Abstract
Cucurbitacins (Cucs) have been classified as signal transducer and activator of transcription 3 inhibitors. Kinase inhibition has been a validated drug target in multiple types of malignancies. B-RAF mutations are highly expressed in the melanoma. Our hypothesis is the Cucs can be a potential candidate to inhibit the signaling kinase pathway. The research presented is the evaluation of Cucs, as B-RAF and MEK1 kinase inhibitors. Virtual screening methods were employed to identify lead compounds. The hypothesis was tested on mutant B-RAF cell lines, A-375 and Sk-Mel-28 cell lines to determine the activity toward melanoma. A series of natural Cucs show an improved activity toward Sk-Mel-28 and A-375 cell lines. Cucs show potential inhibition for the total and phosphorylated ERK using ELISA kits. Cucs could be potential candidate for inhibiting cell growth.
Introduction
Natural products represent an important source for drug discovery. Natural products are considered to be a significant source for new drugs over 30 yearsCitation1. Typically, natural products can be used directly or indirectly via semi-synthetic structural modifications to their structural skeletons to potentiate their activity toward treatment and solving of different clinical problems. Natural products were found out to be potential drug candidates as anti-hypertensive, anti-diabetics, anti-bacterial, anti-fungal, anti-microbial and anti-cancer agentsCitation1.
In this paper, we describe the evaluation of cucurbitacins (Cucs) analogs as kinase inhibitors for treatment of malignant melanoma. The choice to study was based on previous knowledge of the Cuc’s biological activityCitation2–6. The molecular targets were also chosen based on previous knowledge of the Cucs’ activity as well as similar pharmacophores between Cucs and the receptors.
Cancers
Melanoma is the most lethal form of skin cancer. About 1.3 million Americans are diagnosed with skin cancer each year, and melanoma accounts for about 4% of these cases. It has increased in incidence and mortality over the past three decades. Standard chemotherapy produces response rates on the order of 10%Citation7,Citation8. Melanoma is a kind of skin cancer that forms in the melanocytes, the pigmenting cells that give skin its color. The Skin Cancer Foundation reports more than 68 000 cases of melanoma were diagnosed in 2010. Of all the skin cancers, melanoma is the most deadly, causing more than 75% of all skin cancer deaths. It is not fully clear how all melanomas develop, but exposure to ultraviolet radiation plays a pivotal role, especially in fair-skinned people.
Cucurbitacins
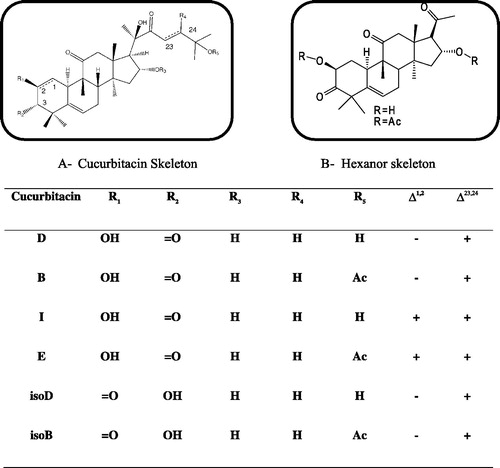
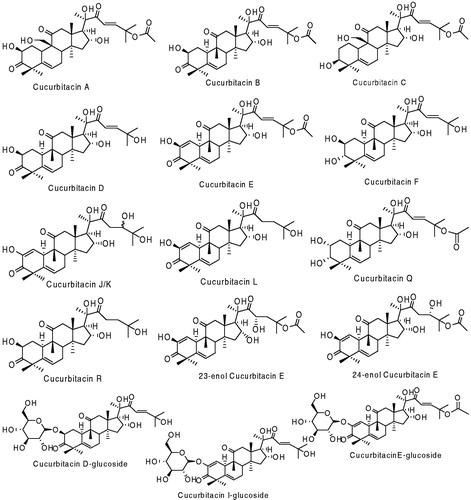
Cucs are highly oxygenated tetracyclic triterpenes that are mainly found in the Cucurbitaceae family, e.g. squash, pumpkin, cucumber and melons. Plants in which the Cucs are found have been used for centuries in various folk medicinesCitation2–4. There have been 19 different Cucs extracted and characterized from plants, as shown in Citation5.
