Abstract
Protein expression data suggests that the SPANX gene family is expressed throughout spermatogenesis, including post-meiotic expression, consistent with a potential role in sperm development. The genomic architecture of this region is unstable and past studies have found evidence of variation in this gene family. This study used a novel assay to evaluate copy number variation (CNV) in SPANX gene family, in fertile and infertile men. The case group was comprised of 50 oligozoospermic and 50 azoospermic men, and the control group was comprised of 67 normozoospermic men with children. The assay, real-time quantitative PCR, evaluated CNV of the entire gene cluster containing all four SPANXA-E genes and with SPANXB, found exclusively in maturing sperm. While variation was found in both groups, average CNV patterns did not differ between fertile and infertile males. As this was a targeted assay, it was limited in scope to detect further CNV at a genome-wide level which is an area of increasing interest in the field of genomics.
| Abbreviations | ||
| CNV: | = | copy number variation |
| bp: | = | base pairs |
| QRT-PCR: | = | quantitative real-time polymerase chain reaction |
| LC480: | = | LightCycler 480 |
| arrayCGH: | = | array comparative genomic hybridization |
INTRODUCTION
Male infertility is a significant health concern whose etiological basis is currently poorly understood [Ferlin et al. Citation2006]. Established genetic causes of male infertility include point mutations and polymorphisms of genes relevant to spermatogenesis, micro-deletions of the Y chromosome, unbalanced trans-locations, and chromosomal loss or gain [Kuroda-Kawaguchi et al. Citation2001; Carrell et al. Citation2006]. Furthermore, evidence of infertility resulting from chromosomal abnormalities, such as inversions and translocations, suggest a possible role for additional copy number variants like deletion or insertion events [Martin Citation2008]. By improving our understanding of potential genetic influences on fertility, clinicians and fertility specialists can help identify specific classes of infertility to better target medical treatment and inform prevention. One exciting new area of genetic analysis focuses on the role of copy number variation (CNV) in complex disease risk [Lee et al. Citation2007]. CNV analysis has yet to be applied to the analysis of male infertility. This study investigates a possible association between copy number variation in the SPANX gene cluster at Xq27 and male infertility. SPANX gene expression has been linked with spermatogenesis [Salemi et al. Citation2004; Citation2006; Westbrook et al. Citation2000; Citation2006] making the gene cluster a plausible candidate region for fertility and related phenotypes.
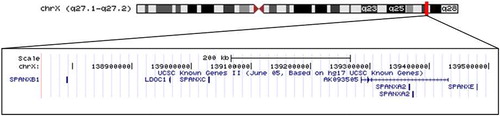
The SPANX gene cluster is comprised of four highly similar genes (A, B, C, and E) on the X chromosome (). These genes lie in a ∼700 kb wide region composed of several large duplicated units (segmental duplications) with high sequence identity (>95%) that are prone to genomic rearrangement potentially causing partial or complete deletion or duplication of the SPANX cluster [Lupski Citation1998; Stankiewicz and Lupski Citation2002; Sharp et al. Citation2005]. Each gene is 1,000 base pairs (bp) in length and contains two small exons that encode a unique protein, which is between 97–103 amino acids in length. The SPANX genes are homologous, approximately 96–98% identical; SPANXB contains an 18-bp insert in the first exon that renders it distinct from the other genes in the cluster.
FIGURE 1 The SPANXA-E gene family is located at Xq27.1-.2 across a 700 kb region at chrX:139,875,000–139,500,000. This UCSC Genome Browser image downloaded in August 2009 using the July 2003 human genome track and Known Gene II function.
SPANX genes encode the cancer-testis class of antigens [Westbrook et al. Citation2004] and extensive protein antibody analysis provides evidence that expression of the human gene family is limited to the testis and malignant tumors [Salemi et al. Citation2004, Citation2006; Westbrook et al. Citation2000; Citation2004; Citation2006; Zendman et al. Citation2003]. Little is understood, however, about the exact function of the individual SPANX genes. All SPANX proteins, except SPANXC, are found exclusively in germ cells, and all the SPANX proteins, except SPANXB, are found at some level in various tumor cells. Current understanding of the role of the SPANX protein during spermatogenesis is still incomplete [Martínez-Heredia et al. Citation2006], but there is consensus that SPANX genes are expressed throughout spermatogenesis, from sperm cell precursors to ejaculated spermatozoa [Salemi et al. Citation2004, Citation2006; Westbrook et al. Citation2000; Citation2006]. To date the role of SPANX gene variation with male fertility has not been evaluated.
