Abstract
Welding fumes include agglomerated particles built up of primary nanoparticles. Particles inhaled through the nose will to some extent be deposited in the protein-rich nasal mucosa, and a protein corona will be formed around the particles. The aim was to identify the protein corona formed between nasal lavage proteins and four types of particles with different parameters. Two of the particles were formed and collected during welding and two were manufactured iron oxides. When nasal lavage proteins were added to the particles, differences were observed in the sizes of the aggregates that were formed. Measurements showed that the amount of protein bound to particles correlated with the relative size increase of the aggregates, suggesting that the surface area was associated with the binding capacity. However, differences in aggregate sizes were detected when nasal proteins were added to UFWF and Fe2O3 particles (having similar agglomerated size) suggesting that yet parameters other than size determine the binding. Relative quantitative mass spectrometric and gel-based analyses showed differences in the protein content of the coronas. High-affinity proteins were further assessed for network interactions. Additional experiments showed that the inhibitory function of secretory leukocyte peptidase inhibitor, a highly abundant nasal protein, was influenced by particle binding suggesting that an understanding of protein function following particle binding is necessary to properly evaluate pathophysiological events. Our results underscore the importance of including particles collected from real working environments when studying the toxic effects of particles because these effects might be mediated by the protein corona.
Introduction
It is estimated that more than one million workers worldwide perform welding as a part of their work (Sjogren & Langard, Citation2004), and studies have shown that welders experience an increased risk of respiratory illness (Antonini, Citation2003). Welding fumes consist of a mixture of gas and agglomerated particles (networks of interacting particles) that are built up of primary nanoparticles (Jenkins & Eagar, Citation2005). A correlation between exposure to nanoparticles in urban air and adverse health effects, such as cardiovascular disease and pulmonary diseases such as bronchial asthma, has been suggested (Nawrot et al., Citation2006). The welding particles are small enough to remain airborne and are easily inhaled and deposited within the respiratory tract (Oberdorster et al., Citation2005). The effects of these deposited particles are probably dependent on particle parameters, e.g. chemical composition, size, surface properties, and shape, which influence solubility and the site of particle deposition in the airways (Maynard & Kuempel, Citation2005).
When particles encounter a biological fluid, molecules such as proteins, lipids, and carbohydrates will, in competition with each other, bind to the particle's surface leading to the formation of a dynamic particle–biomolecule interaction (Cedervall et al., Citation2007). The protein corona that is formed can have a significant impact on the biological effects of the particles (Saptarshi et al., Citation2013; Tenzer et al., Citation2011). The composition of the protein corona will constantly change until equilibrium is reached (Dell'orco et al., Citation2010), but some proteins in the corona can remain bound to the particle as it travels from one environment to another (Monopoli et al., Citation2011; Tenzer et al., Citation2013). Most previous investigations of the protein corona of nanoparticles have been performed with blood plasma proteins and only rarely in other fluids (Tenzer et al., Citation2011, Citation2013).
Human nasal lavage fluid forms a defense against foreign microbes or particulates being deposited in the nasal cavity (Casado, Citation2004; Casado et al., Citation2005). It contains a variety of proteins from plasma, mucus, serous fluids, and secretions from epithelial and immunological cells (Mygind & Dahl, Citation1998). There is a strong resemblance between the proteins identified in nasal secretions and in bronchoalveolar lavage (Bartlett et al., Citation2013; Benson et al., Citation2009; Kosanam et al., Citation2012) making nasal lavage suitable for investigation of the protein corona formed in the lower airways. Proteomic analysis can provide essential information to help understand the pathophysiologic pathways of the inhaled particles (Casado et al., Citation2005).
In this study, welding fume particles were generated, collected on membranes, extracted, and finally incubated with nasal lavage proteins with the purpose of studying the particle–protein interactions. The aim of the study was to identify the protein corona formed in vitro between welding fume particles and nasal lavage proteins and to understand how parameters such as particle size and chemical composition affect the composition and functionality of the protein corona.
Methods
Welding particles
In situ characterization of airborne welding particles
The source of the welding fumes was generated by metal active gas welding in mild steel according to the principle developed by Isaxon et al. (Citation2013). The size distribution (mobility diameter) of the agglomerated airborne welding particles (interacting particles built up of primary nanoparticles) was measured using a scanning mobility particle sizer (SMPS, CPC model 3010, TSI Inc., Shoreview, MN). The SMPS covered a size range of 10–700 nm and had a time resolution of ∼3 min. The mass concentration was monitored online using a tapered element oscillating microbalance (Rupprecht & Patashnic Co. Inc., Albany, NY).
Fractionation and collection of welding particles
Welding particles were collected from a 22 m3 stainless steel chamber using a high volume cascade impactor (HVCI; BGI 900 LPM, BGI Incorporated, Waltham, MA). The HVCI had a flow of 0.9 m3/min, and particles were collected in fractions of ultrafine welding fume (UFWF) particles < 0.1 μm in diameter and fine welding fume (FWF) particles ranging from 0.1 µm to 2.5 μm in diameter. The UFWF fraction was collected on a polytetrafluoroethylene filter (PTFE), and the FWF fraction was collected on polyurethane foam (PUF; Demokritou et al., Citation2002). Welding particles were recovered from the PTFE and PUF using a methanol extraction protocol. The filters were washed repeatedly with methanol, and the resulting solution was decanted into a 50 mL sample tube. The solution was vacuum dried. The extracted particle mass was determined. All weighing was performed in a climate-controlled environment and after a minimum 24-h acclimatization period. The extracted particles were weighed three separate times, and the average mass and the standard deviation were calculated. The chemical composition of the two fractions, FWF and UFWF, was analyzed by particle-induced X-ray emission.
