Abstract
Common aspects of infertility can be seen across several species. In humans, dairy cows, and mares there is only a 25-35% chance of producing a live offspring after a single insemination, whether natural or artificial. Oocyte quality and subsequent embryo development can be affected by factors such as nutrition, hormonal regulation, and environmental influence. The objective of this study was to identify genes expressed in oocytes and/or cumulus cells, across a diverse range of species, which may be linked to the ability an oocyte has to develop following fertilization. Performing a meta-analysis on previously published microarray data on various models of oocyte and embryo quality allowed for the identification of 56 candidate genes associated with oocyte quality across several species, 4 of which were identified in the cumulus cells that surround the oocyte. Twenty-one potential biomarkers were associated with increased competence and 35 potential biomarkers were associated with decreased competence. The upregulation of Metap2, and the decrease of multiple genes linked to mRNA and protein synthesis in models of competence, highlights the importance of de novo protein synthesis and its regulation for successful oocyte maturation and subsequent development. The negative regulation of Wnt signaling has emerged in human, monkey, bovine, and mouse models of oocyte competence. Atrx expression was linked to decreased competence in both oocytes and cumulus cells. Biological networks and transcription factor regulation associated with increased and decreased competence were also identified. These genes could potentially act as biomarkers of oocyte quality or as pharmacological targets for manipulation in order to improve oocyte developmental potential.
Keywords:
Introduction
Common aspects of infertility can be seen across several species. In humans, dairy cows, and mares there is only a 25–35% chance of producing a live offspring after a single insemination, whether natural or artificial [Lucy Citation2001; Shah et al. Citation2003; Squires et al. Citation1998]. Several assisted reproductive technologies (ARTs) have been developed for use in the human reproduction field in order to aid fertility, including in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI). It is estimated that over 3 million babies have been born worldwide using ART since 1978, when the first IVF baby was born [de Mouzon et al. Citation2009]. However, the ARTs available today are both expensive and invasive and often yield inadequate results. A study performed by the International Committee for Monitoring Assisted Reproductive Technologies (ICMART) in 2002 showed that only 40% of the 601,243 initiated cycles for fertility procedures that took place in 53 countries worldwide that year, resulted in babies being born [de Mouzon et al. Citation2009]. Thus, there is clearly room for improvement.
One strategy currently in focus is single embryo transfer in order to limit superovulation performed on women and to decrease multiple births which are associated with IVF [Ledee et al. Citation2008]. The superovulation regime is not without health implications for a number of patients [Rizk and Smitz Citation1992] and has been associated with increased frequencies of imprinting disorders [Allen and Reardon Citation2005; Market-Velker et al. Citation2010]. However, the inability to predetermine an oocytes’ potential to produce a viable embryo remains a major obstacle to overcome. Oocyte selection prior to fertilization would reduce the number of embryos available for transfer and subsequently, storage. The identification of biological markers (biomarkers) indicative of either increased or decreased oocyte competence would facilitate oocyte selection. Global gene expression studies that identify differentially expressed transcripts between competent and incompetent oocytes or their associated cells could potentially lead to the identification of such biomarkers.
Multiple microarray studies have been performed in order to identify biomarkers associated with either increased or decreased oocyte quality and embryo development. Several factors are known to affect oocyte quality and subsequent embryo development, such as nutrition, hormonal regulation, and environmental influence [Fair Citation2010]. This is emphasized by the vast range of models used in microarray studies performed to date. However, global transcript profiling can lead to the identification of hundreds, if not thousands of differentially expressed transcripts, making it difficult to identify the best potential candidate biomarkers. Furthermore, several major issues impede human oocyte and embryo research, namely the small number of often compromised samples available and the ethical issues associated with working with such samples [Cahill Citation2000; Macklin Citation2000].
Bovine oocyte research provides a valuable source of transferable, relevant information for human fertility research. Routine in vitro embryo production (IVP) has been well established in cattle [Sirard and Coenen Citation2006]. However, in contrast to routine human IVF, in vitro oocyte maturation (i.e., the transition from a germinal vesicle (GV) stage oocyte to a metaphase II (MII) oocyte) is common practice in bovine IVP which often undergo maturation in vitro (IVM). The maturation environment can affect oocyte developmental potential; bovine oocytes matured in vitro are less competent than their in vivo derived counterparts. However, the difference in developmental competence is not initially apparent. Both conditions give similar rates of maturation, fertilization, and cleavage [Blondin et al. Citation1996], but, morula-blastocyst stage development is severely diminished in in vitro (∼30%) matured oocytes compared to in vivo (∼85%) matured oocytes [Brackett and Zuelke Citation1993; Dominko and First Citation1997; Gordon and Lu Citation1990]. Microarray experiments have been carried out in monkey and bovine comparing the transcriptomes of in vivo matured MII oocytes to that of in vitro matured MII oocytes. Transcripts that are uniquely or differentially expressed between in vivo matured and in vitro matured oocytes that are common to bovine and monkey may be critical for the high rate of blastocyst formation seen in in vivo matured oocytes. A comparable human dataset, which also considers the maturation environment exists in an analysis of transcriptome profiles of oocytes from patients exhibiting polycystic ovarian syndrome (PCOS) resulting in the oocyte being subjected to an altered microenvironment within the follicle, versus oocytes from healthy women [Kenigsberg et al. Citation2009]. Additional human data has been generated in a transcriptome comparison of morphologically normal MII oocytes from a patient who had undergone three successive total fertilization failures (TFFs) by intracytoplasmic sperm injection (ICSI) with fertile cohorts of oocytes from control patients [Gasca et al. Citation2008]. Interestingly, the two populations of oocytes were morphologically indistinguishable from each other prior to IVF. These studies may reveal abnormalities on a molecular level which affect the quality of the oocyte and its potential to develop successfully as an embryo.
Potentially relevant datasets are available from two mouse models of oocyte competence. The first was generated from a comparison of oocyte transcriptomes from aged (60-70 weeks of age) versus young (6-12 weeks of age) mice [Pan et al. Citation2008], aging has detrimental consequences for oocyte competence [Malhi et al. Citation2007; Pan et al. Citation2008; Tatone Citation2008]. The second compares the transcriptomes of developmentally competent oocytes that display a ring of chromatin around the nucleolus (SN) versus non surrounded nucleolus (NSN) oocytes, which cease development at the 2 cell stage [Bellone et al. Citation2009].
