Abstract
Background: Auto-immunity may contribute to the pathogenesis of chronic obstructive pulmonary disease (COPD), particularly to the presence of emphysema. Auto-immune diseases are characterized by an abnormal distribution of HLA class II alleles (DR and DQ). The distribution of DRB1 and DQB1 alleles has not been investigated in COPD. Methods: To this end, HLA medium-low resolution typing was performed following standardized protocols in 320 clinically stable COPD patients included in the PAC-COPD study. Results were compared with controls of the same geographical and ethnic origin, and potential relationships with the severity of airflow limitation and lung diffusing capacity impairment were explored in patients with COPD. Results: The distribution of DRB1 and DQB1 alleles in COPD was similar to that of controls except for a significantly higher prevalence of DRB1*14 in patients with severe airflow limitation and low diffusing capacity. Conclusions: By and large, HLA distribution was similar in COPD patients and controls, but the HLA class II allele DRB1*14 may contribute to the pathogenesis of severe COPD with emphysema.
Keywords :
| Abbreviations | ||
| COPD | = | chronic obstructive pulmonary disease |
| HLA | = | Human leukocyte antigens |
| DLCO | = | Diffusing capacity of the lung for carbon monoxide |
| FEV1 | = | Forced vital capacity exhaled in the first second |
| HLA-DRB1 | = | Human leukocyte antigen class II DR, beta chain locus 1 |
| HLA-DQB1 | = | Human leukocyte antigen class II DQ,, beta chain locus 1 |
| PAC-COPD study Phenotypic Characterization and Course of COPD study | = | |
Introduction
In patients with chronic obstructive pulmonary disease (COPD), the presence of an enhanced inflammatory response in the lungs mediated by T cells (Citation1, 2) that persist after quitting smoking (Citation3–5) supports an auto-immune response to pan-pulmonary antigen(s) (Citation6–8). Potential self-antigens, such as elastin peptides, have been reported in COPD patients, particularly in those with emphysema (Citation9). Besides, an auto-immune model of emphysema has been described in experimental animals (Citation10).
Human leukocyte antigen (HLA) molecules are highly polymorphic cell surface glycoproteins involved in the presentation of peptides to T lymphocytes (Citation11,12). The recognition of self-peptides bound to HLA molecules is central to the pathogenesis of autoimmune diseases (Citation13). Some particular HLA alleles, mainly HLA class II (Citation14), are often associated with auto-immune diseases (Citation15,16). HLA class II alleles are also involved in other mechanisms, such as the response to infection or the regulation of allergy (Citation14), of potential pathogenic relevance in COPD.
The relationship between HLA and COPD is unclear. In the early eighties it was suggested that some HLA alleles were related to COPD (Citation17), and a small study of 15 patients found that HLA-Bw60 alleles were indeed more common in COPD patients with reduced lung diffusing capacity (DLCO) (Citation18), a well-established marker of emphysema in these patients. By contrast, a more recent study concluded that HLA-B7 does not contribute to the rate of decline of lung function (FEV1) in COPD patients (Citation19). Of note, the HLA genes assessed in these previous studies are mostly involved in the susceptibility to adenovirus infection. So far, no study has assessed the role of those HLA class II genes (e.g. DR and DQ) that are involved in autoimmune responses (Citation14).
We hypothesized that the distribution of HLA-DRB1 and HLA-DQB1 alleles may be different in COPD patients than in controls, particularly in those with more severe lung function impairment and/or emphysema. To test this hypothesis, we compared the distribution of HLA-DRB1 and HLA-DQB1 alleles in a large and well characterized sample of patients with COPD to that observed in controls of the same ethnic and geographical origin, and we investigated if any of the alleles was associated with the severity of airflow limitation and/or DLCO impairment, a well-established surrogate marker of pulmonary emphysema, in patients with COPD.