Various biological activities have been associated with Cucs and their glycosyl derivativesCitation6,Citation9. Cucs E, Q, B and D exhibit anti-proliferative activity on various cancer cell lines, Cuc R shows an anti-inflammatory activity and Cuc B has also shown hepatocurative and hepatoprotective effectsCitation9,Citation10. Research conducted by the Halaweish lab employing a semi-synthetic approach coupled with in vitro testing and quantitative structure activity relationships indicated that cytotoxicity could be altered with slight modifications to the Cuc skeletonCitation11. The study also concluded the hepatoprotective activity as well as anti-proliferative activity could be optimized through generation of analogs. A strong correlation between different types of Cucs and melanoma has also been reportedCitation12. Recently, Chan et al.Citation13 reported the inhibitory effect of Cuc B on STAT3 and Ras/Raf/MEK/ERK signaling pathwayCitation13.
Cucs as signal transducer and activator of transcription 3 inhibitors
Cucs have arguably become best known for their ability to target Janus kinase/signal transducer and activator of transcription 3 (JAK/STAT3). The Cucs have been shown to inhibit this pathway in multiple studiesCitation14,Citation15. STATs are signaling proteins that work to provide a signal from the cell surface proteins to the cytoplamsic proteins as well as translocate into the nucleus in order to initiate DNA synthesis. The STATs are also responsible for the regulation of genesCitation16–20. The STATs function via tyrosine phosphorylation from a number of cytokines and growth factors. Because of their regulation of gene expression and involvement in DNA synthesis, they are an obvious target for the treatment of cancer. Niu et al. and Catlett-Falcone et al. have shown evidence of blocking the phosphorylation of STAT3 reduces tumor growth via apoptosis with very little effect on the normal cells Citation21,Citation22. Due to the fact that Cucs are known to inhibit STAT3 tyrosine phosphorylation and that the Cucs activity can be altered through varying structural motifs, we have proposed that Cucs could be developed to affect multiple targets as tyrosine kinase inhibitors, both selectively and as a multi-faceted drug approach. With the knowledge of the Cucs varying activity and multiple targets, it is conceivable that Cucs could be designed as drug candidate targeting kinases.
Mitogen-activated protein kinase pathway as a target for the potential treatment of melanoma
Mitogen-activated protein kinase (MAPK) pathway is one of the kinase receptor pathways that have been heavily studied for the treatment of melanoma due to its major contribution in multiple cellular processes such as proliferation, differentiation, survival and apoptosisCitation22. MAPK pathway consists of the Ras/B-Raf/MEK/ERK signaling cascadeCitation23. It is stimulated at the cell membrane, either by receptor tyrosine kinase binding its respective ligand or integrin adhesion to the extracellular matrix, which transmits activation signals via the RAS GTPase on the inner surface of cell membraneCitation23. The binding of RAS to GTP leads to the subsequent recruitment of the serine/threonine kinase RAF to be translocated to the membraneCitation23. Once it becomes activated, it then phosphorylates the dual specificity kinase MEK restricting inactive ERK to the cytosolCitation23. This signaling leads to proliferation and differentiationCitation23. RAF is the primary link between RAS and the MAPK pathway. Its activation is necessary and sufficient to activate the cascade. Activated RAF, in turn, phosphorylates and activates MAPK/ERK kinase (MEK 1/2)Citation24. The RAF family includes three proteins: A-Raf, B-Raf and C-Raf.
B-Raf is a Ras-activated serine/threonine protein kinase that has been studied for its relationship between mutations and various types of cancer for the development of potential drug candidates. B-Raf mutations show high incidence, about 66%, in melanoma. The most common mutation (∼90%) is the change of Val600 to Glu in the activation loopCitation25–27. This mutation keeps the kinase pathway constitutively active leading to over-growth of the cells. In light of this, inhibition of the MAPK signaling cascade at the level of B-Raf can provide an approach toward the treatment of melanoma. Also, inhibition of MEK, the direct substrate of RAF, has been demonstrated preclinically to induce apoptosis causing inhibition of tumor growthCitation28. Currently, there are a number of drugs being investigated in clinical trials that target the MAPK pathway at different levels working via competitive and non-competitive ATP inhibition. PD0325901, Pfizer, is a non-competitive ATP MEK inhibitor with high activity on melanoma cell linesCitation27–29.