The goal of the current study was to perform a meta-analysis on datasets from several microarray experiments in order to identify biomarkers that are characteristic of oocyte quality and vital for embryonic survival in several different model systems across multiple species. Vallee et al. [2006] reported a high conservation of genes in oocytes across species. Genes that have been conserved through evolution are likely to play an important role within the oocyte [Vallee et al. Citation2006] implying that genes associated with developmental competence in one species may be of similar importance in other species. The cumulus cells surrounding and supporting the oocyte may also provide a more tractable biomarker than the oocyte itself and thus several studies have analysed differentially expressed transcripts in the cumulus cells of models of developmental competence representing a possible source of non-invasive predictors of oocyte quality [Bettegowda et al. Citation2008; van Montfoort et al. Citation2008].
Results
Conservation of the mammalian oocyte transcriptome
A cross species transcriptome analysis of GV and MII oocytes highlighting the conservation of genes across several species is presented in . Comparing two bovine datasets 88% and 90% of transcripts were conserved in MII in vitro matured and MII in vivo matured oocytes, respectively. Bovine and mouse GV oocytes showed a 51% conservation of transcripts while in vivo matured MII oocytes from human, mouse, monkey, and bovine showed a 46% conservation of transcripts. Thus, as expected a high degree of gene conservation was detected in mammalian oocytes of different stages and the degree of conservation is in line with the overall conservation of the genome across mammalian species.
Table 1. Transcriptomic analysis of oocytes across several species.
Gene ontology networks associated with oocyte developmental competence and incompetence
We performed Ingenuity Pathway Analysis on four oocyte datasets and three cumulus cell datasets (). In order to maintain compatibility between studies only data generated using the Affymetrix GeneChip were incorporated into the meta-analysis. We asked whether the differentially expressed transcripts from these studies would link specific networks or pathways to developmental competence or incompetence. The top network functions associated with oocyte competency include reproductive system development and function (WNT, Notch, and β-catenin signalling), embryonic development, gene expression, and cell death (A). Cytoskeleton rearrangement, cell cycle regulation, IL-6 signalling, and cell adhesion are among the overpopulated networks associated with competence in the cumulus cell datasets (B).
Figure 1. Overpopulated GO Process Networks for A) genes differentially expressed in models of developmental competency in oocytes and B) genes differentially expressed in models of developmental competence in cumulus cells.
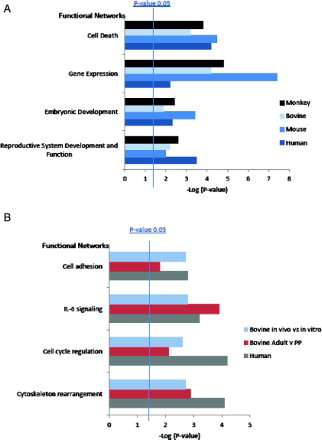
Table 2. Microarray studies performed on models of developmental competence in several species in both oocytes and cumulus cells.
Oocytes at different stages of maturation and matured in different environments have distinct molecular profiles
Differences in the transcriptome of bovine GV, in vivo matured (IVV) MII and in vitro matured (IVM) MII oocytes are represented in a Venn diagram of the absolute gene numbers (A). All groups uniquely expressed a subset of transcripts, the exclusivity of several transcripts to the IVM oocytes suggests the presence of specific transcripts in response to the in vitro environment. This could be due to differential transcription prior to dissolution of the oocyte nucleus, and/or inappropriate polyadenylation of transcripts [Su et al. Citation2007]. The IVV oocytes had 837 transcripts in common with the GV oocytes that the IVM oocytes did not contain; this would suggest that these transcripts may have been wrongly degraded during in vitro maturation. In addition, the IVM oocytes had 547 transcripts in common with the GV oocytes that the IVV oocytes did not contain. This is indicative of a loss in the required degradation of certain transcripts during in vitro maturation and may reflect the reduced potential of these oocytes.
Figure 2. Venn diagramrepresentation of A) the transcriptome of GV stage bovine oocytes [Mamo et al. Citation2011] compared to in vivo matured and in vitro matured bovine oocytes [Kues et al. Citation2008] and B) oocytes matured in vitro and in vivo in bovine [Kues et al. Citation2008] and Monkey [Lee et al. Citation2008].
![Figure 2. Venn diagramrepresentation of A) the transcriptome of GV stage bovine oocytes [Mamo et al. Citation2011] compared to in vivo matured and in vitro matured bovine oocytes [Kues et al. Citation2008] and B) oocytes matured in vitro and in vivo in bovine [Kues et al. Citation2008] and Monkey [Lee et al. Citation2008].](/cms/asset/916e27ce-2d26-44ae-bc08-d9a7a10929bf/iaan_a_656217_f0002_b.jpg)
In order to identify species conserved differences between IVV and IVM MII oocytes the bovine data was compared to a similar monkey dataset (B). All four of the datasets had 6,183 transcripts in common between them, highlighting the commonality between the two different maturation environments and the different species. This number may prove to be higher as the annotation of the bovine genome continues. The IVV oocyte datasets from the two species had 7,034 transcripts in total in common between them, with 60 of these transcripts uniquely common between just the IVV datasets. The two IVM datasets had 6,740 transcripts in common between them with 15 of these transcripts uniquely common to the two IVM datasets.
In a human study the transcriptome of total fertilization failure (TFF) oocytes was found to be more similar to that of GV oocytes than stage MII control oocytes indicating that oocyte maturation had a particular molecular fingerprint, and failure was possibly due to inadequate degradation or utilization of these transcripts in the TFF MII oocytes [Gasca et al. Citation2008]. We extended this analysis by comparing the human TFF transcriptome to the bovine GV transcriptome, identifying a number of factors common to human MII TFF and bovine GV –oocytes (). The overlapping transcripts include Growth Differentiation Factor 9 (Gdf9). This correlates with a previous study performed in PCOS patients where Gdf9 expression in the cumulus cells was associated with decreased developmental potential of the patients’ oocytes [Zhao et al. Citation2010]. In addition, Fbxo5 a gene with a known role in oocyte maturation, specifically germinal vesicle breakdown (GVBD) and cytostatic factor arrest, was identified [Marangos et al. Citation2007; Reimann and Jackson Citation2002].
Table 3. Transcripts upregulated in oocytes from TFF patients and bovine GV oocytes.