Material and Methods
Study design
The PAC-COPD study is a longitudinal observational study conducted in 9 university teaching hospitals in Spain that sought to get a better understanding of the phenotypic heterogeneity of the disease (Citation20). A complete description of the design and methodological details of the study has been published elsewhere (Citation21). Here we analyzed the cross-sectional results of 320 COPD patients at recruitment from the 9 involved hospitals. Because the PAC-COPD study does not include healthy controls, we compared this data with that of external controls, including DRB1 frequencies from 941 cord blood samples obtained in Barcelona (93% Spanish origin) (Citation22), and DQB1 frequencies from a cohort of 118 healthy Spanish Caucasians recruited in Barcelona (Citation23). To match the origin of the patients and control samples, we complemented the published control data with actual determinations of HLA-DRB1 distribution in 175 healthy and HLA-DQB1 in 126 blood donors from Hospital Son Dureta (Mallorca, Spain), one of the centres participating in the PAC-COPD study. There were no differences in allelic frequency between both groups of controls (supplementary ).
Table 1. Anthropometric clinical, functional and imaging data (mean ± SD) or n (%) of participants by ATS/ERS stages of disease severity
Supplementary . Distribution of DRB1 and DQB1 alleles in published controls and additional controls recruited in Mallorca, number of alleles (n) and allelic frequency (AF%)
Population and ethics
The diagnosis and severity of COPD was established according to the ATS/ERS guidelines (Citation24). Patients younger than 45 years of age, with cancer, residual extensive tuberculous lesions of more than 1/3 of the pulmonary parenchyma, pneumonectomy and/or pneumoconiosis were excluded. All patients signed the informed consent after being fully aware of the nature and objectives of the study, which had been previously approved by the Ethics Committee of all participating institutions more information about recruitment and ethics has been previously published (Citation20).
Characterization of the COPD patients
Anthropometric data and information regarding relevant clinical aspects of the medical history of the patient were obtained using structured questionnaires. Forced spirometry and DLCO were measured according to international guidelines (Citation25) in all patients, using local reference values (Citation26,27).
HLA typing
A venous blood sample (10 ml) was obtained in EDTA containing tubes by peripheral venipuncture. Genomic DNA was extracted using a salting-out technique, and HLA-DRB1 and HLA-DQB1 low-medium resolution typing was performed by polymerase chain reaction sequence-specific oligonucleotide probes (PCR-SSOP) using commercial kits (Invitrogen Ltd). Amplification and detection were carried out according to the manufacturer´s instructions. PCR-SSO cycling parameters were: 35 cycles at 96ºC for 15 s, 58ºC for 45 s, 72ºC for 15 s, followed by a hold program at 72ºC for 5 min. Hybridization was performed at 50ºC for 30 min, while washing and detection was carried out using an streptavidin-HRP conjugate and substrate.
Statistical analysis
Allele frequencies were determined for patients and controls as the total number of copies of the allele in the population sample (Alleles/2n) and expressed as percentage. The overall association for each locus was assessed using a Chi2-test. The frequency distribution of HLA alleles in patients and controls were compared using Chi2-test analysis with Yates correction. If one allelic group had five or fewer individuals, the comparison was done by Fisher's 2-tailed exact test. The p-value was corrected (pc) by the number of comparisons performed (Bonferroni correction). Thus we considered statistically significant a p-value of 0.004 for DRB1 alleles (13 comparisons) and of 0.01 for DQB1 (5 comparisons). The degree of association was estimated using the odds ratio (OR) and the 95% CI. Analysis was done with GraphPad Prism software package (GraphPad Software, Inc. San Diego, US). DRB1-DQB1 haplotypes were estimated using Arlequin 3.1 (Citation28). Results are shown as mean (SD) or percentage, as appropriate.
Results
Characteristics of COPD patients
presents the main characteristics of the COPD patients studied, by ATS/ERS stages of disease severity (Citation24). Most of them had moderate to severe COPD and only a small number of patients had mild or very severe disease. Age was similar between groups, and there was a clear male predominance. Patients with stage IV had a lower BMI than the other groups. Cumulative smoking exposure (pack-yr.) and the percentage of current smokers was similar in all groups. By definition, airflow limitation worsened with increased ATS/ERS severity, and DLCO did so too ().
