxyphenyl)-3,5-heptanediolCitation33 (23) by spectroscopic methods using 2D-NMR ().
All the isolates obtained in this investigation were evaluated for their ability decreasing lipase activity against the porcine pancreatic lipase. Fifteen compounds from the active subfractions of D. opposita extract revealed in vitro lipase inhibitions at the IC50 values of less than 50 µM (). In particular, 3,3′,5-trihydroxy-2′-methoxybibenzyl (3) and (4E,6E)-1,7-bis(4-hydroxyphenyl)-4,6-heptadien-3-one (20) effectively reduced lipase activity with the IC50 of less than 10 µM and dose-dependently in the concentration range 5–100 µM (). Of the 15 active constituents, 8 contain dihydrostilbene skeleton, more commonly known as bibenzyl, and 3,3′,5-trihydroxy-2′-methoxybibenzyl showed the highest inhibition activity with an IC50 value of 8.77 ± 2.03 µM (p < 0.05) and Ki value of 1.82 ± 0.43 µM. These results clearly suggest that phenolic compounds with stilbenoid are considered to play important roles in the lipase inhibition of the D. opposita extract.
Figure 3. A dose-dependent inhibition of lipase activity by 3,3′,5-trihydroxy-2′-methoxybibenzyl (A), (4E,6E)-1,7-bis(4-hydroxyphenyl)-4,6-heptadien-3-one (B) and Orlistat (C). Values are means of triplicate assays ± standard error.
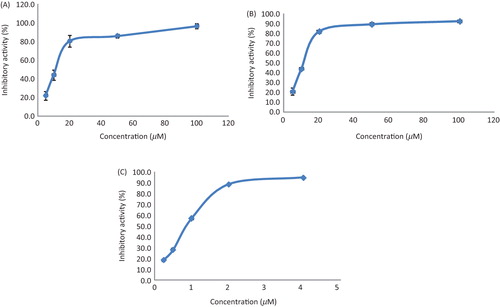
Table 1. Inhibitory effects of compounds isolated from D. opposita against porcine pancreatic lipase†.
Compounds 1–6 with a hydroxyl group of C-3 were more potent than 7–10 which have the methoxy group at the same position. Especially, the stilbenoids possessing 3,5-dihydroxybibenzyl moiety showed higher inhibitory potencies than the others. The lipase inhibitory activities of stilbenoids depend on the presence of the hydroxyl group in the C-3 position. The structural difference also influences the inhibitory effect of diarylheptanoids on pancreatic lipase. Three diarylheptanoids (19–21) with the carbonyl group reduced pancreatic lipase activity at low concentrations but C-3 hydroxylation (22 and 23) had no effect on the lipase inhibition. This indicates that the carbonyl group at C-3 contributed to the inhibitory activity of diarylheptanoids against pancreatic lipase.
To date, there were a few attempts to reveal the pancreatic lipase inhibition for Dioscorea species and saponin-type constituents such as dioscin and diosgenin were found to be lipase inhibitors as Dioscorea secondary metabolitesCitation16. There has been no previous research associated with the beneficial effects of phenolics from the D. opposita extract against pancreatic lipase. Current data suggested that the phenolic constituents from D. opposita were effective in inhibiting pancreatic lipase in vitro. Based on the results of all isolates, dihydrostilbene, phenanthrene and diarylheptanoid structures were deemed to be responsible for lipase inhibition. Particularly, 3,3′,5-trihydroxy-2′-methoxybibenzyl including dihydrostilbene moiety exhibited the most potent inhibition as a new lipase inhibitor.
Declaration of interest
The authors declare no conflict of interest.