Model particles
Magnetite Fe3O4 and Fe2O3 were selected as model particles because iron oxides are the main components of welding fumes. Magnetite Fe3O4 (8 nm) in 30 wt% aqueous suspension was purchased from PlasmaChem GmbH (Berlin, Germany). Fe2O3 (20–40 nm, 99% purity) was purchased in the form of dry powder from SkySpring Nanomaterials (Houston, TX).
Particle characterization in fluid
Each particle type was suspended in 20 mL vials (Scint-Burk glass pp-lock-Alu-foil, Wheaton Industries, Inc., Millville, NJ) to a total concentration of 2.56 mg/mL in milliQ water. The suspension was sonicated for 16 min at 400 W and 10% amplitude using a Vibra-Cell sonifier (Soniucs & Materials, Danbury, CT) according to the Nanogenotox protocol (Jensen, Citation2011). The size of the particles in the suspension was measured by dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA). DLS measurements were performed with a Malvern Zeta NANO S (Malvern Instruments Ltd, Worcestershire, UK) equipped with a 532 nm red laser and operating with a 90 ° scattering angle. The measurements were evaluated with the Zetasizer software version 6.20 (Malvern Instruments Ltd, Worcestershire, UK). All measurements were performed at 25 °C. Each sample was measured at least three times with a delay of 15 s between the measurements, and the stability of the suspension was confirmed by re-measurement after 20 min. The mean hydrodynamic diameter was measured for the agglomerated particles as well as the aggregated form (complexes consisting of several particles each with a protein corona) after incubation with nasal lavage proteins. For some samples, the diameters could be determined by NTA using a NanoSight LM10 (Nanosight Ltd., Amesbury, UK) equipped with a 635 nm laser. All samples were measured at room temperature three times for 60 s.
Transmission electron microscopy (TEM)
Particles were collected from the air and from the liquid particle suspensions onto carbon-coated copper grids for TEM analysis. The particles were collected using an electrostatic precipitator (NAS Model 3089, TSI Inc., Shoreview, MN).
Endotoxin quantification
To test how much endotoxin the particles contained, a kinetic chromogenic Limulus Amebocyte Lysate (LAL) assay (Lonza, Walkersville, MD) was used according to the protocol of the manufacturer. A 7-point standard curve was generated over the concentration range of 0.005–50 endotoxin unit/mL, and a control standard endotoxin Escherichia coli O55:B5 was used for reference values. The samples were assayed in a 96-well pyrogen-free microplate (TPP, Hannover, Switzerland), and spectrophotometric measurements were taken at 405 nm.
Collection of nasal lavage
Nasal lavage was collected by flushing the nasal cavity of 20 healthy volunteers with 18 mL of saline solution. The samples were evaporated to dryness, dissolved in 500 µL of milliQ water, pooled, and then desalted in Amicon Ultra-0.5 centrifugal filters (3 kDa cut-off) (Millipore, Carrigtohill, Ireland) at 14 000 × g for 30 min. The samples were washed with 300 µL water and centrifuged again for 30 min at 14 000 × g. The total protein content was determined with a bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL). The solution of proteins obtained from the pooled nasal lavages was used in all incubations described below.
Particle–nasal lavage protein incubation
For each type of particle suspension, three different amounts of particles were added to a final concentration of 800 μg nasal lavage protein/mL to yield 400, 200, and 100 μg particles/mL, and these were incubated for 6 h at 32 °C (Scheme 1). All incubations were performed in triplicate. Unbound proteins were separated from the particles by centrifugation for 1 h at 16 000 × g. The supernatant was stored for further analysis, and the particle pellet was washed three times with milliQ water followed by a second and third centrifugation. The supernatant was collected and further sampled in triplicate. The amount of bound protein was determined by measuring the amount of proteins that did not bind to the particles using the BCA protein assay kit. Proteins from the different incubations were analyzed by two different methods.
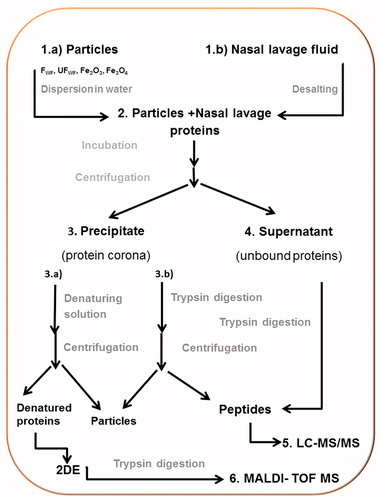
Scheme 1. Overview over the experiment. (1) (a) The particles (FWF, UFWF, Fe2O3 and Fe3O4) were suspended in water. (b) Nasal lavage fluids were concentrated and desalted. (2) Each particle type was incubated with nasal lavage proteins for 6 h. (3) Proteins bound to the particles (the protein corona) were separated from unbound proteins by centrifugation. The pellet was either (a) dissolved in water and the proteins were trypsin digested or (b) incubated with denaturing solution. (4) The proteins in the supernatant were trypsin digested. (5) The tryptic peptides from steps 4 and 3b were analyzed with LC-MS/MS. (6) The denatured proteins were separated on 2DE, gel slices were cut out, the proteins were digested with trypsin, and the peptides were identified with MALDI-TOF MS.