HLA distribution in COPD patients and controls
(top panel) presents the frequency distribution of the different DRB1 alleles, ordered from the most to the less prevalent ones, in COPD patients (grey columns) and controls (white columns). The overall distribution of DRB1 and DQB1 alleles among patients and controls was not dissimilar (p = 0.061 and p = 0.614). The alleles most frequently found in controls were DRB1*07, DRB1*11 and DRB1*13 (16.3%, 14.7% and 13.9%), whereas these were DRB1*13, DRB1*07 and DRB1*03 (15.78%, 15% and 13.59%) in COPD patients. Overall, the distribution of DRB1 alleles in COPD patients was characterized by a higher allelic frequency of DRB1*14, DRB1*01, DRB1*03 and DRB1*13 and a lower one of DRB1*11, DRB1*10, DRB1*04 and DRB1*07 (, top panel).
Figure 1. Prevalence of the different DRB1 (top panel) and DQB1 (bottom panel) allelic frequencies, ordered from most to less prevalent in controls (white columns), compared to those determined in patients with COPD (grey columns).
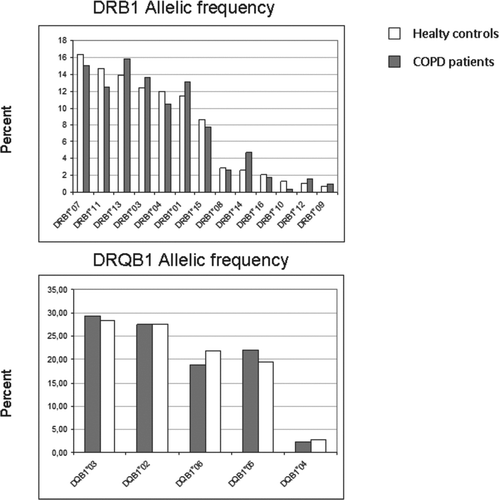
presents the absolute number of alleles and the allelic frequencies of the sixteen DRB1 alleles determined in controls and in COPD patients. HLA DRB1*14 (4.69 vs. 2.6%, p = 0.013; OR: 1.81; 95%CI: 1.16-2.84) was more abundant in patients with COPD than in controls, whereas DRB1*10 was more abundant in controls (0.31 vs. 1.3%, p = 0.03; OR: 0.23; (95%CI: 0.05-0.96) though these differences were not statistical significant after correction for multiple comparisons ().
Table 2. Number of DRB1 and DQB1 alleles (n), allelic frequency (AF%) and Odds Ratio (OR) in controls and COPD patients
(bottom panel) presents the frequency distribution of the five DQB1 alleles determined in controls and in COPD patients, whereas shows their absolute number and allelic frequencies. We did not find statistically significant differences between COPD patients and controls. Yet, it was reassuring that, DQB1*05 was more abundant in patients with COPD because it is found in haplotype (combination of adjacent loci in the chromosome that are transmitted together) (Citation29) with DRB1*14 which, as discussed above, was also more abundant in patients with COPD ().
Table 3. Haplotype frequency (HF%) of HLA DRB1-DQB1 haplotypes in COPD patients
HLA distribution in COPD patients by severity of airflow limitation
Because the number of COPD patients with ATS/ERS stage I or IV was too small to allow independent comparisons (), to investigate the relationship between HLA and the severity of airflow limitation we grouped patients with mild-moderate COPD (ATS/ERS stage I and II) and patients with severe-very severe COPD (stage III and IV) (). The overall distribution analysis for DRB1 showed no differences among controls and ATS I-II patients p = 0.862, but was highly significant for controls vs. III-IV patients p = 0.0018. In the two-by-two contingency tests we found that the frequency of DRB1*14 was significantly higher in III-IV stage than in controls (OR: 2.60, 95% CI = 1.53–4.42, p = 0.001) and just failed to reach statistical significance with respect to patients with stage I-II (OR: 2.18, 95% CI = 1.02-4.68 p = 0.062). By contrast, DQB1 allelic frequencies were similar in COPD patients irrespective of the severity of airflow limitation ().