Cucs as kinase inhibitors
Kinase inhibition at different levels can be a platform toward development of drugs for treatment of melanoma. It is hypothesized that the Cucs’ overall rigid structure may play a role in surpassing current kinase inhibitors, which are generally much more flexible than the Cucs. The rigid backbone of the structure along with flexible side-chain, which can be viable for synthetic transformation, allows for versatility while maintaining the appropriate lipophilicity for a small molecule kinase inhibitor. Therefore, Cucs can be potential candidates targeting MAPK pathway at different levels inhibiting cell growth.
Materials and methods
Molecular modeling
All calculations were performed on a Gateway computer running windows XP with a 2.13 Intel Core2 CPU and 1.97 GB RAM. All OpenEye software uses the MMF94 force field.
Molecular docking calculations
A virtual library of 900 analogs including Cucs, Cuc glycosides, hexanor structures and known B-RAF as well as MEK inhibitors were prepared and minimized using Chem Office 2004 with PM3 minimization along with the standard drugs of choice for the treatment of melanoma. These files were then converted into .pdb files maintaining all heavy atoms. Each .pdb file was concatenated into one continuous .pdb file to be used as an input for Omega. The Omega utility creates multiple conformations for each small molecule in the library in order to induce ligand flexibility in an otherwise rigid model. The receptor Protein Data Bank (PDB) file was taken from the PDB website (MEK1 PDB ID 3EQC, Raf PDB ID 3OG7) and was prepared using OpenEye’s Fred Receptor program. This program defines the space in which the search algorithm will perform and also defines the shape potentials for calculation. The shape potentials define the space in which the ligand must sit to avoid heavy atom clashes while maintaining a close enough proximity to allow for appropriate interactions. Finally, both the ligand input file and the receptor input file were passed into FRED to perform the molecular docking simulations. Multiple scoring functions were employed in order to obtain a consensus structure and score in the final output. The scoring functions include shapegauss, chemgauss3, Oechemscore, Screenscore and PLP. See OpenEye’s FRED manual for details of each scoring function (www.eyesopen.com/products). Snap shots could be obtained using VIDA application showing the main interaction forces between the Cucs and the receptor of interest.
Absorption, distribution, metabolism and elimination calculations
To determine whether or not the compounds in the virtual library pass certain drug like criteria, OpenEye’s Filter program was employed. Filter’s default settings fit into three different categories: physical properties, atomic and functional group content and molecular graph topology. Physical properties include molecular weight, topological polar surface area, LogP and aqueous solubility. Atomic functional group content addresses absolute and relative amount of heteroatoms and limits on the number of wide variety of functional groups. Graph topology filters address the number and size of ring systems, flexibility of the molecule and the size and shape of non-ring chains.
Isolation of Cucs
The isolation of Cucs was performed as described by Bartalis and HalaweishCitation11. Briefly, the ripe fruits from Cucurbita andreana were cut into small pieces and homogenized in methanol for 5 min. The homogenate was filtered through cheesecloth and the insoluble residue was re-homogenized in methanol and re-filtered. The organic solvent was removed on the rotoevaporator and the aqueous phase was freeze-dried. The freeze-dried extracts were then purified using flash column chromatography with a gradient elution. Fractions of 250 mL were collected and analyzed using TLC. Similar fractions were pooled together and final purification was performed using RP-HPLC and preparative TLC. Cucs B, D and E were isolated and fully structurally elucidated using NMR and high-resolution mass spectroscopy.
Cell viability assay
In order to determine the IC50 of the Cucs and analogs, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was employed. The Cuc analogs that were investigated were based on the results of the molecular docking study. PD-0325901 (MW 482.19; C16H14F3IN2O4) were used as a standard for melanoma. All compounds were dissolved in 1% DMSO.