Meta-analysis of transcriptomes from models of oocyte developmental competence and incompetence
Five oocyte datasets and three cumulus cell datasets were used to perform the meta-analysis (). In order to maintain compatibility between studies only data generated using the Affymetrix GeneChip were incorporated into the meta-analysis. A comparative analysis was performed to identify genes commonly differentially regulated in two or more of these datasets. A total of 56 differentially expressed genes associated with oocyte and embryo developmental competence were identified (). The upregulation of 18 of these genes were associated with increased competence, 3 of which were specific to the cumulus cells. The upregulation of 35 genes were associated with decreased competence, one of which was also identified in the cumulus cells. These datasets were then analyzed using an Ingenuity Pathway Analysis (IPA)-Biomarker program (http://www.ingenuity.com/products/ipa-biomarker.html) in order to identify the most promising and relevant biomarker candidates. This identified 5 genes; Hmga1, Foxm1, Map3k12, Tgfbr3, and Sfrp1.
Table 4. Transcripts associated with increased developmental competence in oocytes in two or more datasets.
Aberrant gene expression due to an altered maturation environment
Interestingly 19 out of the 56 genes identified were common between the two in vivo vs. in vitro matured oocytes dataset. This indicates that in vitro maturation causes similar aberrant gene expression in both the bovine and the monkey models. Nine transcripts were commonly upregulated and 10 transcripts were commonly downregulated between the two datasets. Of the 9 genes associated with increased competence three are associated with cell cycle and two are ubiquitin regulators. One of these genes, Hmga1, was also the only gene associated with increased competence that was identified in the IPA-Biomarker analysis. It is known to indirectly regulate the G2/M transition [Fedele et al. Citation2001] and is also involved in apoptosis, DNA binding, transcription regulation of genes, and 3’ mRNA processing and chromatin remodeling [Adair et al. Citation2007; Farnet and Bushman Citation1997; Herdegen and Leah Citation1998; Seth et al. Citation2006], which are key events in progression through oocyte meiotic maturation [Masui and Clarke Citation1979]. An interesting candidate gene associated with decreased competence is Atrx, which is involved in chromatin binding and DNA repair [Garrick et al. Citation2006; Picketts et al. Citation1998]. Of the 10 genes whose downregulation was associated with decreased competence four were growth factors, their receptors or regulators i.e., a decrease in insulin-like growth factor binding protein 3, the secreted signalling protein Bmp4, its receptor TGF beta receptor III (Tgfbr3), and a soluble Wnt receptor the secreted frizzled related I (Sfpr1). This points to a link between decreased Bmp4 signalling and an increase in Wnt signaling (due to loss of a negative regulator) and oocyte developmental competence. Tgfbr3 and Sfrp1 were also identified in the IPA-biomarker analysis. Sfrp1 was also upregulated in the polycystic ovarian syndrome (PCOS) dataset [Kenigsberg et al. Citation2009]. PCOS patients are believed to have poor oocytes due to a detrimental oocyte maturation environment, therefore genes common between this dataset and the in vitro matured oocyte dataset may be particularly sensitive to the oocyte maturation environment. Thus negative regulation of Wnt signalling may be correlated with decreased oocyte developmental competence. Four additional genes associated with increased or decreased competence were identified in binary comparisons of the human PCOS data to either the monkey or bovine data. Med1 is a transcriptional regulator known to be essential for embryonic development and Med1 gene knockout in mice impairs embryonic cell growth by regulating Aurora A kinase gene expression [Udayakumar et al. Citation2006]. One of the two genes associated with increased competence is Cugbp1 an mRNA binding protein that regulates translation.
Mouse models of competence
A study compared the oocyte transcriptome in young and aged mice revealing significantly more differentially regulated transcripts in young oocytes compared to aged oocytes, leading to the hypothesis that transcripts are degraded with age [Pan et al. Citation2008]. An additional mouse dataset identified differentially expressed genes in the developmentally defective non surrounded nucleolus model [Zuccotti et al. Citation2009]. These two data sets had 22 differentially expressed genes in common. We identified 5 transcripts that were upregulated in the two models of increased competence () and 17 transcripts that were upregulated in the two models of reduced competence (). Transcripts associated with increased competence included a cell cycle regulator, two signal transduction regulators and two proteases. Kif23, a mitotic kinesin motor protein required for mitosis and cytokinesis, is also a component of male and female embryonic intercellular bridges [Greenbaum et al. Citation2009]. The upregulated genes also included an SH3 binding protein and a GTPase activator. Of the two proteases Metap2 has a probable role in cotranslational removal of N-terminal methionine in protein synthesis and may regulate eIF-2 during protein synthesis [Datta Citation2000].
Table 5. Transcripts associated with decreased developmental competence in oocytes in two or more datasets.
A number of the down regulated genes are DNA and RNA interactors some of which are linked to RNA and protein synthesis and processing (DNA binding histone and transcription factor Foxm1 (UniProt: Q08050), RNA polymerase Pol21 and RNA helicase Ddx55 (UniProt: Q8NHQ9), the regulator of protein elongation Rsp18 and Sec61 (UniProt: P61619) that targets proteins to the ER). Two mitochondrial proteins are downregulated, a component of the respiratory chain complex I - Ndufa1 [Au et al. Citation1999] and the methyl transferase Shmt2 (UniProt: P34897). Two genes, Prkg1 (UniProt: Q6P5T7) and Tmsb10 (UniProt: P63313), are linked to cytoskeletal reorganization. Fank1 is linked to spermatogenesis and meiosis in the testis [Zheng et al. Citation2007].
A comparison of the differentially expressed transcripts from the SN vs. NSN mouse model to those from the bovine in vivo vs. in vitro model identified four genes that were commonly associated with increased competence () and eight with decreased competence across the two models in the two different species (). Interestingly we identified a single gene, the methionine aminopeptidase, which was linked to developmental competence in young mice oocytes, surrounded nucleolus oocytes and bovine in vivo matured oocytes. The RNA helicase Ddx52 was also linked to increased competence which correlates with a decrease in RNA helicase expression in the aged and NSN decreased competence models.
Of the 8 transcripts associated with decreased competence four were linked to signal transduction; a tyrosine phosphatase, a MAP kinase, an interferon induced receptor signaling protein, and Inversin an inhibitor of the canonical Wnt signaling pathway. Map3k12 and Ifitm1 play a role in cellular development [Deblandre et al. Citation1995; Robitaille et al. Citation2005] and Ptpn1 has been shown to negatively regulate oocyte maturation [Cicirelli et al. Citation1990].