Table 4. Number of alleles (n) and DRB1 allelic frequency (AF%) in COPD patients by severity of airflow limitation (ATS/ERS stages of disease severity)
HLA distribution in COPD patients by severity of DLCO impairment
To investigate the relationship between HLA and DLCO, we grouped COPD patients above or below a DLCO value of 60% of reference, a clinically relevant threshold (Citation25–30). The overall DRB1 distribution analysis for controls vs. DLCO > 60% ref. was not significant p = 0.338, while for DLCO ≤ 60% ref. it was significant p = 0.046.
In the two-by-two comparisons we found that the frequency of DRB1*14 was significantly more frequent in patients with DLCO ≤ 60% ref. than in controls (OR 2.41 (95%CI: 1.3–4.32); p = 0.004), whereas no differences were found between the latter and patients with DLCO > 60% of ref. (3.9% vs. 2.6%, respectively) (). In keeping with the results discussed here for the entire COPD population (), the frequency of DRB1*11 tended to be lower in patients with DLCO <60% ref. (9.43%) than in controls (14.7%; OR 0.61 (95%CI: 0.39-0.95); p = 0.033), a tendency that was not observed when patients with DLCO > 60% ref. were compared to controls (14.7% vs. 14.61%).
Table 5. Number of alleles (n) and DRB1 allelic frequency (AF%) in COPD patients by DLCO impairment
On the other hand, we did not find any significant difference in the DQB1 allelic frequency distribution between COPD patients with DLCO values below or above 60% ref. and controls (). Yet, in keeping with the higher frequency of DRB1*14 observed in patients with DLCO <60% ref. discussed previously, the frequency of DQB1*05 was also higher in this group of patients, at variance with those with DLCO values >60% of the reference value, whose DRB1 allele frequency distribution was similar to that of controls.
Discussion
To our knowledge, this is the first study that investigates HLA class II alleles, known to be associated with autoimmune diseases (Citation14), in COPD. We found that, by and large, their distribution was similar in COPD patients and controls of the same ethnic and geographical origin. Yet, we found a significantly higher prevalence of DRB1*14 in patients with severe airflow limitation and low DLCO, a well-established clinical marker of emphysema.
Previous studies
Only a few previous studies have investigated the role of HLA in COPD. A small study of 15 patients showed that HLA-Bw60 alleles were more common in patients with low DLCO levels (Citation18). Another study concluded that HLA-B7 does not contribute to COPD or rate of decline of FEV1 in smokers (Citation19), a finding that has been recently confirmed (Citation31). Finally, a genome-wide association study (GWAS) has reported that some single nucleotide polymorphisms (SNPs) in the extended HLA class I and class II regions of chromosome 6 were associated to COPD (Citation32).
Interpretation of findings
Our results show that the overall distribution of the HLA class II alleles DR and DQ in COPD is not dissimilar to that of controls of the same geographical and ethnic origin, except for a significantly higher prevalence of DRB1*14 in patients with severe lung function impairment. The later can be a coherent finding if the role of HLA class II is related to the severity of lung function impairment rather than to the presence or absence of the disease.
COPD is a very heterogeneous disease and, as recommended by current guidelines (Citation24, Citation33), the only inclusion criteria in the PAC-COPD study was the presence of airflow limitation, which is known to fail to capture the complexity of the disease (Citation34). It is not surprising, therefore, that the association of HLA distribution with the presence of COPD so defined is not striking. However, when more specific COPD phenotypes were considered, some interesting associations emerged. For instance, we observed a higher prevalence of the DRB1*14 allele in patients with more severe airflow limitation (OR: 2.60, 95% CI = 1.53–4.42, p = 0.0005) and lower DLCO (OR 2.41 (95%CI: 1.3–4.32); p = 0.004). Importantly, these associations were independent among them. Because allelic HLA class II (DR and DQ) variations are often reported in auto-immune diseases (Citation16), our observation of a higher prevalence of DRB1*14 in patients with severe airflow limitation and low DLCO fits with the hypothesis that there is an auto-immune component in the pathogenesis of COPD, particularly in its emphysema component (Citation6, Citation8). Likewise, since HLA class II alleles are also involved in the presentation of infectious antigens (Citation14), our findings can potentially be related to the episodes of acute exacerbations that these patients often suffer.