Sk-Mel-28 and A-375 cells were purchased from the American Type Culture Collection to study melanoma. Monolayer cultures of Sk-Mel-28/A-375 cells were grown in Eagle’s minimum essential medium/Dulbecco’s modified Eagle medium supplemented with 10% FBS and penicillin-G and incubated at 37 °C and 5% CO2. Cells were seeded in 96-well plates at 30 000 cells/well and incubated for 24 h. Compounds were added to a specified final concentration (100, 20, 4, 0.8 and 0.16 μM) through five-serial dilution. Finally, the cells were treated with 5 mg/mL MTT in PBS (Sigma–Aldrich) for four hours and lysed with 0.01N HCl containing 10% sodium dodecyl sulfate. Plates were read on a Biotech plate reader at 570 nm for four hours after lysingCitation30. All experiments were performed in triplicate. The inhibitory concentration at 50% (IC50) was determined from the exponential curve of viability versus concentration. The viability was calculated by
where Adrug is the absorbance at 570 nm for the drug, ANC the absorbance for the negative control (no cells) and APC the absorbance of positive control (cells with no drug).
ERK1/2 in cell-based ELISA kit
Target specific in cell-based ELISA kits are complete and effective cell-based assays for identification of specific proteins in the cells, where it provides better data and higher throughput than quantitative western blotting. Each kit contains the detection reagents, specific antibodies and buffers. The assay is performed in 96-well plates. Moreover, the assay is amenable to automation and statistical analysis for drug screening.
Pierce ERK1/2 Colorimetric In-Cell ELISA Kit, purchased from Thermo scientific, including buffers, target-specific primary antibodies (anti-ERK 1/2 (Thr202/Tyr204) antibody and anti-ERK1/2 antibody) and a horseradish peroxidase (HRP) conjugated detection reagent.
The colorimetric methods allow multiple targets to be analyzed in a single experiment when using the colorimetric detection method; different proteins are measured in separate wells at 450 nm and then the data is normalized by the cell number in each well using whole cell stain Janus Green at 615 nm.
A-375 cells were seeded in 96-well plates at 20 000 cells/well in the corresponding media. The cells were incubated at 37 °C for 24 h. The cells were treated with the IC50 doses of Cuc B, D, E and PD0325901 for 10 and 30 min then stimulated with 100 ng/mL of epidermal growth factor (EGF) prior to the fixation of the cells using 4% formaldehyde. Followed by, addition of permeabilization buffer, then quenching solution and blocking buffer before the addition of the primary antibody to be incubated overnight. Then, HRP conjugate was added followed by 3, 3′, 5, 5′ tetramethylbenzidine (TMB) substrate and TMB stop solution. Absorbance was measured using ELISA plate reader at 450 nm and then the data was normalized by staining the whole cell using Janus Green to be measured again at 615 nm.
Results and discussions
Absorption, distribution, metabolism and elimination results
The filter study revealed that standard Cucs had passed the criteria set for the filter study according to Lipinski’s rule of five including log P, number of hydrogen bond acceptors, hydrogen bond donors, number of heavy atoms, number of heteroatoms and solubility parametersCitation31. This indicates that the proposed analogs satisfy the absorption, distribution, metabolism and elimination requirements using the computational chemistry approach.
Flexible molecular docking results
Molecular docking results revealed that the Cucs show a significant binding toward the crystal structure of RAF and MEK compared to the standard B-RAF and MEK inhibitors. Where, the two-dimensional chemical structures of the top scoring analogs are listed in .
As shown in , Cuc B and E exhibit stronger binding affinity via filling the entire hydrophobic pocket of the RAF receptor compared to PD0325901 via hydrophobic–hydrophobic interactions.
Figure 3. (A) Overlay of Cuc B (Green) and PD0325901 (Red) at B-RAF through hydrophobic–hydrophobic interactions predominates; (B) Overlay of Cuc B (Green) and Cuc E (yellow) along with B-RAF, showing the potential of the Cucs to bind at the same binding pocket of PD0325901.
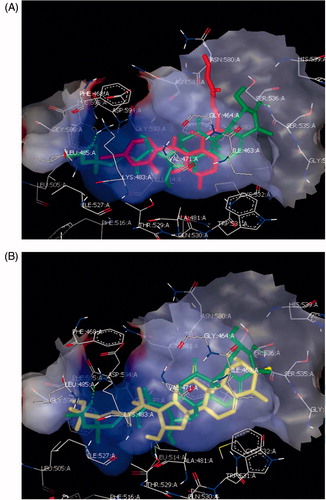
In addition to formation of two hydrogen bonds formed by the two carbonyl groups at C-2 and C-22 along with Ser 536 and Lys 483, respectively, as shown in the supporting information.