Somatic cell markers of developmental competence
Comparative analysis of the differentially expressed transcripts from cumulus cell studies identified a small list of transcripts associated with either increased or decreased developmental competency (). Overexpression of Atrx was found to be associated with decreased developmental competence here while decreased expression of Atrx was linked to increased competence in oocytes. Wasl, Erg1, and Hsp90β1 were identified as being associated with increased competence in cumulus. WASL plays a role in actin-depolymerization [Miki et al. Citation1998], EGR1 is a transcription factor involved in mitogenesis and differentiation [Liu et al. Citation1996] and HSP90β1 is a chaperone protein involved in the heat-shock response [Richter et al. Citation2007].
Table 6. Transcripts associated with either increased or decreased competence in cumulus cells from several models of developmental competence.
Transcription factor regulation of oocyte developmental competence
Transcription factors that regulate the differentially expressed transcripts in the various models of oocyte competency were analyzed using the program MetaCore from GeneGo (Mississippi, USA). Several transcription factors were found to regulate transcripts involved in both the increased and the reduced competence models. These transcription factors must play an important role in the oocyte, independent of specifically developing competence. Interestingly, many transcription factors were found to segregate their regulation to either increased or decreased competence models ().
Table 7. Transcription regulation of transcripts differentially expressed in oocytes.
The cumulus cell experiments were also analyzed to determine the transcription factors that regulate a significant number of differentially expressed transcripts. The cumulus cells represent a separate cell population to the oocyte and transcription continues in these cells while the oocyte undergoes maturation. As with the oocyte analyses several transcription factors were found to regulate a significant number of transcripts in both the increased and reduced competency models. However, the analyses also identified transcription factors unique to either screen ().
Table 8. Transcription regulation of transcripts differentially expressed in cumulus cells. Datasets from different models of developmental competence were used; bovine in vivo vs. in vitro, human early vs. late cleaved blastocysts, and bovine adult vs. prepubertal MII oocytes.
Discussion
The first component of the present study was directed at the identification of genes related to oocyte developmental competence in both the oocyte itself and the cumulus cells that surround it. This was achieved through the use of several different models of competence from bovine, human, monkey, and murine datasets. A cross study comparison was performed in order to identify candidate genes associated with several factors that affect oocyte competence (such as maturation environment, genetic implications, and aging) in these species.
The methods by which RNA is processed prior to sample analysis can affect the resulting transcriptomic data [Mamo et al. Citation2011]. Although, we cannot take account of personnel or laboratory variability, the datasets that were chosen for the current analysis were derived from samples which were processed in a standardized manner using the GeneChip® Expression 3’-Amplification Two-Cycle cDNA Synthesis kit (Affymetrix) and were subjected to the same quality control checks. Several existing transcriptome data sets were omitted from the present study as their microarray platforms were incompatible with our preferred platform, Affymetrix. The ability to merge datasets from multiple platforms would permit further validation of potential candidate genes and possibly increase the number of candidate genes identified.
One of the most striking aspects of the above comparisons is the upregulation of a gene linked to protein synthesis in competence models and the decrease of multiple genes linked to mRNA and protein synthesis in models of incompetence, thus underlining the importance of de novo protein synthesis and its regulation for successful oocyte maturation and subsequent development. The negative regulation of Wnt signalling has emerged in four different models of oocyte developmental incompetence across different species; in vitro matured bovine and monkey oocytes, non surrounded nucleolus mouse oocytes and PCOS human oocytes. A number of secreted proteins that impact on developmental signalling pathways were identified here, possibly providing an avenue to manipulating oocyte competence via the external environment. For example, the secreted frizzled receptor might be targeted with an antagonist to improve oocyte competence. Alternatively, the increased expression of factors by oocytes of lower developmental potential might be exploited as a biomarker of reduced oocyte competency. Although further research is required to determine the most relevant WNTs and BMPs, the addition of soluble WNT and/or BMP signalling factors to IVM or IVF culture media may promote positive Wnt signalling and enhance oocyte and embryo developmental potential. Studies in rhesus monkeys support a role for Wnt signalling during oocyte growth or maturation [Zheng et al. Citation2006].
Several genes that are known to be involved in oocyte maturation and/or embryonic developmental competence such as Gdf9, Emi1, and Ptpn1 were identified. This verifies the ability of the meta-analysis to identify biomarkers of oocyte quality. Many genes identified in this meta-analysis have never been linked to oocyte quality before, these genes represent putative novel biomarkers of oocyte quality and embryonic development.
One gene, Atrx, was found to be associated with decreased developmental competence in both the oocyte and the cumulus cells. Atrx has been shown to be critical for mouse extraembryonic trophoblast formation and subsequent embryo survival 9.5 d postcoitus and is also involved in developmental silencing of imprinted genes [Kernohan et al. Citation2010] and may prove to be a non-invasive marker of an oocyte's developmental competence. Markers in the cumulus cells that surround the oocyte are of particular interest allowing for a non-invasive method of detecting oocyte quality without destroying the oocyte in the process.
In the second component of the study we identified overpopulated pathways expressed in both oocytes and their cumulus cells which were associated with acquisition of oocyte competence. The Notch and Wnt signalling networks were associated with increased developmental competence in oocytes. Work in Drosophila polar follicles has linked Notch to the regulation of the G2 cell cycle arrest [Shyu et al. Citation2009]. It has been shown in mice that when lunatic fringe, an important regulator of Notch signalling, is knocked down that oocytes from these mutants failed to undergo meiotic maturation leaving the mice infertile [Hahn et al. Citation2005]. This is an example of where a defect in the somatic granulosa cells surrounding the oocyte leads to a defect in the oocyte itself. It has been shown that Jagged 1, a Notch ligand, stimulates Notch receptors in the cumulus cells [Johnson et al. Citation2001]. This interaction may prove important for oocyte maturation, fertilization, and embryonic development. The association of Wnt signalling with increased competency correlates well with the fact that negative regulators of Wnt signalling were associated with decreased competency in multiple models.
Apoptosis was another network found to be associated with increased oocyte developmental competence. The transcripts that populated this network were mostly anti-apoptotic and therefore pro-survival. It has been shown previously that the bovine oocyte prevents apoptosis of its surrounding cumulus cells by secreting anti-apoptotic factors such as BMP15 and BMP6 [Hussein et al. Citation2005]. It has also been shown that apoptosis in the compact cumulus–oocyte complex is negatively correlated to the developmental competence of oocytes [Yuan et al. Citation2005].