One of the difficulties in assessing HLA genes is the presence of multiple variants and the need of performing multiple comparisons, which may result in some false positive associations. First, for all comparisons we did an overall distribution assessment. In our study only DRB1*14 showed to be associated beyond the threshold of multiple comparisons. The fact that DRB1*14 had been previously associated with several human diseases characterized either by chronic pulmonary inflammation (sarcoidosis (Citation35)), auto-immunity (ulcerative colitis and Pemphigus vulgaris (Citation36–37)) or the capacity to resolve infections (Citation38), all of them pathogenic mechanisms postulated in COPD (Citation5, Citation8, Citation39–40), provides biological plausibility to the possibility that DRB1*14 is a novel genetic risk factor for COPD severity.
Strengths and limitations of the study
Two strengths of our study are worth mentioning. First, although the sample size of our cohort may appear small for current standards of genetic studies, it was much larger and better characterized (Citation20) than previous studies on HLA in COPD (Citation17–19). Second, this is the first investigation of HLA class II alleles (DR and DQ) in COPD. We acknowledge, however, that our study has several limitations that also deserve discussion. First, the lack of well characterized controls in the PAC-COPD study (Citation21), particularly smokers without COPD, is a limitation of our study. To address it we used as controls actual measurements and published data from the same geographical and ethnic origin (Citation22–23); importantly, results were unchanged if only the former were used for analysis.
And, second, to account for multiple comparisons, we increased our level of statistical significance requirement by using the Bonferroni correction. Finally, we believe that, if bias by population admixture would have occurred, it would have most likely affected several HLA variants, rather than the only one we found in our study. In any case, our results need to be confirmed in a different population since candidate gene studies often fail to be replicated (Citation41).
Conclusions
Our study shows that HLA class II alleles (DR and DQ) distribution in COPD is not vastly different from that of controls. Yet, the HLA allele DRB1*14 appears to be more prevalent in a specific COPD phenotype characterized by severe airflow limitation and impaired diffusing capacity, most likely reflecting the presence of emphysema.
Declaration of Interest
The work was supported, in part, by Fondo de Investigación Sanitaria (FIS PI020541, FIS PI052082, FIS PI020541, PI052486, PI052302), Ministry of Science, Spain; ABEMAR; Agència d'Avaluació de Tecnologia i Recerca Mèdiques (AATRM 035/20/02), Catalonia Government; Spanish Society of Pneumology and Thoracic Surgery (SEPAR 2002/137); Catalan Foundation of Pneumology (FUCAP 2003 Beca Marià Ravà); Red RESPIRA (RTIC C03/11); Red RCESP (RTIC C03/09); Fundació La Marató de TV3 (num. 041110); DURSI (2005SGR00392); and an unrestricted educational grant from Novartis Farmacèutica, Spain. CIBERESP and CIBERES are funded by the Instituto de Salud Carlos III, Ministry of Science, Spain. Judith Garcia-Aymerich has a researcher contract from the Instituto de Salud Carlos III (CP05/00118), Ministry of Health, Spain. Any financial/commercial conflicts of interests have been disclosed by authors.
Contributions: conception and design: AA, JMA, JGA, JS; analysis and interpretation: RF, BN, JRG, JGA, AA; drafting the manuscript for important intellectual content: AA, RF, JGA, JMA, JS. The authors are responsible for the content and writing of this paper.
Acknowledgments
Authors thank the participants in the PAC-COPD study for their willingness to contribute to medical research, as well as all the local investigators (listed in the Appendix), nursing and technical personnel involved in the study for their support and contribution.
References
- Finkelstein R, Fraser RS, Ghezzo H, Cosio MG. Alveolar inflammation and its relation to emphysema in smokers. Am J Respir Crit Care Med 1995; 152:1666–1672.
- Saetta M, Di Stefano A, Maestrelli P, Activated T-lymphocytes and macrophages in bronchial mucosa of subjects with chronic bronchitis. Am Rev Respir Dis 1993; 147(2):301–306.