MEK has two hydrophobic pockets, one where ATP and the other one is allosteric site where PD0325901 binds giving the inhibitory activity via non-ATP competitive inhibition mechanismCitation28,Citation29,Citation32. This study shows that Cucs exhibit stronger binding affinity to an allosteric site adjacent to the ATP binding region via hydrophobic interactions compared to PD0325901 as shown in the supporting information. However, other data suggests on a slight change on the Cuc skeleton will change its conformation to bind toward the ATP binding pocket of the receptor.
Cell viability results
The results from the cell viability, MTT assay show a clear cytotoxicity can be seen for the natural Cucs. They exhibit a much larger cytotoxic effect on the Sk-Mel-28 cell line than on the A-375 cell line. This can be due to the expression of different isozymes in both of the cell lines, where Sk-Mel-28 has other isozymes such as glucose-6-phosphate dehydrogenase and adenyl kinase-1Citation33. Both experiments were run in comparison with PD0325901 as a standard MEK inhibitor for treatment of melanomaCitation27. The data show the potential of the Cucs to inhibit cell growth and induce cytoxicity, as presented in .
Table 1. Cell viability data showing IC50 (the half maximal (50%) IC of a substance).
ELISA assay data
EGF was used to stimulate the induction of ERK in the A-375 cell lines, MAPK and MEK inhibitors are typically evaluated using ELISA techniquesCitation34. The data show the ability of the Cucs to inhibit total and phosphorylated ERK compared to the stimulation induced by EGF, where Cuc E shows up to twofold decrease in the signaling induced by EGF in case of the total ERK and phosphorylated ERK as shown in the supporting information. The data show the potential activity for the Cucs to inhibit the MAPK kinase pathway by inhibiting total and phosphorylated ERK. Furthermore, the ability of the Cucs to inhibit the total ERK can validate the ability of the Cucs to binds toward earlier stages at the MAPK pathway either at the RAF or the MEK to inhibit the cascade either upstream or downstream. This data suggests the ability of the Cucs to bind to the ERK to be well correlated along with the docking study and the cell study, where better binding affinity of the Cucs toward the receptors will enhance the cytotoxic activity of the Cucs over the cell lines as well as the proper binding toward ERK. This is the first report for the Cucs on A-375 cell lines using in cell-based ELISA kit to monitor its effect on ERK.
Conclusions
The docking study of B-Raf and MEK revealed that the native Cucs score better than PD032590. This indicates that the binding mode may be more critical for improving the activity on this receptor. The IC50 values indicate that the compounds binding in the same pocket as PD0325901 will have greater activity. Moreover, in cell-based ELISA, data revealed the potential inhibitory effect of Cucs toward the total and the phosphorylated ERK, inhibiting the whole MAPK pathway cascade. These findings can be a viable route toward further investigations for structural modification of the Cucs skeleton and validates the potential of the Cucs as a drug candidate is high.
Declaration of interest
The authors report no declaration of interest.
The authors thank the SD Board of Regents 2010 Center for Biological Control and Analysis by Applied Photonics (BCAAP) for funding this work.
Acknowledgements
The authors thank the OpenEye molecular modeling software for supporting an academic license.