Two-dimensional gel electrophoresis (2DE) analysis/matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF MS)
Prior to 2DE analysis, proteins from the coronas formed by the four different particles (three replicates each and a positive control) were suspended in 150 µL of sample buffer containing 9 M urea, 65 mM DTT, 2% Pharmalyte (GE Healthcare, Bioscience, Uppsala, Sweden), 4% CHAPS, and 1% bromophenol blue and then centrifuged at 23 000 × g for 30 min at 4 °C. The supernatant containing denatured proteins released from the particles (50 µg protein) was then mixed with a rehydration buffer consisting of 8 M urea, 4% CHAPS, 0.5% immobilized pH gradient, IPG, buffer 3-10NL (GE Healthcare, Bioscience, Uppsala, Sweden), 19 mM DTT, and 5.5 mM Orange G to a final volume of 350 µL. The 2DE was performed in a horizontal 2DE set up IPGphor. The first dimension was performed by in-gel rehydration on pH 3–10 non-linear IPG strips (GE Healthcare, Bioscience, Uppsala, Sweden) and then proteins were transferred to homogenous home-cast gels (0.5 × 180 × 245 mm, 14% T, 1.5% C) running at 20–40 mA, 40–800 V, and 10 °C overnight.
Proteins were silver stained according to Shevchenko (Citation1996) with minor modifications. The protein patterns were then visualized using a cooled charged-coupled device camera digitizing at 1340 × 1040 pixels resolution (Fluor-S Multi-Imager, Bio-Rad, Hercules, CA) in combination with a computerized imaging 12-bit system designed for evaluation of 2DE patterns (PDQuest version 7.1.1, Bio-Rad, Hercules, CA).
The protein spots were excised and destained according to Ghafouri et al. (Citation2007). About 25 µL of 20 µg/mL trypsin in 25 mM ammonium bicarbonate was added to the dried residue, and the samples were incubated overnight at 37 °C. Tryptic peptides were dissolved in trifluoroacetic acid (TFA) and mixed 1:1 with matrix solutions consisting and 1 µL of the solution was spotted on the target plate. Analysis of peptides was performed using MALDI-TOF MS (Voyager-DE PRO, Applied Biosystems, Foster City, CA) equipped with a 337 nm laser and delayed extraction and operated in a reflector mode. Data processing of the spectra was performed with Data Explorer™ (version 4.0, Applied Biosystems, Foster City, CA).
The mass list (mass + H+) generated from the 40 most abundant peaks of the MALDI spectra was submitted to a database search (NCBI, or SWISS-PROT, PIR, Newark, DE). Restrictions were placed on species (Human), mass tolerance ( ± 50 ppm), maximum missed cleavages by trypsin (up to 1), and cysteine modification by carbamidomethylation.
Liquid chromatography coupled to mass spectrometry (LC-MS/MS) analysis
The protein corona samples from four different particles with three different particle concentrations and the supernatant, containing unbound proteins, were reduced by the addition of 2 µL of 50 mM DTT and incubated for 1 h at 60 °C. Proteins were alkylated by the addition of 1 µL of 200 mM iodoacetamide and incubated in darkness at room temperature for 30 min. All experiments were performed in triplicate. To each sample, 10 µL of sequencing-grade trypsin (1:10 trypsin to protein ratio, Roche Diagnostics, Mannheim, Germany) and 1 µL of 100 mM calcium chloride were added. The samples were then incubated overnight at 37 °C. The trypsin-digested samples were centrifuged for 1 h at 16 000 × g, and the supernatants containing the trypsin-digested peptides were evaporated to dryness and dissolved in 200 µL milliQ water.
The MS platform was a 5500QTRAP hybrid triple quadropole/linear ion trap mass spectrometer equipped with a TurboIonSpray source (Applied Biosystems, Foster City, CA/MDS Sciex, Framingham, MA) coupled to an online liquid chromatography (LC) system (UFLCXR; Shimadzu Corporation, Kyoto, Japan). A targeted LC-MS/MS method developed specifically for nasal lavage proteins with a total of 245 different proteins was used (Mörtstedt et al., Citation2013). Briefly peptides were separated on a C18-column (2.1 mm × 50 mm, 3 µm pore size, VisionHT C18 CL; Grace Vydac, Hesperia, CA) using 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) in a gradient from 5% B to 10% B in the first 10 min, from 10% B to 30% B over 15 min, and from 30% B to 99% B over 5 min. Each sample was analyzed twice. Results were integrated in Skyline free software (Skyline, MacCoss Lab at University of Washington, Washington, DC) and manually reviewed. To estimate the degree of binding of each individual protein in the nasal lavage protein solution to the different particle types, a ratio was determined between bound and unbound proteins (Rb/u), according to the equations below. The mean peak values were calculated for each transition for which the two plus and three plus ions were detected (Equation (1)). Peptide fold changes were calculated by relating the mean peptide peak area from the bound to the mean peptide peak area from the supernatant (Equation (2)). The logarithmic ratio was used from Equation (2) to calculate the median ratio (Bantscheff et al., Citation2012) for each protein, Rb/u, in Equation (3).