IL-6 signalling and the immune pathway were found to be over-represented GO networks associated with increased oocyte competence in cumulus cells. It has been shown in oocytes from mice that when IL-6 is added during IVM it induces cumulus expansion and improves oocyte competence measured by delivery of live pups [De Matos et al. Citation2008]. IL-6 is produced by granulosa and cumulus cells in response to the activation of Toll-like receptors 2 and 4 which are innate immune cell-related surveillance proteins [Shimada et al. Citation2006]. Ovulation has the characteristics of an inflammatory process and activates an immune response within the follicle and cumulus-oocyte-complex [Richards et al. Citation2008], this may be a contributing factor to the over-population of this specific network.
Cytoskeleton regulation, cell adhesion, and cell cycle regulation networks were overpopulated in cumulus cells from the models of developmental competence. Meiotic maturation of oocytes is accompanied by remodelling of the cumulus cell cytoskeleton [Allworth and Albertini Citation1993]. The GO ontology analysis shows that regulation of this process is important for oocyte developmental competence. It has been shown that cumulus cell contact with the oocyte during maturation regulates the spatial organization and function of the meiotic spindle through actin-dependent mechanisms to enhance oocyte quality and developmental competence [Barrett and Albertini Citation2010]. This illustrates the importance of cell adhesion molecules as well as actin filaments involved in cytoskeleton regulation.
The final component of the current study was the analysis of transcription factor regulation and regulatory networks. Transcription plays a key role during oogenesis but once the nucleus degrades during oocyte maturation, gene expression is silenced until the zygotic genome is activated. Therefore oocyte maturation, fertilization, and the early stages of embryo development rely on the translational activation of maternally derived mRNAs. Cytoplasmic polyadenylation is probably the most well defined mechanism for translational activation in the maturing oocyte and early stage embryo. This involves the interaction of regulatory RNA-binding proteins with the 3’ UTR region of activated mRNAs [Bettegowda and Smith Citation2007]. A number of RNA regulating genes were identified in the meta-analysis, with a high proportion of these genes being associated with decreased competence (). This may indicate improper transcriptional regulation occurs in the various models of reduced developmental competence. This corresponds with the fact that transcriptional regulation was identified as an overpopulated network associated with oocyte competence (A).
During oocyte maturation transcription stops once the oocyte resumes meiosis and undergoes GVBD. However, it has been shown that residual transcriptional activity can still be observed mainly in the perinucleolar ring [Bouniol-Baly et al. Citation1999]. This residual transcription was specifically linked to the NSN mouse model discussed earlier. Transcription was found to occur in this model of reduced developmental competence and was attributed to RNA polymerase I activity. It has been suggested that the NSN oocyte is at a less advanced stage of differentiation than the SN oocyte [Bouniol-Baly et al. Citation1999]. Data from cattle indicate that the perinucleolar ring is formed on the completion of the oocyte growth phase and corresponds to mRNA and rRNA transcriptional quiescence [Crozet Citation1989].
Heat shock factor 1 (Hsf1) was identified as a transcription factor that regulates 38 transcripts associated with increased oocyte developmental competence. HSF null female mice produce embryos that undergo early developmental arrest. Metaphase II arrest in mature oocytes, cortical granule exocytosis, and formation of pronuclei in zygotes are all impaired in Hsf1−/− mutants and the oocytes undergo irreversible cell death [Bierkamp et al. Citation2010]. The Hsf1 knockout also resulted in Hsp90 depleted oocytes. Hsp90b1 in cumulus cells was associated with the two models of increased competence of bovine oocytes. The HSP90 family of proteins has many functions including cell signalling, transcription, kinase regulation, protein folding, DNA replication and repair, and protection against cell death [Richter et al. Citation2007]. It has been shown that the addition of phorbol myristate acetate during in vitro maturation of bovine oocytes lead to an increase in blastocyst formation [Ali and Sirard Citation2005]. Subsequent microarray analysis performed on the cumulus cells from the oocytes associated with this increased blastocyst formation showed that HSP90B1 was upregulated [Assidi et al. Citation2008]. Perhaps its association with competence is due to its regulation by Hsf1.
Nanog was identified as a transcription factor that regulates 34 transcripts in cumulus cells associated with increased developmental competence [Hatano et al. Citation2005]. It is known to be crucial in early postimplantation embryos by maintaining them in an undifferentiated state. Studies in mice using the SN versus NSN model show that Oct-4 and Stella are expressed only in SN oocytes (associated with increased competence) and the function of Oct-4 is directed at the Nanog locus, regulating the expression of Stella [Zuccotti et al. Citation2009].
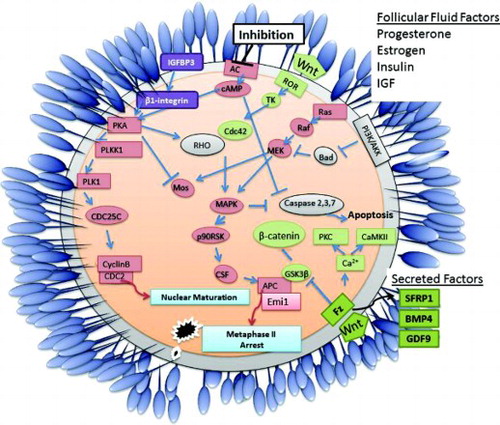
The development of a high quality oocyte is a complex process which involves many interlinking biological pathways as indicated in . Nuclear oocyte maturation is initiated following extracellular inhibition of adenylate cyclase (AC) which induces the translation of specific mRNAs (such as Mos) leading to the activation of maturation promoting factor (CyclinB/Cdc2). However, as already discussed nuclear maturation alone is not sufficient to produce a high quality oocyte. This meta-analysis identified several biological pathways including Wnt signaling and the anti-apoptotic PI3K/Akt pathway as being key to the maturation of a high quality oocyte. Regulation of the canonical Wnt/beta-catenin pathway interlinks with Anaphase Promoting Complex (APC) regulation and metaphase II arrest of an oocyte, the secreted frizzled related protein 1 involved in this pathway is also a potential non-invasive biomarker of oocyte quality. The non-canonical Wnt pathway works through tyrosine kinase signaling to activate the Mapk pathway, inducing oocyte maturation and inhibiting caspase activation and subsequent apoptosis. Additionally, factors secreted by the oocyte and/or cumulus cells and components of the follicular fluid (such as progesterone, estrogen, insulin, and IGF) can also impact on oocyte development and quality.