RESULTS
SPANXB Copy Number Analysis
There are two tandem, identical copies of SPANXB located between chrX: 139,810,215–139,823,500. Each is 1,115 bp in length with two exons. Therefore, the RT-PCR assay is expected to identify two copies in hemizygous males. Males with more than two copies of the SPANX gene cluster will register values greater than a normal male with two copies, but less than or equivalent to a female's signal from four copies of the cluster on two X chromosomes. A male missing one or both SPANXB genes would have little to no signal. The custom microarray chips contain dozens of probes across the SPANXA-E region that can be averaged to generate a value that can be interpreted to make a call of either duplication, deletion, or no copy number change. These data were used to calibrate the SYBR Green assay by comparing the two different values for a set of individuals interrogated by the arrayCGH chips, in this case 7 females and one male from the HapMap panel (4 Yorubans, 1 Han Chinese, 1 Japanese, 2 CEPH Utah). contains the relative copy number values from the oligonu-cleotide array and SYBR assays, and the correlation coefficient for both primer pairs, SPANXB () and SPANXA-E () exceeded the threshold value of .85. Copy number values were calculated for each individual from the infertility and fertility panels. Two infertile individuals were extreme outliers with copy number values exceeding 5, but were very noisy and did not validate on an SNP-based platform and thus were removed from evaluation [Aston unpublished]. Infertile men were sorted into two different groups, oligozoospermic and azoospermic, and compared to the fertile group for the SPANXB assay. Copy number (CN) was calculated for 96 infertile males and 51 fertile men. The average value for the infertile group was .52 (.03–1.96), and the average value for the fertile group was .47 (.01–1.7). These values are consistent with the expected value of .5 indicating one X in a male in comparison to a female with two X chromosomes. The azoospermic group reported an average mean of .51 (), and the oligozoospermic group reported an average mean of .44 (). In general, oligo and azoospermic men showed similar patterns of variation in SPANXB copy number to the fertile men. The reference individual is female with a relative copy number of 1, and the expected value of a male should be .5.
FIGURE 2 SPANXA-E copy number values for oligozoospermic men (black dots, N = 44) compared to fertile controls (grey triangles, N = 48). The first data point represents the baseline female control, and the lines represent the combined standard deviation of both the control primer and test primer amplifications. The lines represent the combined standard deviation of both the control primer and test primer amplifications. The average means between both groups are approximately the same.
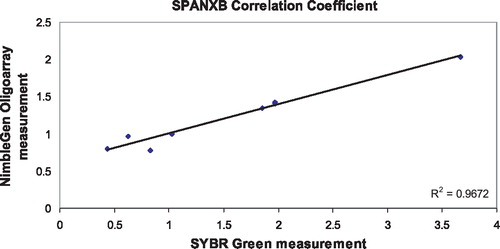
FIGURE 3 SPANXA-E copy number values for azoospermic men (black dots, N = 49) compared to fertile controls (grey triangles, N = 48). The first data point represents the baseline female control, and the lines represent the combined standard deviation of both the control primer and test primer amplifications. The lines represent the combined standard deviation of both the control primer and test primer amplifications. The average means between both groups are approximately the same.
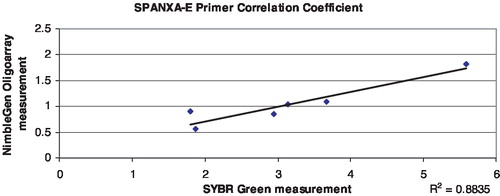
FIGURE 4 Comparison of NimbleGen oligoarray and SYBR Green real-time quantification values for 7 HapMap individuals using the SPANXB primers.
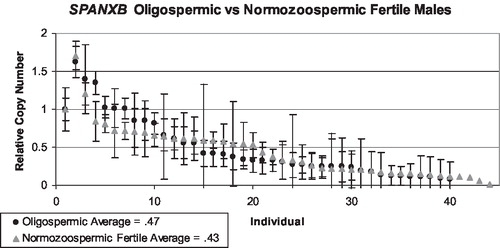
FIGURE 5 Coefficient chart comparing NimbleGen oligoarray and SYBR green real-time quantification values for six HapMap individuals using the SPANXA-E primers.