(1)
(2)
(3)
Statistical evaluation
All statistical analyses were performed with the SPSS software (SPSS Inc., Chicago, IL). Size differences between FWF and UFWF were statistically evaluated using the Mann–Whitney test. The Jonckheere–Terpstra trend test was used for proteins that showed a decreased Rb/u trend with decreasing particle concentrations, and p values ≤ 0.05 were considered significant (Bewick et al., Citation2004; Simpson & Margolin, Citation1986). False discovery rate validation of the statistically significant results was pursued with a sequential goodness of fit test using the free software SGoF + (Software AG, Reston, VA) (De Uña-Alvarez & Carvajal-Rodriguez, Citation2010; Diz et al., Citation2011).
Pathway analysis of significantly stable proteins
Proteins that had high affinity for the particles were further analyzed with Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Redwood City, CA, www.ingenuity.com) to map interacting networks. Default settings were used except for species, which was set to human, and only experimentally observed relationships were considered.
ELISA (HNE substrate inhibition)
Detection of secretory leukocyte peptidase inhibitor (SLPI, antileukoproteinase) functionality was performed with a Human Neutrophil Elastase (HNE) Immunocapture Activity Assay Kit (EMD Chemicals, Darmstadt, Germany) that uses a monoclonal antibody to HNE immobilized on a 96-well microplate (EMD Chemicals, Darmstadt, Germany). A total of 100 µL of the 20 ng/mL HNE solution was added to the plates and incubated for 1 h at room temperature. The plate was washed four times with sample buffer followed by addition of SLPI standard, nasal lavage proteins, and different protein coronas for SLPI inhibitor functionality assessment. SLPI standard solutions ranging from 0.01 nM to 7.5 nM along with the SLPI:particle aggregates (diluted to the same SLPI concentrations as for SLPI alone), nasal lavage proteins, particles, nasal lavage protein:particle corona, and a blank with just assay buffer were added in separate wells and incubated for 4 h at 32 °C. SLPI was added to the particles at a concentration similar to the SLPI concentration in nasal lavage, thus 26.4 ng/mL SLPI was incubated with 400 µg/mL particles corona. Material that did not interact with HNE was washed away four times. The activity of HNE was detected with the fluorogenic substrate MeOSuc-Ala-Ala-Pro-Val added to the wells, and the plates were incubated for 4 h at 37 °C. Fluorescence was measured at an excitation wavelength of 370 nm and an emission wavelength of 460 nm and analyzed with a FlexStation 3 Microplate Reader (Molecular Devices, Silicon Valley, CA).
Results
Nanoparticles
We focused on welding fume particles and two model iron oxide particles, Fe2O3 and Fe3O4, which differ in particle size and in oxidation number.
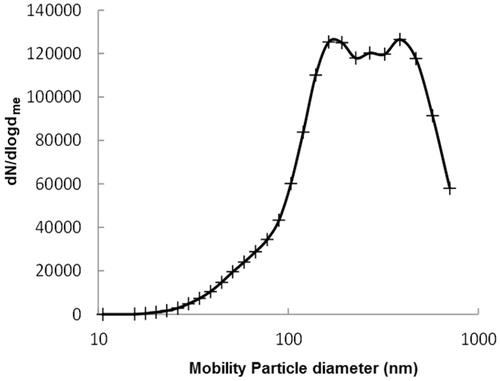
The properties of welding fume particles in air
The concentration of welding fume particles in the chamber during collection was ∼105/cm3. The average number size distribution is shown in . The distribution is, as is typical for distributions of welding fumes in air, lognormal with a geometrical mean diameter of 270 nm. The average mass concentration in the air during collection was, according to the tapered element oscillating microbalance, 2.2 ± 1.2 mg/m3. The chemical composition was analyzed and the major constituents of the welding particles after fractionation were iron (75%) and manganese (15%) equally distributed in both FWF and UFWF fractions (Figure S1). Iron was expected to be in the form of the ferrous oxides FeO, Fe2O3, and Fe3O4 (Jenkins & Eagar, Citation2005). The welding particles are built up by chain-like structures of a few up to several hundred primary particles ranging in size from 2 nm to 70 nm (Figure S2) (Isaxon et al., Citation2013).
Endotoxin levels in particle suspensions
Particles collected from the air can adsorb different compounds. Endotoxins are common in the air, and certain proteins that are present in the airway bind to or have a defense activity against endotoxins. Endotoxin contamination of the particles might influence the pattern of nasal lavage proteins binding to the particles; therefore, the endotoxin levels in the particles were determined. The results showed that the endotoxin levels before incubation in nasal lavage protein solution were low (Table S1). Because the concentrations of endotoxins were three orders of magnitude lower than the particle levels, they were expected to have a minor effect, if any effect at all, on the protein-binding capacity of the particles.
Particle characterization in fluid
The shape and the number of primary particles in agglomerated particles change when collected from air and suspended in liquid depending on the collection method and the dispersion protocol used. Characterization of the agglomerated particles size after sonication in water was determined by DLS and NTA ( and Figures S3 and S4). Significant differences in mean size (p < 0.01, Mann–Whitney test) between FWF and UFWF were observed, but were not as large as expected. This was likely because large agglomerates can break up and form smaller or new agglomerates after they are suspended in liquid and sonicated. FWF and UFWF have a broad size distribution compared with Fe2O3 and Fe3O4 (Figure S3), probably because of the more complex composition of the primary particles both with respect to size and chemical composition. The manufactured Fe3O4 and Fe2O3 particles have larger particle diameters than specified by the manufacturer (), which probably can be explained by the primary particles forming agglomerates. The agglomerated structure was confirmed by TEM (Figures S5 and S6).