Figure 3. Schematic diagram detailing primary events and pathways associated with acquisition of competence in mature vertebrate oocytes. Oocyte maturation is initiated following extracellular inhibition of adenylate cyclase (AC) which induces the translation of specific mRNAs (such as mos) leading to the activation of maturation promoting factor (cyclinB/cdc2). The activity of Oocyte maturation, Wnt signaling and the PI3K/Akt apoptosis pathways are key to the maturation of a high quality oocyte. Factors produced by the cumulus cells and/or components of the follicular fluid such as progesterone, estrogen, insulin, and IGF may impact on oocyte development and quality. Secreted factors such as BMP4, GDF9, and SFRP1 are potential non-invasive biomarkers of oocyte quality.
TK: tyrosine kinase, Fz: frizzled receptor
In summary, we identified 63 candidate genes in oocytes and 4 candidate genes in cumulus cells that are putative biomarkers of an oocyte's ability to develop successfully following fertilization by performing a meta-analysis of published data. Secreted proteins and markers in cumulus cells give a possible non-invasive method of determining and possibly manipulating oocyte quality. Biological networks and transcription factor regulation associated with increased and decreased competence were also identified.
Methods
Selection and analysis of oocyte developmental competency resources
Datasets assessing developmental competence () were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) [Barrett et al. Citation2009]. For all oocyte datasets selected for the comparison total RNA was extracted, subjected to linear two-round amplification, cRNA was biotinylated and fragmented for hybridization to the appropriate array. The raw data obtained from the GEO was normalized using an invariant set normalization algorithm [Li and Hung Wong Citation2001] using dCHIP software. Differentially expressed genes were identified comparing parameters of interest ( and ). For gene list generation, the filtration criteria used was a combination of fold change, difference in expression and Welch modified two-sample t-test for unequal variance. Transcripts were considered to be significantly differentially expressed when the following criteria were met: t-test p-value ≤ 0.05, fold change > 1.2 and difference of means > 100. For one study [Gasca et al. Citation2008], comparing the human transcriptome of total fertilization failure oocyte to normal oocytes, expression data tables were obtained from within the publication. This data was independently compared to the bovine GV transcriptome [Mamo et al. Citation2011] and was not incorporated into the rest of the meta-analysis due to the fact that the raw data was unavailable for normalization and standardization to the rest of the datasets used.
Annotation, categorization, and processing of data
We used Ensembl (version 57) as the annotation reference database (www.ensembl.org). Homology between the different species was determined using BioMart and using the ortholog information from NetAffyTM (www.affymetrix.com). Gene lists were then compared for common genes across two or more datasets. Microsoft Access was used to create links between the orthologs and to identify common genes in two or more datasets. In addition Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to compare two or more gene lists and create Venn diagrams for the same [Oliveros Citation2007].
In order to perform the transcriptomic comparative analysis in A the data from two separate microarray studies were combined (; [Kues et al. Citation2008; Mamo et al. Citation2011]).
Identification of developmental competency related networks
MetaCore™ (GeneGo Inc., Mississippi, USA) (www.genego.com/metacore.php) was used to perform a Gene Ontology (GO) network analysis on the differentially expressed transcripts obtained from the datasets from the models of oocyte developmental competence indicated in . Overpopulated GO Networks associated with oocyte competence were identified in both oocytes and cumulus cells (). Transcription factor regulation analysis was also assessed using the MetaCore program.
Declaration of interests: The authors report no conflicts of interest. This work was supported by Science Foundation Ireland (grant number 07/SRC/B1156) (the opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the Science Foundation Ireland).
Abbreviations
| ART: | = | assisted reproductive technologies |
| IVF: | = | in vitro fertilization |
| ICSI: | = | intracytoplasmic sperm injection |
| IVP: | = | in vitro embryo production |
| GV: | = | germinal vesicle |
| MII: | = | metaphase II |
| IVM: | = | in vitro maturation |
| PCOS: | = | polycystic ovarian syndrome |
| TFF: | = | total fertilization failure oocytes |
| SN: | = | surrounded nucleolus |
| NSN: | = | non surrounded nucleolus |
| IVV: | = | in vivo matured |
| GO: | = | gene ontology. |
References
- Adair, J.E., Maloney, S.C., Dement, G.A., Wertzler, K.J., Smerdon, M.J. and Reeves, R. (2007) High-mobility group A1 proteins inhibit expression of nucleotide excision repair factor xeroderma pigmentosum group A. Cancer Res 67:6044–6052.
- Ali, A. and Sirard, M.A. (2005) Protein kinases influence bovine oocyte competence during short-term treatment with recombinant human follicle stimulating hormone. Reproduction 130:303–310.
- Allen, C. and Reardon, W. (2005) Assisted reproduction technology and defects of genomic imprinting. BJOG 112:1589–1594.
- Allworth, A.E. and Albertini, D.F. (1993) Meiotic maturation in cultured bovine oocytes is accompanied by remodeling of the cumulus cell cytoskeleton. Dev Biol 158:101–112.
- Assidi, M., Dufort, I., Ali, A., Hamel, M., Algriany, O., Dielemann, S., (2008) Identification of potential markers of oocyte competence expressed in bovine cumulus cells matured with follicle-stimulating hormone and/or phorbol myristate acetate in vitro. Biol Reprod 79:209–222.
- Au, H.C., Seo, B.B., Matsuno-Yagi, A., Yagi, T. and Scheffler, I.E. (1999) The NDUFA1 gene product (MWFE protein) is essential for activity of complex I in mammalian mitochondria. Proc Natl Acad Sci U S A 96:4354–4359.
- Barrett, S.L. and Albertini, D.F. (2010) Cumulus cell contact during oocyte maturation in mice regulates meiotic spindle positioning and enhances developmental competence. J Assist Reprod Genet 27:29–39.
- Barrett, T., Troup, D.B., Wilhite, S.E., Ledoux, P., Rudnev, D., Evangelista, C., (2009) NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 37:D885–890.
- Bellone, M., Zuccotti, M., Redi, C.A. and Garagna, S. (2009) The position of the germinal vesicle and the chromatin organization together provide a marker of the developmental competence of mouse antral oocytes. Reproduction 138:639–643.
- Bettegowda, A., Patel, O.V., Lee, K.B., Park, K.E., Salem, M., Yao, J., (2008) Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: functional and diagnostic implications. Biol Reprod 79:301–309.