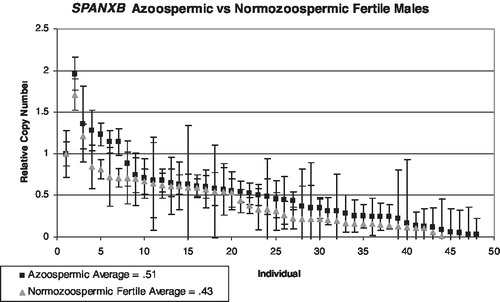
TABLE 1 Copy Number Estimations for 7 Individuals from the HapMap Panel that Were Evaluated Using Both NimbleGen Oligoarray and SYBR Green Real-time Quantification for Both SPANXB and SPANXA-E.
SPANX Gene Cluster Copy Number Analysis
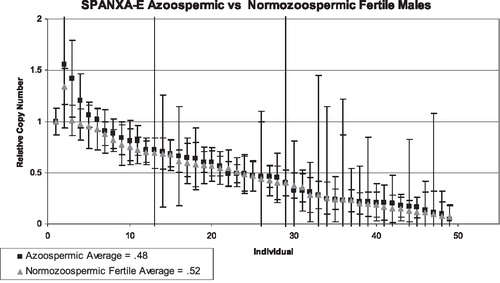
The SPANX gene cluster contains four homologous genes—SPANXA, SPANXB (two identical tandem copies of each), SPANXC, and SPANXE. Copy number values were calculated for each individual using a PCR that simultaneously targeted a sequence common to all four genes. As with the SPANXB analysis, infertile men were sorted into two different groups, oligozoospermic and azoospermic, and compared to the fertile group for each gene target. CN was calculated for 96 infertile males and 44 fertile men. The average value for the infertile group was .49 (.05–2.6), and the average value for the fertile group was .44 (.35–1.34). These values are consistent with the expected value of .5 indicating one X in a male in comparison to a female with two X chromosomes. The oligozoospermic group reported an average mean of .46 (), while the azoospermic group reported an average mean of .5 (). In general, oligo and azoospermic men showed similar patterns of variation to the fertile men.
FIGURE 6 SPANXB copy number values for oligozoospermic men (black dots, N = 43) compared to normozoospermic fertile controls (grey triangles, N = 43). The first data point represents the baseline female control, and the lines represent the combined standard deviation of both the control primer and test primer amplifications. The average means between both groups are approximately the same.
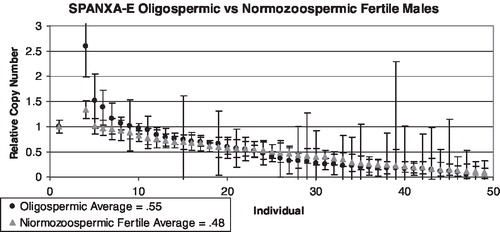
FIGURE 7 SPANXB copy number values for azoospermic men (black dots) compared to fertile controls (grey triangles). The first data point represents the baseline female control, and the lines represent the combined standard deviation of both the control primer and test primer amplifications. The lines represent the combined standard deviation of both the control primer and test primer amplifications. The average means between both groups are approximately the same.
DISCUSSION
Male factor infertility is a complex disease caused by a number of environmental, genetic, chemical, and developmental factors. Current estimates suggest that genetic causes account for 10% of male factor infertility with an identifiable cause [Ferlin et al. Citation2006]. Approximately 60% of male infertility is of an unknown origin. To date, the majority of genetic studies have primarily focused on point mutations in candidate genes and a well-characterized microdeletion of the AFZ region at Yq11 which is rich in spermatogenic genes and strongly associated with male factor infertility [Lin et al. Citation2007; Giachini et al. Citation2005, Citation2008]. A recent study of genomic copy number varition of Asian men with congenital bilateral absence of the vas deferens (CBAVD) but without the most common CFTR-associated mutant did identify CNV in genes possibly associated with the disorder [Lee et al. Citation2009]. Initial studies of CNVs have shown associations in complex disorders such as systemic lupus, HIV acquisition, and breast cancer [Yang et al. Citation2007; Shao et al. Citation2007; Frank et al. Citation2007].