References
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 2012;75:311−35
- Spits-Tanin T, Grossman S, Dovrat S, et al. Growth inhibitory activity of cucurbitacin glucosides isolated from Citrillus colocynthis on human breast cancer cells. Biochem Pharmacol 2007;73:56–67
- Lavie D, Glotter E. The cucurbitacins, a group of tetracyclic triterpenes. In: Herz W, Grisebach H, Kirby GW, eds. Progress in the chemistry of organic natural products. New York: Springer-Verlag; 1971:307–62
- Miro M. Cucurbitacins and their pharmacological effects. Phytother Res 1995;9:159–68
- Chen JC, Chiu MH, Nie RL, et al. Cucurbitacins and cucurbitane glucosides. Structure and biological activities. Nat Prod Rep 2005;22:386–99
- Huang Y, de Bruyne T, Apers S, et al. Complement-inhibiting cucurbitacin glycosides from Picriea fel-terrae. J Nat Prod 1998;61:757–61
- Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev 2006;20:2149–82
- Petermann KB, Rozenberg GI, Zedek D, et al. CD200 is induced by ERK and is a potential therapeutic target in melanoma. J Clin Invest 2007;12:3922–7
- Sun J, Blaskovich MA, Jove R, et al. Cucurbitacin Q: a selective STAT3 activation inhibitor with potent anti tumor activity. Oncogene 2005;24:3236–45
- Bernard SA, Olayinka OA. Search for a novel antioxidant, anti-inflammatory/analgesic or anti-proliferative drug: cucurbitacins hold the ace. J Med Plants Res 2010;4:2821–6
- Bartallis J, Halaweish FT. In vitro and QSAR studies of cucurbitacins on HepG2 and HSC-T6 liver cell lines. Bioorg Med Chem 2011;19:2757–66
- Oh H, Mun Y-J, Im S-J, et al. Cucurbitacins from Trichosanthes kirilowii as the inhibitory components on tyrosinase activity and melanin synthesis of B16/F10 melanoma cells. Planta Med 2002;68:832–3
- Chan KT, Li K, Liu SL, et al. Cucurbitacin B inhibits STAT3 and the Raf/MEK/ERK pathway in leukemia cell line K562. Cancer Lett 2010;289:46–52
- Blaskovich MA, Sun J, Cantor A, et al. Discover of JIS-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res 2003;63:1270–9
- Fujita M, Zhu X, Sasaki K, et al. Inhibition of STAT3 promotes the efficacy of adoptive transfer therapy using type-1 CTLs by modulation of the immunological microenvironment in a murine intracranial glioma. J Immunol 2008;180:2089–98
- Sun J, Blaskovich MA, Jove R, et al. Cucurbitacin Q: a selective STAT3 activation inhibitor with potent antitumor activity. Oncogene 2005;24:3236–45
- Stark GR, Kerr IM, Williams BR, et al. How cells respond to interferons. Annu Rev Biochem 1998;67:227–64
- Horvath CM, Darnell JE. The state of the STATs: recent developments in the study of signal transduction to the nucleus. Curr Opin Cell Biol 1997;9:233–9
- Ihle JN, Kerr IM. JAKs and STATs in signaling by the cytokine receptor super family. Trends Genet 1995;11:69–74
- Schindler C, Darnell, JE Jr. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem 1995;64:621–51
- Niu G, Heller R, Catlett-Falcone R, et al. Gene therapy with dominant-negative STAT3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res 1999;59:5059–63
- Catlett-Falcone R, Landowski TH, Oshiro MM, et al. Constitutive activation of STAT3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 1999;10:105–15
- Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol 2007;25:1606–20
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54
- Pollock PM, Meltzer PS. A genome-based strategy uncovers frequent BRAF mutations in melanoma. Cancer Cell 2002;2:5–7
- Thompson N, Lyons J. Recent progress in targeting the Raf/MEK/ERK pathway in cancer drug discovery. Curr Opin Pharmacol 2005;5:350–6
- Ciuffreda L, Bufalo DD, Desideri M, et al. Growth-inhibitory and antiangiogenic the MEK inhibitor PD0325901 in malignant melanoma with or without BRAF mutations. Neoplasia 2009;8:720–31
- Wang JY, Wilcoxen KM, Nomoto K, Wu S. Recent advances of MEK inhibitors and their clinical progress. Curr Top Med Chem 2007;7:1364–78
- Noble MEM, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science 2004;303:1800–5
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55–63
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3–26
- García-Echeverría C. Protein and lipid kinase inhibitors as targeted anticancer agents of the Ras/Raf/MEK and PI3K/PKB pathways. Purinergic Signal 2009;5:117–25
- Pollack MS, Heagney SD, Livingston PO, Fogh J. HLA-A,B,C and DR alloantigen expression on forty-six cultured human tumor cell lines. J Natl Cancer Inst 1981;66:1003–12
- Iverson C, Larson G, Lai C, et al. RDEA119/BAY 869766: a potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer. Cancer Res 2009;17:6829–47