References
- Shi Y, Burn P. Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat Rev Drug Discov 2004;3:695–710
- Verger R. Pancreatic lipase. In: Borgstrom B, Brockman HL, eds. Lipase. Amsterdam (The Netherlands): Elsevier; 1984:83–105
- Ramírez M, Amatea L, Gilb A. Absorption and distribution of dietary fatty acids from different sources. Early Hum Dev 2001;65:S95–101
- Embleton JK, Pouton CW. Structure and function of gastrointestinal lipases. Adv Drug Deliv Rev 1997;25:15–32
- Birari RB, Bhutani KK. Pancreatic lipase inhibitors from natural sources: unexplored potential. Drug Discov Today 2007;12:879–89
- Harborne JB. Methods in plant biochemistry. Vol. 1: Plant phenolics. London: Academic Press; 1989:1–28
- Gülçin İ. Antioxidant activity of food constituents: an overview. Arch Toxicol 2012;86:345–91
- McDougall GJ, Kulkarni NN, Stewart D. Berry polyphenols inhibit pancreatic lipase activity in vitro. Food Chem 2009;115:193–9
- Kurihara H, Shibata H, Fukui Y, et al. Evaluation of the hypolipemic property of Camellia sinensis var. ptilophylla on postprandial hypertriglyceridemia. J Agric Food Chem 2006;54:4977–81
- Chen HL, Wang CH, Chang CT, Wang TC. Effects of Taiwaness yam (Dioscorea japonica Thunb var. pseudojaponica Yamamoto) on upper gut function and lipid metabolism in Balb/c mice. Nutrition 2003;19:646–51
- Ireland CR, Schwabe WW, Coursey DG. The occurrence of batatasins in the Dioscoreaceae. Phytochemistry 1981;20:1569–71
- Ofelia E, Jesús CL, Helgi J, Francisco G. Spirostanic diosgenin precursors from Dioscorea composita tubers. Phytochemistry 1982;21:413–16
- Gao X, Li B, Jiang H, et al. Dioscorea opposita reverses dexamethasone induced insulin resistance. Fitoterapia 2007;78:12–15
- Yang MH, Yoon KD, Chin YW, et al. Phenolic compounds with radical scavenging and cyclooxygenase-2 inhibitory activities from Dioscorea opposita. Bioorg Med Chem 2009;17:2689–94
- Kawaguchi K, Mizuno T, Aida K, Uchino K. Hesperidin as an inhibitor of lipases from porcine pancreas and Pseudomonas. Biosci Biotechnol Biochem 1997;61:102–4
- Kwon C-S, Sohn HY, Kim SH, et al. Anti-obesity effect of Dioscorea nipponica Makino with lipase-inhibitory activity in rodents. Biosci Biotechnol Biochem 2003;67:1451–6
- Cheng YC, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 1973;22:3099–108
- Leong YW, Harrison L, Powell AD. Phenanthrene and other aromatic constituents of Bulbophyllum vaginatum. Phytochemistry 1999;50:1237–41
- Fagboun DE, Ogundana SK, Adesanya SA, Roberts MF. Dihydrostilbene phytoalexins from Dioscorea rotundata. Phytochemistry 1987;26:3187–9
- Majumder PL, Guha S, Sen S. Bibenzyl derivatives from the orchid Dendrobium amoenum. Phytochemistry 1999;52:1365–9
- Takasugi M, Kawashima S, Monde K, et al. Antifungal compounds from Dioscorea batatas inoculated with Pseudomonas cichorii. Phytochemistry 1987;26:371–5
- Kaganda NG, Adesanya SA. A new dihydrostilbene from diseased Dioscorea mangenotiana. J Nat Prod 1990;53:1345–6
- Jarevang T, Nilsson MC, Wallstedt A, et al. A bibenzyl from Empetrum nigrum. Phytochemistry 1998;48:893–6
- Hashimoto T, Tajima M. Structures and synthesis of the growth inhibitors batatasins IV and V, and their physiological activities. Phytochemistry 1978;17:1179–84
- Leong YW, Kang CC, Harrison LJ, Powell AD. Phenanthrenes, dihydrophenanthrenes and bibenzyls from the orchid Bulbophyllum vaginatum. Phytochemistry 1997;44:157–65
- Coxon DT, Ogundana SK, Dennis C. Antifungal phenanthrenes on yam tubers. Phytochemistry 1982;21:1389–92
- Takagi S, Yamaki M, Inoue K. Antimicrobial agents from Bletilla striata. Phytochemistry 1983;22:1011–15
- Kaewamatawong R, Ruangrungsi N, Likhitwitayawuid K. Chemical constituents of Polyalthia parviflora stem. J Nat Med 2007;61:349–50
- Darwish FMM, Reinecke MG. Ecdysteroids and other constituents from Sida spinosa L. Phytochemistry 2003;62:1179–84
- Nakayama R, Tamura Y, Yamanaka H, et al. Two curcuminoid pigments from Curcuma domestica. Phytochemistry 1993;33:501–2
- Ali MS, Tezuka Y, Awale S, et al. Six new diarylheptanoids from the seeds of Alpinia blepharocalyx. J Nat Prod 2001;64:289–93
- Moon SS, Cho SC, Lee JY. Tsaokoarylone, a cytotoxic diarylheptanoid from Amomum tsao-ko fruits. Bull Korean Chem Soc 2005;26:447–50
- Yokosuka A, Mimaki Y, Sakagami H, Sashida Y. New diarylheptanoids and diarylhelptanoid glucosides from the rhizomes of Tacca chantrieri and their cytotoxic activity. J Nat Prod 2002;65:283–9