- Bettegowda, A. and Smith, G.W. (2007) Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front Biosci 12:3713–3726.
- Bierkamp, C., Luxey, M., Metchat, A., Audouard, C., Dumollard, R. and Christians, E. (2010) Lack of maternal Heat Shock Factor 1 results in multiple cellular and developmental defects, including mitochondrial damage and altered redox homeostasis, and leads to reduced survival of mammalian oocytes and embryos. Dev Biol 339:338–353.
- Blondin, P., Coenen, K., Guilbault, L.A. and Sirard, M.A. (1996) Superovulation can reduce the developmental competence of bovine embryos. Theriogenology 46:1191–1203.
- Bouniol-Baly, C., Hamraoui, L., Guibert, J., Beaujean, N., Szollosi, M.S. and Debey, P. (1999) Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol Reprod 60:580–587.
- Brackett, B.G. and Zuelke, K.A. (1993) Analysis of factors involved in the in vitro production of bovine embryos. Theriogenology 239:43–64.
- Cahill, L.S. (2000) Social ethics of embryo and stem cell research. Womens Health Issues 10:131–135.
- Cicirelli, M.F., Tonks, N.K., Diltz, C.D., Weiel, J.E., Fischer, E.H. and Krebs, E.G. (1990) Microinjection of a protein-tyrosine-phosphatase inhibits insulin action in Xenopus oocytes. Proc Natl Acad Sci U S A 87:5514–5518.
- Crozet, N. (1989) Nucleolar structure and RNA synthesis in mammalian oocytes. J Reprod Fertil Suppl 38:9–16.
- Datta, B. (2000) MAPs and POEP of the roads from prokaryotic to eukaryotic kingdoms. Biochimie 82:95–107.
- De Matos, D.G., Tran, C.A., Kagan, D., Nataraja, S.G. and Palmer, S. (2008) Interleukin-6 (IL-6) Induces Cumulus Expansion and Improves Oocyte Competence When Present During Mouse In Vitro Oocyte Maturation (IVM). . Biology of Reproduction 78:
- de Mouzon, J., Lancaster, P., Nygren, K.G., Sullivan, E., Zegers-Hochschild, F., Mansour, R., (2009) World collaborative report on Assisted Reproductive Technology, 2002. Hum Reprod 24:2310–2320.
- Deblandre, G.A., Marinx, O.P., Evans, S.S., Majjaj, S., Leo, O., Caput, D., (1995) Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J Biol Chem 270:23860–23866.
- Dominko, T., First, N.L. (1997) Timing of meiotic progression in bovine oocytes and its effect on early embryo development. Mol Reprod Dev 456–467.
- Fair, T. (2010) Mammalian oocyte development: checkpoints for competence. Reprod Fertil Dev 22:13–20.
- Farnet, C.M. and Bushman, F.D. (1997) HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483–492.
- Fedele, M., Pierantoni, G.M., Berlingieri, M.T., Battista, S., Baldassarre, G., Munshi, N., (2001) Overexpression of proteins HMGA1 induces cell cycle deregulation and apoptosis in normal rat thyroid cells. Cancer Res 61:4583–4590.
- Garrick, D., Sharpe, J.A., Arkell, R., Dobbie, L., Smith, A.J., Wood, W.G., (2006) Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet 2:e58.
- Gasca, S., Reyftmann, L., Pellestor, F., Reme, T., Assou, S., Anahory, T., (2008) Total fertilization failure and molecular abnormalities in metaphase II oocytes. Reprod Biomed Online 17:772–781.
- Gordon, I., andLu, H.K. (1990) Production of embryos in vitro and its impact on livestock production. Theriogenology 33:77–78.
- Greenbaum, M.P., Iwamori, N., Agno, J.E. and Matzuk, M.M. (2009) Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol Reprod 80:449–457.
- Hahn, K.L., Johnson, J., Beres, B.J., Howard, S. and Wilson-Rawls, J. (2005) Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development 132:817–828.
- Hatano, S.Y., Tada, M., Kimura, H., Yamaguchi, S., Kono, T., Nakano, T., (2005) Pluripotential competence of cells associated with Nanog activity. Mech Dev 122:67–79.
- Herdegen, T. and Leah, J.D. (1998) Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev 28:370–490.
- Hussein, T.S., Froiland, D.A., Amato, F., Thompson, J.G. and Gilchrist, R.B. (2005) Oocytes prevent cumulus cell apoptosis by maintaining a morphogenic paracrine gradient of bone morphogenetic proteins. J Cell Sci 118:5257–5268.
- Johnson, J., Espinoza, T., McGaughey, R.W., Rawls, A. and Wilson-Rawls, J. (2001) Notch pathway genes are expressed in mammalian ovarian follicles. Mech Dev 109:355–361.
- Kenigsberg, S., Bentov, Y., Chalifa-Caspi, V., Potashnik, G., Ofir, R. and Birk, O.S. (2009) Gene expression microarray profiles of cumulus cells in lean and overweight-obese polycystic ovary syndrome patients. Mol Hum Reprod 15:89–103.
- Kernohan, K.D., Jiang, Y., Tremblay, D.C., Bonvissuto, A.C., Eubanks, J.H., Mann, M.R., (2010) ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell 18:191–202.
- Kocabas, A.M., Crosby, J., Ross, P.J., Otu, H.H., Beyhan, Z., Can, H., (2006) The transcriptome of human oocytes. Proc Natl Acad Sci U S A 103:14027–14032.
- Kues, W.A., Sudheer, S., Herrmann, D., Carnwath, J.W., Havlicek, V., Besenfelder, U., (2008) Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc Natl Acad Sci U S A 105:19768–19773.
- Ledee, N., Lombroso, R., Lombardelli, L., Selva, J., Dubanchet, S., Chaouat, G., (2008) Cytokines and chemokines in follicular fluids and potential of the corresponding embryo: the role of granulocyte colony-stimulating factor. Hum Reprod 23:2001–2009.
- Lee, Y.S., Latham, K.E. and Vandevoort, C.A. (2008) Effects of in vitro maturation on gene expression in rhesus monkey oocytes. Physiol Genomics 35: 145–158.
- Li, C. and Hung Wong, W. (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2: RESEARCH0032.
- Liu, C., Calogero, A., Ragona, G., Adamson, E. and Mercola, D. (1996) EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit Rev Oncog 7:101–125.