Table 1. Size increase (mean diameter) between particles suspended in water and following addition of nasal lavage proteins or secretory leukocyte peptidase inhibitor (SLPI).
Protein corona formation
Significant changes in the diameter of the particles were detected after incubation of the particles with nasal lavage proteins ( and Figure S3). The experiment showed that the protein-driven particle aggregation is time dependent and reproducible data were only obtained by following a strictly time-controlled experimental procedure. The DLS measurement was performed directly following the sonication of the particles. All incubations with particles and nasal lavage proteins were performed within 20 min after the end of particle sonication, which is the time frame in which the particle agglomerates were shown to have a stable size distribution, followed by an additional DLS measurement. Large increases in size following incubation in nasal lavage protein solution were detected for all particles (). The relative increase in size differed between particle types. For example, Fe2O3 and UFWF particles were of equal size in suspension, but the diameters increased by a factor of 2.6 and 1.5, respectively, after incubation in nasal lavage protein solution. The amount of bound proteins per particle mass unit also differed between particles (Table S2). Plotting the relative increase in aggregate size versus the amount of bound protein per particle mass unit showed that these two parameters are correlated (Figure S7).
Identification of proteins in the corona
Three different concentrations of each particle suspended in water were incubated with nasal lavage proteins (). Proteins bound to the particles (the protein corona) were identified and relatively quantified by 2DE/MALDI-TOF MS and with targeted LC-MS/MS in selected reaction monitoring (SRM) mode.
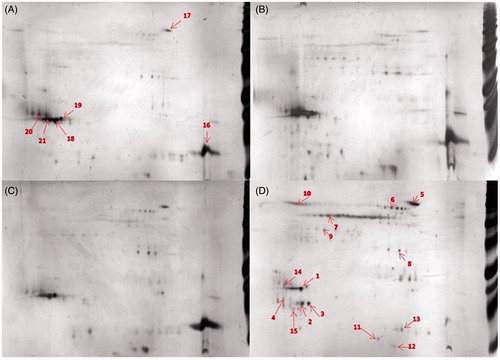
Representative 2DE separations of the proteins in the corona showed similar protein patterns for FWF, UFWF, and Fe2O3 (, Figure S8, and Table S3). Proteins that bound to these particles to a high degree were lysozyme C and palate lung and nasal epithelium clone (PLUNC). These proteins bound to a lower extent to Fe3O4. This particle showed an overall different protein composition in its corona, for example, lipocalin 1 was identified in the corona of the Fe3O4 particles but not in the coronas of any of the other particles.
Figure 2. Comparison of protein-corona patterns from different particle, 50 μg of the bound proteins were separated on 2DE. (A) FWF, (B) UFWF, (C) Fe2O3, and (D) Fe3O4. (1) PLUNC, (16) lysozyme C, (7) keratine and (2) lipocalin 1. Each sample was analyzed in triplicates. The most representative gels are presented here. Numbers refer to identified proteins listed in Table S3.
The protein coronas were further analyzed with a complementary method using a label-free targeted LC-MS/MS (Mörtstedt et al., Citation2013). To estimate the degree of binding of each individual protein in the nasal lavage protein solution to the different particle types, a ratio was determined between bound and unbound proteins (Rb/u). In a first step, the Rb/u was calculated for particle concentrations of 400 µg/mL. Proteins with a Rb/u ≥ 1 are listed in Table S4 along with the calculated isoelectric point (pI) and molecular weight (kDa) for each protein.
The protein coronas of FWF and UFWF were quite similar (Table S4), but a few proteins were only found bound to one or the other particle. The Rb/u ratios for FWF and UFWF were similar indicating that individual proteins bound to the particles with a similar affinity. Fe2O3 particles bound to the same set of proteins as FWF and UFWF. In addition, about 14 unique proteins were observed to bind only to Fe2O3. The protein corona of the smallest particles, Fe3O4, was distinct from the coronas of the three other particles. However, the protein coronas formed on the four particles cannot be categorized based on a distinct size, net charge, or hydrophobicity of proteins. The protein binding did not correlate to protein abundance for any of the four particles suggesting that the affinity between protein and particle is important.
Proteins binding with high affinity to the particles
In order to determine which proteins occupy the particle surface with a high probability, proteins with a high affinity to the particle surface were identified using Rb/u ratios. Proteins with Rb/u ≥ 1 were considered to be proteins with high affinity for binding to the particle surface, and these were included for further analysis (Table S4). The Rb/u at 400 µg/mL ranged from 1 to 1000 suggesting that the affinity of the proteins for the particles differed by a magnitude of approximately 103. The Rb/u was also determined for each protein and particle at two lower particle concentrations of 200 µg/mL and 100 µg/mL. At lower particle concentrations, less surface area is available for binding, and this leads to increased competition between binding proteins. It was then possible to compare affinities between the binding proteins. Proteins with significantly decreasing ratios (tested with the Jonckheere–Terpasta statistical test) at particle concentrations lower than at 400 µg/mL were excluded (including proteins at 100 µg/mL and 200 µg/mL with Rb/u ≥ 1) because these proteins were considered to have a low affinity for the particles. The remaining proteins are listed in . These are considered to bind the particles with high affinity and are probably the most interesting proteins in a realistic exposure situation.