- Rutgers SR, Postma DS, ten Hacken NH, Ongoing airway inflammation in patients with COPD who do not currently smoke. Thorax 2000; 55(1):12–18.
- Willemse BWM, ten Hacken NHT, Rutgers B, Lesman-Leegte IGAT, Postma DS, Timens W. Effect of 1-year smoking cessation on airway inflammation in COPD and asymptomatic smokers. Eur Respir J 2005; 26(5):835–845.
- Hogg JC, Chu F, Utokaparch S, The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med 2004; 350(26):2645–2653.
- Agusti A, MacNee W, Donaldson K, Cosio M. Hypothesis: Does COPD have an autoimmune component? Thorax 2003; 58(10):832–834.
- Sullivan AK, Simonian PL, Falta MT, Oligoclonal CD4+ T cells in the lungs of patients with severe emphysema. Am J Respir Crit Care Med 2005; 172(5):590–596.
- Cosio M, Saetta M, Agusti A. Immunological aspects of COPD. N Engl J Med 2009; 360:2445–2454.
- Lee SH, Goswami S, Grudo A, Antielastin autoimmunity in tobacco smoking-induced emphysema. Nat Med 2007; 13(5):567–569.
- Taraseviciene-Stewart L, Scerbavicius R, Choe KH, An Animal Model of Autoimmune Emphysema. Am J Respir Crit Care Med 2005; 171(7):734–742.
- Davis SJ, Ikemizu S, Evans EJ, Fugger L, Bakker TR, van der Merwe PA. The nature of molecular recognition by T cells. Nat Immunol 2003; 4(3):217–224.
- van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol 2003; 21:659–684.
- Flavell RA, Hafler DA. Autoimmunity. What is the turning point? Curr Opin Immunol 1999; 11(6):635–637.
- Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med 2000; 343(11):782–786.
- Thorsby E, Lie BA. HLA associated genetic predisposition to autoimmune diseases: Genes involved and possible mechanisms. Transpl Immunol 2005; 14(3–4):175–182.
- Heard R. HLA and autoimmune diseases. In: Lechler R, editor. HLA and Disease. London, UK: Academic Press Limited; 1994:123–151.
- Kauffmann F, Kleisbauer JP, Cambon-De-Mouzon A, Genetic markers in chronic air-flow limitation. A genetic epidemiologic study. Am Rev Respir Dis. 1983; 127(3):263–269.
- Maranetra N, Chandanayingyong D, Bovornkitti S. HLA antigen and ventilatory drive in Thais with chronic obstructive pulmonary disease. Asian Pac J Allergy Immunol 1990; 8(2):137–140.
- Kasuga I, Ruan J, Connett JE, Anthonisen NR, Sandford AJ. Lack of association of human leukocyte antigen-B7 with COPD and rate of decline in lung function. Respir Med 2005; 99(12):1528–1533.
- Garcia-Aymerich J, Gomez FP, Benet M, Identification and prospective validation of clinically relevant chronic obstructive pulmonary disease (COPD) subtypes. Thorax 2011; 66:430–437.
- Garcia-Aymerich J, Gomez FP, Anto JM. Phenotypic characterization and course of chronic obstructive pulmonary disease in the PAC-COPD study: Design and methods. Arch Bronconeumol 2009; 45(1):4–11.
- Vidal S, Morante MP, Moga E, Molecular analysis of HLA-DRB1 polymorphism in north-east Spain. Eur J Immunogenet 2002; 29(1):75–77.
- Simeon CP, Fonollosa V, Tolosa C, Association of HLA class II genes with systemic sclerosis in Spanish patients. J Rheumatol 2009; 36(12):2733–2736.
- Celli BR, MacNee W, Agusti AG, Standards for the diagnosis and treatment of patients with COPD: A summary of the ATS/ERS position paper. Eur Respir J. 2004; 23(6):932–946.
- Clausen JL. Pulmonary function testing. Guidelines and controversies. Equipment, Methods and Normal Values. 1 ed. Orlando: Grune & Stratton, Inc.; 1984.