In this study, a novel assay was developed to assess copy number variation (CNV) of the SPANXA-E cluster, a gene family located within a complex region of chromosome X shown previously to be prone to genomic rearrangements. Although there were good prior reasons to suspect that CNV of the gene cluster might contribute to male infertility, two different assays, including one targeted specifically to the SPANXB gene and one designed to measure CNV across the SPANXA-E gene cluster, showed no difference in copy number between fertile and infertile study samples. Either CNV of the SPANX region is not a major contributor to infertility in the samples examined here or the chosen assay was not sensitive enough to detect possible copy number differences. There are a few key limitations in the SYBR Green QRT-PCR approach. First, this assay cannot measure exact copy number in duplicated regions; therefore, CNV calls are limited to simple descriptions as a deletion or duplication. A male with a value equivalent to a female value is considered to have a duplication, but an exact number cannot be ascertained from this value. Second, as with any study, an additional assay would be performed to validate any extreme findings. For this study, the amount of genomic DNA was extremely limited and unavailable for additional testing. There is a certain level of noise involved in the SYBR assay so a commonly used assay, oligoarray technology, could be used to verify potential duplications and deletions seen in this study, particularly the two outliers. One final limitation is the lack of data available on population frequency of duplications and deletions of this region. The vast majority of CNV studies have limited analysis to autosomes, therefore no data is available on the population frequency of duplication or deletion events in the SPANXA-E region. Without this data and additional validation of outliers, it is not possible to determine what falls within a normal range of variation and what events are unique in this data set.
This study represents one of the few evaluations of a potential association between copy number of a candidate gene and male infertility. Previous studies have found strong associations between male infertility and microdeletions on the Y chromosome. Future studies evaluating genome-wide copy number variation in association with male or female infertility should be conducted to identify additional regions and genes.
MATERIALS AND METHODS
Participant Sample
The population sample for this study consisted of 50 azoospermic men, 50 severely oligozoospermic men (<5 million sperm/mL), and 67 fertile, normozoo-spermic men, as defined by WHO [Citation1999] criteria. Patients with known Y-chromosome microdeletion, cystic fibrosis, varicocele, Klinefelter's syndrome, or prior exposure to chemotherapeutics or radiation were excluded. Subjects were screened and consented to participate by the Andrology Laboratories at University of Utah following Institutional Review Board (IRB) approval. Donors provided blood for genetic research and DNA was extracted from peripheral lymphocytes with the use of the Puregene DNA extraction kit (Gentra Systems, Minneapolis, MN) at the University of Utah. The IRB at the University of Washington determined it was unnecessary to obtain Human Subjects approval for this CNV analysis because de-identified genetic samples were used.
Identification of SPANX Gene Cluster CNV by Real-Time Quantitative PCR
Two different real-time quantitative PCR (RT-PCR) assays were developed to assess CNV in the SPANX gene cluster. The first involved PCR amplification of the SPANXB gene, using primers designed to amplify a 96-bp region of single-copy DNA nested uniquely within the locus: Forward: 3′CATTGCCGAAGTTT-GAGGGATG-5′, Reverse: CCAACCCCGACCAAATG-GAGG; annealing temperature 58˚C. The second assay involved PCR amplification of a 129-bp fragment homologous to a sequence found in each of the four SPANX genes: Forward: CCTTTGATATGTC-CATTCAGCAGAACTATG, Reverse: CTGGGGGAGG-ACTGATTCCTAAAG; annealing temperature 60˚C. The latter assay provides an indication of copy number variation as it may involve the entire gene cluster, as opposed to single gene. The reference amplification product targeted the well-known housekeeping gene human albumin (ALB), located at chr4q13.3, and was initially reported for use in CNV evaluation using a SYBR Green Dye assay by Brasch-Anderson et al. [Citation2004]. The forward primer is 5′-GCTGTCATCTCTTGTGGGCTGT-3′ and the reverse primer is 5′-ACTCATGGGAGCTGCTGGTTC-3′. To quantify DNA copy number, each fragment was amplified using SYBR Green Dye and genomic copy number was calculated from a real-time analysis of dye-tagged DNA fragments produced during amplification in a Roche LightCycler 480 (Roche Applied Sciences, Indianapolis, IN, USA). The value used to determine copy number is generated by a laser scanner measuring the exponential doubling of SYBR Green fluorescence as it binds to double-stranded DNA throughout the PCR process, which provides a cycle count value.