Table 2. Proteins with high affinity for the particles.
For some of the proteins in the corona, the Rb/u values were high indicating that a large fraction of the protein was bound to the particle and thus leaving only a small fraction unbound. If the binding affects the function of the protein this might have clinically relevant implications.
Network protein analysis by IPA of the high-affinity proteins for FWF and Fe2O3 in shows common connections for nuclear factor kappa beta (NFκB) and tumor necrosis factor alpha (TNF-α), while inerleukin-6 (IL-6) showed a connection for UFWF (Figures S9–S11). This indicates that such binding can influence an inflammatory process. No such connections were found for Fe3O4 (Figure S12).
Effect of particle–protein interaction on protein functionality
SLPI bound to UFWF and Fe2O3 particles at an Rb/u between 16 and 28 () but bound only weakly to FWF and Fe3O4. This protein was selected for a functionality testing due to the high abundancy in the nasal lavage and high ratio bound to the particles. Before the experiment, the size of the SLPI:particle aggregates was determined (). The diameter of the formed aggregates was large but could only be determined for UFWF and Fe2O3. No reproducible results could be obtained for the other particle types.
The potential effect on SLPI function due to the particle binding was tested by measuring the protease activity of HNE on elastine using a fluorogenic substrate (instead of elastine) that gives a fluorescent signal when cleaved (Figure S13A). HNE activity is inhibited by SLPI, (Figure S13B), and we found that increasing concentrations of SLPI alone correlated with a decreasing fluorescent signal demonstrating inhibition of HNE (). The addition of particles without SLPI had no effect on the HNE activity (data not shown). Low concentrations of SLPI:UFWF and SLPI:Fe2O3 aggregates eliminated the inhibitory effect of SLPI ( and Figure S13C), and this indicated that SLPI function was lost upon binding to the UFWF and Fe2O3 particles. At high concentrations of these aggregates, the inhibitory effect was equal to that of SLPI alone, and this was probably due to maximum inhibition being reached. The SLPI:FWF aggregates showed the opposite effect, however, and inhibition of HNE by SLPI was actually increased in these particles. The decreased signal can be explained either by SLPI binding to FWF particles in a way that improves the protein's function or by steric hindrance for the substrate to reach to HNE due to large SLPI:FWF aggregates.
Figure 3. HNE substrate inhibition. Detection of SLPI inhibition of HNE activity was performed with (A) free SLPI compared with SLPI:FWF, SLPI:UFWF, and SLPI:Fe2O3 protein corona complexes and (B) nasal lavage protein (NL, which contains endogenous SLPI) compared with NL:FWF, NL:UFWF, and NL:Fe2O3 protein corona complexes.
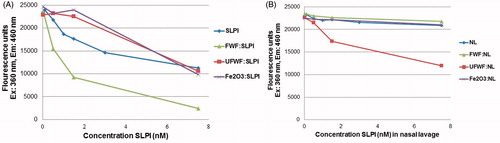
We next measured the endogenous SLPI inhibition of HNE in the nasal lavage protein background containing endogenous SLPI (). No inhibition of HNE was observed in nasal lavage protein alone indicating that SLPI primarily inhibits free endogenous HNE present in nasal lavage protein solution or that the effect of SLPI on HNE is delayed due to the presence of other nasal lavage protein proteins. Therefore, no particle effect was expected, and this was the case for FWF and especially for Fe2O3 because it binds to endogenous HNE (P08246, ). However, adding UFWF particles to nasal lavage decreased the HNE activity. It is possible that differences in the protein corona formation influenced the function of bound SLPI. Thus, it is difficult to predict the influence of protein function induced by particle binding, but changes in activity are clearly possible and are likely to be dependent on particle type.
Discussion
The characteristics of the agglomerates depend on the welding process temperature and on the vapor pressure of the elements. Therefore, the particles were collected as two fractions, FWF and UFWF, according to the aerodynamic size of the particles.
DLS is sensitive to larger particles since the intensity of the scattered light is proportional to the sixth power of the particle diameter. It has a low peak resolution, and particles can only be resolved if the size differs by a factor of 3. This makes it difficult to measure an exact size of the agglomerates. But it is fast, easy to operate, and delivers a large amount of statistical data. The shape and the structure of the agglomerates were confirmed with TEM.
Measurement of the formation of the protein corona revealed that although the hydrodynamic particle mean size was similar for Fe2O3 and UFWF particles, Fe2O3 bound twice the amount of proteins than the UFWF particles. Thus the chemical composition of the primary particle along with the primary particle size of the agglomerates might determine the specific surface area available for binding (surface area per mass unit).
Both 2DE-MALDI-TOF MS and targeted LC-MS/MS revealed distinct differences in the protein coronas between Fe3O4 and the other particles. But conflicting results were obtained regarding, for example, lactotransferrin. Targeted analysis showed that three of the particles bound this protein to a high degree, but the 2DE separation and MALDI-TOF MS analysis did not confirm this finding. Prolactin-inducible protein was detected by the 2DE method but not with SRM. Discrepancies between proteins identified by the two methods might arise from a number of methodological differences, e.g. that targeted peptides must be unique for the identified protein or that parameters such as hydrophobicity and abundance might affect the results of 2DE. Furthermore, digestion by trypsin could be sterically inhibited when the protein is bound to the particle. This emphasizes the importance of combining methods.