- Lucy, M.C. (2001) Reproductive loss in high-producing dairy cattle: where will it end? J Dairy Sci 84:1277–1293.
- Macklin, R. (2000) Ethics, politics, and human embryo stem cell research. Womens Health Issues 10:111–115.
- Malhi, P.S., Adams, G.P., Mapletoft, R.J. and Singh, J. (2007) Oocyte developmental competence in a bovine model of reproductive aging. Reproduction 134:233–239.
- Mamo, S., Carter, F., Lonergan, P., Leal, C.L., Al Naib, A., McGettigan, P., (2011) Sequential analysis of global gene expression profiles in immature and in vitro matured bovine oocytes: potential molecular markers of oocyte maturation. BMC Genomics 12:151.
- Marangos, P., Verschuren, E.W., Chen, R., Jackson, P.K. and Carroll, J. (2007) Prophase I arrest and progression to metaphase I in mouse oocytes are controlled by Emi1-dependent regulation of APC(Cdh1). J Cell Biol 176:65–75.
- Market-Velker, B.A., Zhang, L., Magri, L.S., Bonvissuto, A.C. and Mann, M.R. (2010) Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet 19:36–51.
- Masui, Y. and Clarke, H.J. (1979) Oocyte maturation. Int Rev Cytol 57:185–282.
- Miki, H., Sasaki, T., Takai, Y. and Takenawa, T. (1998) Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature 391:93–96.
- Oliveros, J.C. (2007) An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Pan, H., Ma, P., Zhu, W. and Schultz, R.M. (2008) Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol 316:397–407.
- Picketts, D.J., Tastan, A.O., Higgs, D.R. and Gibbons, R.J. (1998) Comparison of the human and murine ATRX gene identifies highly conserved, functionally important domains. Mamm Genome 9:400–403.
- Reimann, J.D. and Jackson, P.K. (2002) Emi1 is required for cytostatic factor arrest in vertebrate eggs. Nature 416:850–854.
- Richards, J.S., Liu, Z. and Shimada, M. (2008) Immune-like mechanisms in ovulation. Trends Endocrinol Metab 19:191–196.
- Richter, K., Hendershot, L.M. and Freeman, B.C. (2007) The cellular world according to Hsp90. Nat Struct Mol Biol 14:90–94.
- Rizk, B. and Smitz, J. (1992) Ovarian hyperstimulation syndrome after superovulation using GnRH agonists for IVF and related procedures. Hum Reprod 7:320–327.
- Robitaille, H., Proulx, R., Robitaille, K., Blouin, R. and Germain, L. (2005) The mitogen-activated protein kinase kinase kinase dual leucine zipper-bearing kinase (DLK) acts as a key regulator of keratinocyte terminal differentiation. J Biol Chem 280:12732–12741.
- Seth, R.B., Sun, L. and Chen, Z.J. (2006) Antiviral innate immunity pathways. Cell Res 16:141–147.
- Shah, K., Sivapalan, G., Gibbons, N., Tempest, H. and Griffin, D.K. (2003) The genetic basis of infertility. Reproduction 126:13–25.
- Shimada, M., Hernandez-Gonzalez, I., Gonzalez-Robanya, I. and Richards, J.S. (2006) Induced expression of pattern recognition receptors in cumulus oocyte complexes: novel evidence for innate immune-like functions during ovulation. Mol Endocrinol 20:3228–3239.
- Shyu, L.F., Sun, J., Chung, H.M., Huang, Y.C. and Deng, W.M. (2009) Notch signaling and developmental cell-cycle arrest in Drosophila polar follicle cells. Mol Biol Cell 20:5064–5073.
- Sirard, M.A. and Coenen, K. (2006) In vitro maturation and embryo production in cattle. Methods Mol Biol 348:35–42.
- Squires, E.L., Brubaker, J.K., McCue, P.M. and Pickett, B.W. (1998) Effect of sperm number and frequency of insemination on fertility of mares inseminated with cooled semen. Theriogenology 49:743–749.
- Su, Y.Q., Sugiura, K., Woo, Y., Wigglesworth, K., Kamdar, S., Affourtit, J., (2007) Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol 302:104–117.
- Tatone, C. (2008) Oocyte senescence: a firm link to age-related female subfertility. Gynecol Endocrinol 24:59–63.
- Udayakumar, T.S., Belakavadi, M., Choi, K.H., Pandey, P.K. and Fondell, J.D. (2006) Regulation of Aurora-A kinase gene expression via GABP recruitment of TRAP220/MED1. J Biol Chem 281:14691–14699.
- Vallee, M., Robert, C., Methot, S., Palin, M.F. and Sirard, M.A. (2006) Cross-species hybridizations on a multi-species cDNA microarray to identify evolutionarily conserved genes expressed in oocytes. BMC Genomics 7:113.
- van Montfoort, A.P., Geraedts, J.P., Dumoulin, J.C., Stassen, A.P., Evers, J.L. and Ayoubi, T.A. (2008) Differential gene expression in cumulus cells as a prognostic indicator of embryo viability: a microarray analysis. Mol Hum Reprod 14:157–168.
- Yuan, Y.Q., Van Soom, A., Leroy, J.L., Dewulf, J., Van Zeveren, A., de Kruif, A., (2005) Apoptosis in cumulus cells, but not in oocytes, may influence bovine embryonic developmental competence. Theriogenology 63:2147–2163.
- Zhao, S.Y., Qiao, J., Chen, Y.J., Liu, P., Li, J. and Yan, J. (2010) Expression of growth differentiation factor-9 and bone morphogenetic protein-15 in oocytes and cumulus granulosa cells of patients with polycystic ovary syndrome. Fertil Steril 94:261–267.
- Zheng, P., Vassena, R. and Latham, K. (2006) Expression and downregulation of WNT signaling pathway genes in rhesus monkey oocytes and embryos. Mol Reprod Dev 73:667–677.
- Zheng, Z., Zheng, H. and Yan, W. (2007) Fank1 is a testis-specific gene encoding a nuclear protein exclusively expressed during the transition from the meiotic to the haploid phase of spermatogenesis. Gene Expr Patterns 7:777–783.
- Zuccotti, M., Merico, V., Sacchi, L., Bellone, M., Brink, T.C., Stefanelli, M., (2009) Oct-4 regulates the expression of Stella and Foxj2 at the Nanog locus: implications for the developmental competence of mouse oocytes. Hum Reprod 24:2225–2237.