The SYBR assay was optimized and calibrated to detect SPANXA-E and SPANXB CNV accurately using previous microarray copy number quantification data. These microarray data were generated through array comparative genomic hybridization (arrayCGH). Array-CGH data are generated by labeling two individuals with different fluorescent dyes and hybridizing them to a chip with targeted DNA. By comparing a test individual to a reference individual, duplicated or deleted sections of the genome can be identified. For the SYBR assay, the reference individual is the same female that has been extensively assayed using microarray and paired-end sequencing to detect CNVs in multiple previous studies. To verify the accuracy of the primer amplification, i.e. to ensure that DNA sequences within the SPANX gene cluster were appropriately targeted by the chosen PCR primers, amplification products of each PCR primer pair were sequenced directly using an ABI3700. The resultant DNA sequence data confirmed that the PCR products were specifically amplifying their intended targets in the SPANX gene cluster.
Statistical Analysis
Estimates of copy number were derived using cycle count methodology, a technique widely used in the analysis of RT-PCR data [Livak and Schmittgen Citation2001]. This methodology requires two types of reference information: a reference individual with a known copy number for the assayed region, and a reference amplification product targeted to a single-copy gene unlinked to the region under investigation. In this study, the reference individual was a HapMap female (GM15510) with copy number determined through deep-resequencing [Kidd et al. Citation2008] and showing no evidence of duplication or deletion of the SPANX gene cluster. As the reference individual is female, the reference copy number is 1; therefore, the expected copy number value of a male would be approximately 0.5. Each individual was run in quadruplicate, with the average cycle count value used for the reference and test sample. Individuals with a standard deviation exceeding .5 were repeated. In total, copy number values with standard deviations below the threshold were calculated for (96) infertile and (49) fertile individuals.
ACKNOWLEDGMENTS
Initial support for this work provided by The Institute of Public Health Genetics, University of Washington. Current funding for this project provided by the UW Multidisciplinary Predoctoral Research Training Program—a Roadmap Initiative from the National Institutes of Health/National Center for Research Resources (T32 RR0203256), P. Mitchell & S. Marshall, Program Directors. Evan E. Eichler is an investigator of the Howard Hughes Medical Institute.
Declaration of Interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- Brasch-Andersen, C., Christiansen, L., Tan, Q., Haagerup, A., Vestbo, J. and Kruse, T. A. (2004) Possible gene dosage effect of glutathione-S-transferases on atopic asthma: Using real-time PCR for quantification of GSTM1 and GSTT1 gene copy numbers. Human Mutation 24:208–214.
- Carrell, D. T., De Jonge, C. and Lamb, D. J. (2006) The genetics of male infertility: A field of study whose time is now. Arch Androl 52(4):269–274.
- Ferlin, A., Arredi, B. and Foresta, C. (2006) Genetic causes of male infertility. Repro Tech 22:133–141.
- Frank, B., Bermejo, J. L., Hemminki, K., Sutter, C., Wappenschmidt, B., Meindl, A., Kiechle-Bahat, M., Bugert, P., Schmutzler, P. K., Bartram, C. R. and Burwinkel, B. (2007) Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis 28(7):1442–1445.
- Giachini, C., Guarducci, E., Longepied, G., Degl'Innocenti, S., Becherini, L., Forti, G., Mitchell, M. J. and Krausz, C. (2005) The gr/gr deletion(s): A new genetic test in male infertility? J Med Genet 42:497–502.
- Giachini, C., Laface, I., Guarducci, E., Balercia, G., Forti, G. and Krausz, C. (2008) Partial AZFc deletions and duplications: Clinical correlates in the Italian population. Hum Genet 124(4)399–10.
- Kidd, J. M., Cooper, G. M., Donahue, W. F., Hayden, H. S., Sampas, N., Graves, T., Hansen, N., Teague, B., Alkan, C., Antonacci, F., . (2008) Mapping and sequencing of structural variation from eight human genomes. Nature 453(7191):56–64.
- Kuroda-Kawaguchi, T., Skaletsky, H., Brown, L. G., Minx, R J., Cordum, H. S., Waterston, R. H., Wilson, R. K., Silber, S., Oates, R., Rozen, S. and Page, D. C. (2001) The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 29:279–286.