- Roca J, Rodriguez-Roisín R, Cobo E, Burgos F, Perez J, Clausen JL. Single-breath carbon monoxide diffusing capacity (DLCO) prediction equations for a mediterranean population. Am Rev Respir Dis 1990; 141:1026–1032.
- Roca J, Sanchis J, Agustí-Vidal A, Spirometric reference values for a mediterranean population. Bull Eur Physiopathol Respir 1986; 22:217–224.
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online 2005; 1:47–50.
- Miretti MM, Walsh EC, Ke X, A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet 2005; 76(4):634–646.
- Fujimoto K, Kitaguchi Y, Kubo K, Honda T. Clinical analysis of chronic obstructive pulmonary disease phenotypes classified using high-resolution computed tomography. Respirology 2006; 11(6):731–740.
- Kasuga I, Hogg JC, Pare PD, Role of genetic susceptibility to latent adenoviral infection and decreased lung function. Respir Med 2009; 103(11):1672–1680.
- Pillai SG, Ge D, Zhu G, A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009; 5(3):e1000421.
- Rabe KF, Hurd S, Anzueto A, Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2007; 176(6):532–555.
- Agusti A, Calverley P, Celli B, Characterisation of COPD heterogeneity in the ECLIPSE cohort. Respir Res 2010; 11(1):122–136.
- Sharma SK, Balamurugan A, Pandey RM, Saha PK, Mehra NK. Human leukocyte antigen-DR alleles influence the clinical course of pulmonary sarcoidosis in Asian Indians. Am J Respir Cell Mol Biol 2003; 29(2):225–231.
- Yamamoto-Furusho JK, Rodriguez-Bores L, Granados J. HLA-DRB1 Alleles are associated with the clinical course of disease and steroid dependence in mexican patients with ulcerative colitis. Colorectal Dis 2010; 12(12):1231–1235.
- Tunca M, Musabak U, Sagkan RI, Koc E, Akar A. Association of human leukocyte antigen class II alleles with pemphigus vulgaris in a Turkish population. J Dermatol 2010; 37(3):246–250.
- Kotb M, Norrby-Teglund A, McGeer A, An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med 2002; 8(12):1398–1404.
- Taylor AE, Finney-Hayward TK, Quint JK, Defective macrophage phagocytosis of bacteria in COPD. Eur Respir J 2010; 35(5):1039–1047.
- Sethi S, Mallia P, Johnston SL. New Paradigms in the pathogenesis of chronic obstructive pulmonary disease II. Proc Amer Thorac Soc 2009; 6(6):532–534.
- Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One 2009; 4:e5639.
Appendix
The “Phenotype and Course of COPD (PAC-COPD)” Study Group
Centre for Research in Environmental Epidemiology (CREAL), Barcelona: Marta Benet, Jordi de Batlle, Ignasi Serra, David Donaire-Gonzalez, Stefano Guerra; Hospital del Mar-IMIM. Barcelona: Eva Balcells, Àngel Gayete, Mauricio Orozco-Levi, Ivan Vollmer; Hospital Clínic - Institut D'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS). Barcelona: Joan Albert Barberà, Carles Paré, Josep Roca, Robert Rodriguez-Roisin, Xavier Freixa, Diego A Rodriguez, Elena Gimeno, Karina Portillo; Hospital General Universitari Vall D'Hebron. Barcelona: Jaume Ferrer, Jordi Andreu, Esther Pallissa, Esther Rodríguez; Hospital de la Santa Creu i Sant Pau. Barcelona: Pere Casan, Rosa Güell, Ana Giménez; Hospital Universitari Germans Trias i Pujol. Badalona: Alicia Marín, Josep Morera; Hospital Universitari de Bellvitge. L'Hospitalet de Llobregat: Eva Farrero, Joan Escarrabill; Hospital de Sabadell. Corporació Parc Taulí. Institut Universitari Parc Taulí (Universitat Autònoma de Barcelona). Sabadell: Antoni Ferrer; Hospital Universitari Son Dureta (currently, Hospital Univ. Son Espases). Palma de Mallorca: Bernat Togores; Hospital de Cruces. Barakaldo: Juan Bautista Gáldiz. Lorena López, José Belda,Universidad de Valencia.