In a real exposure scenario, many factors, such as particle number concentration, the deposited surface area, and the amount and composition of nasal lavage proteins, might influence the formation of the protein corona. The formation of the protein corona is a dynamic process, and it is possible that in the upper airways the protein corona is constantly being partly or completely changed. There will be a continuous secretion and removal of nasal fluids and deposited particles, and it is possible that the nasal fluid composition can change in response to particle deposition (Albanese et al., Citation2014). These factors, together with the time it will take for equilibrium between proteins and particle surface to be reached (Dell'orco et al., Citation2010; Lundqvist et al., Citation2011; Tenzer et al., Citation2013), suggest the possibility that there are many different protein coronas present around particles in the nasal fluid at any given time. Furthermore, in the in vitro situation, the ratio of particles and proteins will be important. A large number of particles, i.e., a large surface area, in diluted protein samples, will enable proteins with low affinity to the particles to interact due to the excess of particle surface area (Dell'orco et al., Citation2010). This might lead to the identification of proteins in the corona that would never occur in a real-life scenario where the surface areas are likely to be limited. In this study, we have identified proteins binding with a high affinity to the particles. These proteins were selected due to an increased probability that they remain in the corona for a long time and thus have an increased probability of either influencing the distribution route of the particle or being subjected to inherent functionality changes.
There were still differences between the overall protein coronas in terms of which high-affinity binding proteins were associated with the different particle types (). The protein corona of Fe3O4 was distinct compared with the three other particles, and this suggests that the composition of the protein corona depends on particle size and/or chemical composition.
A loss of inhibitory function of SLPI was observed when the protein was incubated with UFWF and Fe2O3 particles in an HNE activity assay. The FWF particles, however, appeared actually to not cause any loss of the inhibitory function of SLPI. Thus, it is difficult to predict the potential influence that particle binding has on protein functionality, but this is still a relevant factor to investigate. The majority of the amount of SLPI in nasal lavage was shown to bind to UFWF and Fe2O3 particles leaving little unbound SLPI left. Thus, any functional alteration would have a significant impact on the overall mechanism. In general, binding of proteins to nanoparticles may be an important mechanism underlying a toxicological response due to any functional alteration induced during the particle-protein interaction.
Most of the proteins presented in are associated with acute phase responses and immunological responses, and differences in the protein corona might have implications for the biological response caused by the particles. The somewhat different coronas identified for different particles suggest that the initiation of different biological pathways could be expected upon inhalation of the particles. Example of proteins with potential interest is BPI fold-containing family B member 4, which was found to bind to a much higher degree to Fe3O4 than to the FWF and Fe2O3 particles. This protein has a lipid binding capacity, and it also acts as an odorant carrier through the mucus to receptor sites. Lipocalin-15, a transport protein that enables direct movement of substances into the cell, was found to bind almost exclusively to Fe3O4 particles. Previous studies have shown cellular uptake of nanoparticles through endocytosis (Albanese et al., Citation2014; Chen et al., Citation2011), and it might be possible that nanoparticles with strong affinity for these proteins will be taken up by cells more efficiently. However, this hypothesis needs to be further evaluated, for example, by exposing cells to particles coated with these proteins.
Conclusions
In this study, particles were collected from welding fumes and compared with model particles with a well-defined size distribution and less chemical complexity. Analyses of the protein corona formation and the identification of the protein corona formed between different particle types and nasal lavage proteins were performed. The results showed that chemical composition, physical shape, and agglomeration state of the particle influenced particle–protein interactions and functionality of bound proteins. In addition, the protein pattern of the corona differed between the particle types. Thus, it is important to include particles from real working environments and not just study more homogenous model particles.
In summary we demonstrate that
The amount of protein bound to each particle type correlate with the relative size increase of the aggregates, suggesting that the surface area per mass unit determines the binding capacity.
The relative increase in aggregate size following addition of nasal lavage proteins to particles showed that the increase was not only associated with particle size.
The overall nasal lavage protein corona differed between particle types.
The Fe3O4 particles, composed of the smallest agglomerates, had a very specific protein corona.
The protein corona that formed did not correlate with the abundance of the proteins or with their size or net charge.
Subsets of proteins were identified as binding with high affinity to different particles. These proteins should be investigated in further studies because they might have relevant associations with toxicological responses.
When proteins bind to particles, the function of the protein can be influenced in different ways as shown by SLPI in this study. The inhibitory function of SLPI was decreased when bound to UFWF and Fe2O3 particles, but not when bound to FWF particles.
Supplementary material
Supplementary material is available in Figures S1-S13 and Tables S1-S4.
Supplementary Materials
Download Zip (2.5 MB)Declaration of interest
The authors declare that they have no competing interests. The work presented here was funded by AFA Insurance and Forte, The medical faculty of Lund University and The Nanometer Structure Consortium (nmC) at Lund University.