- Lee, C, lafrate, A. J. and Brothman, A. R. (2007) Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nature Genetics Supplement 39:S48–S54.
- Lee, C.-H., Wu, C.-C. and Chiang, H.-S. (2009) Gene copy number variations in Asian patients with congenital bilateral absense of the vas deferens. Human Reprod 24(3):748–755.
- Lin, Y. W., Hsu, L. C., Kuo, P. L., Huang, W. J., Chiang, H. S., Yeh, S. D., Hsu, T. Y., Yu, Y H., Hsiao, K. N., Cantor, R. M. and Yen, P. H. (2007) Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat 28(5):486–494.
- Livak, K. J. and Schmittgen, T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402.
- Lupski, J. (1998) Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. TIG 14:417–422.
- Martin, R. H. (2008) Cytogenetic determinants of male fertility. Hum Reprod Update 14(4):379–390.
- Martinez-Heredia, J., Estanyol, J. M., Ballesca, J. L. and Oliva, R. (2006) Proteomic identification of human sperm proteins. Proteomics 6:4356–1369.
- Salemi, M., Calogero, A. E., Di Benedetto, D., Cosentino, A., Barone, N., Rappazzo, G. and Vicari, E. (2004) Expression of SPANX proteins in human-ejaculated spermatozoa and sperm precursors. Int J Androl 27:134–139.
- Salemi, M., Calogero, A. E., Di Benedetto, D., Cosentino, A., Barone, N., Rappazzo, G. and Vicari, E. (2006) Expression of SpanX proteins in normal testes and in testicular germ cell tumors. Int J Androl 29:368–373.
- Shao, W., Tang, J., Song, W., Wang, C., Li, Y., Wilson, C. M. and Kaslow, R. A. (2007) CCL3L1 and CCL4L1: Variable gene copy number in adolescents with and without human immunodeficiency virus type 1 (HIV-1) infection. Genes Immun Apr:8(3):224–231.
- Sharp, A. J., Locke, D. P., McGrath, S. D., Cheng, Z., Bailey, J. A., Vallente, R. U., Pertz, L. M., Clark, R. A., Schwartz, S., Segraves, R., . (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77(1):78–88.
- Stankiewicz, P. and Lupski, J. (2002) Genome architecture, rearrangements, and genomic disorders. Trends Genets 18:74–82.
- Westbrook, V. A., Diekman, A. B., Klotz, K. L., Khole, V. V., von Kap-Herr, C., Golden, W. L., Eddy, R. L., Shows, T. B., Stoler, M. H., Lee, C. Y., . (2000) Spermatid-specific expression of the novel X-linked gene product SPAN-X localized to the nucleus of human spermatozoa. Biol Reprod 63:469–181.
- Westbrook, V. A., Schoppee, P. D., Diekman, A. B., Klotz, K. L., Allietta, M., Hogan, K. T., Slingluff, C. L., Patterson, J. W., Frierson, H. F., Irvin, W. P., Jr., . (2004) Genomic organization, incidence, and localization of the SPAN-x family of cancer-testis antigens in melanoma tumors and cell lines. Clin Cancer Res 1:10(1 Pt 1): 101–112.
- Westbrook, V. A., Schoppee, P. D., Vanage, G. R., Klotz, K. L., Diekman, A. B., Flickinger, C. J., Coppola, M. A. and Herr, J. C. (2006) Hominoid-specific SPANXA/D genes demonstrate differential expression in individuals and protein localization to a distinct nuclear envelope domain during spermatid morphogenesis. Mol Hum Reprod 12(11):703–716.
- World Health Organization (1999) WHO laboratory manual for the examination of human and sperm-cervical mucus interactions. Cambridge University Press, Cambridge, UK.
- Yang, Y., Chung, E. K., Wu, Y L, Savelli, S. L., Nagaraja, H. N., Zhou, B., Hebert, M., Jones, K. N., Shu, Y., Kitzmiller, K., . (2007) Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet Jun:80(6): 1037–1054.
- Zendman, A. J., Zschocke, J., van Kraats, A. A., Kurpisz, M., Weidle, U. H., Weiss, E. H. and van Muijen, G. N. (2003) The human SPANX multi-gene family: genomic organization, alignment and expression in male germ cells and tumor cell lines. Gene 309(2):125–133.