References
- Albanese A, Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW. 2014. Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles. Acs Nano 8:5515–26
- Antonini JM. 2003. Health effects of welding. Crit Rev Toxicol 33:61–103
- Bantscheff M, Lemeer S, Savitski MM, Kuster B. 2012. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem 404:939–65
- Bartlett JA, Albertolle ME, Wohlford-Lenane C, Pezzulo AA, Zabner J, Niles RK, et al. 2013. Protein composition of bronchoalveolar lavage fluid and airway surface liquid from newborn pigs. Am J Physiol Lung Cell Mol Physiol 305:L256–66
- Benson LM, Mason CJ, Friedman O, Kita H, Bergen Iii HR, Plager DA. 2009. Extensive fractionation and identification of proteins within nasal lavage fluids from allergic rhinitis and asthmatic chronic rhinosinusitis patients. J Sep Sci 32:44–56
- Bewick V, Cheek L, Ball J. 2004. Statistics review 10: further nonparametric methods. Crit Care 8:196–200
- Casado B. 2004. Proteomics for nasal secretion analysis. Curr Allergy Asthma Rep 4:224–9
- Casado B, Pannell LK, Iadarola P, Baraniuk JN. 2005. Identification of human nasal mucous proteins using proteomics. Proteomics 5:2949–59
- Cedervall T, Lynch I, Lindman S, Berggård T, Thulin E, Nilsson H, et al. 2007. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci USA 104:2050–5
- Chen C-L, Zhang H, Ye Q, Hsieh W-Y, Hitchens TK, Shen H-H, et al. 2011. A new nano-sized iron oxide particle with high sensitivity for cellular magnetic resonance imaging. Mol Imaging Biol 13:825–39
- Dell'orco D, Lundqvist M, Oslakovic C, Cedervall T, Linse S. 2010. Modeling the time evolution of the nanoparticle-protein corona in a body fluid. PLoS One 5:1–8
- Demokritou P, Kavouras I, Ferguson S, Koutrakis P. 2002. Development of a high volume cascade impactor for toxicological and chemical characterization studies. Aerosol Sci Technol 36:925–33
- Diz AP, Carvajal-Rodriguez A, Skibinski DO. 2011. Multiple hypothesis testing in proteomics: a strategy for experimental work. Mol Cell Proteomics 10:M110 004374
- De Uña-Alvarez J, Carvajal-Rodriguez A. 2010. ‘SGoFicance Trace’: assessing significance in high dimensional testing problems. PLoS One 5:e15930
- Ghafouri B, Karlsson H, Mörtstedt H, Lewander A, Tagesson C, Lindahl M. 2007. 2,5-Dihydroxybenzoic acid instead of alpha-cyano-4-hydroxycinnamic acid as matrix in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for analyses of in-gel digests of silver-stained proteins. Anal Biochem 371:121–3
- Isaxon C, Dierschke K, Pagels J, Löndahl J, Gudmundsson A, Hagerman I, et al. 2013. A novel system for source characterization and controlled human exposure to nanoparticle aggregates generated during gas–metal arc welding. Aerosol Sci Technol 47:52–9
- Jenkins NT, Eagar TW. 2005. Chemical analysis of welding fume particles – Airborne particle size is the most important factor in determining the accuracy of a method for chemical analysis. Weld J 84:87S–93
- Jensen KA. 2011. WP 4: Physicochemical Characterisation of Manufactured Nanomaterials (MNs) and Exposure Media (EMs). Available from: http://www.nanogenotox.eu/files/PDF/web%20nanogenotox%20dispersion%20protocol.pdf. Accessed on 29 June 2015
- Kosanam H, Sato M, Batruch I, Smith C, Keshavjee S, Liu M, Diamandis EP. 2012. Differential proteomic analysis of bronchoalveolar lavage fluid from lung transplant patients with and without chronic graft dysfunction. Clin Biochem 45:223–30
- Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, et al. 2011. The evolution of the protein corona around nanoparticles: a test study. Acs Nano 5:7503–9
- Maynard AD, Kuempel ED. 2005. Airborne nanostructured particles and occupational health. J Nanopart Res 7:587–614
- Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB, Dawson KA. 2011. Physical–chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc 133:2525–34
- Mygind N, Dahl R. 1998. Anatomy, physiology and function of the nasal cavities in health and disease. Adv Drug Deliv Rev 29:3–12
- Mörtstedt H, Kåredal MH, Jönsson BAG, Lindh CH. 2013. Screening method using selected reaction monitoring for targeted proteomics studies of nasal lavage fluid. J Proteome Res 12:234–47
- Nawrot T, Nemmar A, Nemery B. 2006. Update in environmental and occupational medicine 2005. Am J Respir Crit Care Med 173:948–52
- Oberdorster G, Oberdorster E, Oberdorster J. 2005. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect 113:823–39
- Saptarshi SR, Duschl A, Lopata AL. 2013. Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle. J Nanobiotechnol 11:26 doi: 10.1186/1477-3155-11-26
- Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–8
- Simpson DG, Margolin BH. 1986. Recursive nonparametric testing for dose-response relaflonships subject to downturns at high doses. Biometrika 73:589–96
- Sjogren B, Langard S. 2004. RE: pulmonary effects of welding fumes: review of worker and experimental animal studies Antonini et al. 2003. Am J Ind Med 43:350–60 (Am J Ind Med 45:478–9)
- Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, et al. 2013. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat Nano 8:772–81
- Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, et al. 2011. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. Acs Nano 5:7155–67