Abstract
Establishing a model for in vitro differentiation of human embryonic stem cells (hESCs) towards the germ cell lineage could be used to identify molecular mechanisms behind germ cell differentiation that may help in understanding human infertility. Here, we evaluate whether a lack of exogenous fibroblast growth factor 2 (FGF2) is supporting spontaneous differentiation of hESCs cultured on human foreskin fibroblast (hFF) monolayers towards germ cell lineage. Additionally to depriving the hESCs of exogenous FGF2, cells were stimulated with all-trans retinoic acid (ATRA). To get a more comprehensive impression on effects of removal of FGF2 and stimulation with ATRA, we combined the results of three cell lines for each experimental setting. When combining gene expression profiles of three cell lines for 96 genes, only 6 genes showed a significant up-regulation in all cell lines, when no FGF2 was added to the media for 12 weeks. None of these genes are related to the germ lineage, whereas genes for neuronal cells (PAX6 and NR6A1) and endothelial cells (FLT-1 and PTF1A) were up-regulated. To induce and support the differentiation towards the germ lineage we stimulated hESCs with different concentrations of ATRA for 7 and 14 days. We observed no significant difference in gene expression on RNA level when combining all cell lines. Whereas, the overall outcome was negative, one of these cell lines demonstrated an up-regulation of DDX4 on RNA and protein level after 7 days of ATRA stimulation. In summary, our data showed that the lack of exogenous FGF2 results in up-regulation of genes crucial for neuronal and endothelial cell differentiation of hESCs, but not in the up-regulation of genes related to germ cell differentiation when cultured on hFFs. Additionally, we demonstrated that ATRA supplementation did not result in a general specific direction of hESCs towards the germ lineage.
Introduction
Infertility is a common problem, affecting 10-15% of all couples, and in around half of the cases the cause is related to males [de Kretser Citation1997]. The cells which give rise to the male gametes (spermatozoa) are the spermatogonial stem cells (SSCs), which originate from the primordial germ cells (PGCs) present in the fetus [Stukenborg et al. Citation2010]. Due to obvious ethical reasons these cells cannot be studied in vivo and little is known about the early developmental phase, due to technical difficulties in vitro. Therefore, providing an alternative source of germ cells would be of interest for studying the early mechanisms of germ cell differentiation and identifying potential causes of infertility.
Human embryonic stem cells (hESCs) are promising candidates for regenerative medicine due to their pluripotency and capability to self-renew [Bongso et al. Citation1994; Thomson et al. Citation1998]. An important factor for hESC self-renewal and pluripotency is fibroblast growth factor 2 (FGF2) [Levenstein et al. Citation2006; Rosler et al. Citation2004]. When deprived of FGF2, hESCs lose their pluripotency and differentiate spontaneously [Greber et al. Citation2011; Laursen et al. Citation2007; Levenstein et al. Citation2006]. Although hESC represent very early stages of differentiation, different hESC lines have been shown to vary in their characteristics and potential to differentiate. Most hESC lines have the tendency to differentiate towards either one of the three germ cell layers [Abeyta et al. Citation2004; Melichar et al. Citation2011]. This difference between hESC lines has furthermore been reported to be laboratory specific [Newman and Cooper Citation2010].
Regarding differentiation potential of these cell lines, hESCs have been shown to differentiate spontaneously to early germ cells when cultured as embryoid bodies [Clark et al. Citation2004] and also in adherent monolayer with mouse embryonic fibroblasts (mEFs) [Bucay et al. Citation2009; Laursen et al. Citation2007; West et al. Citation2011]. In terms of the germ lineage, it has been shown that both mouse embryonic stem cells (mESCs) and hESCs are capable to differentiate to male and female germ cells in vitro but with low efficiency [Clark et al. Citation2004; Hubner et al. Citation2003; Panula et al. Citation2011; Toyooka et al. Citation2003]. Many factors have been suggested to play a role in germ cell development. One of them is all-trans retinoic acid (ATRA), which is generated by a series of oxidative reactions from the dietary vitamin A. ATRA is known to play an important role in spermatogenesis and is believed to be a growth activator of mouse PGCs in vitro [Akmal et al. Citation1997; Koshimizu et al. Citation1995]. Previous studies have used various concentrations of ATRA for stimulating germ cell differentiation from both mESCs and hESCs [Eguizabal et al. Citation2009; Geijsen et al. Citation2004; Richards et al. Citation2010].
In order to optimize a general protocol for early germ cell differentiation for hESCs we asked whether human foreskin fibroblasts (hFFs), which have been shown to be supportive for hESC cultures [Hongisto et al. Citation2012; Hovatta et al. Citation2003] were able to support spontaneous differentiation of different hESCs to germ cell linage when no additional FGF2 was added to the culture media. Additionally, we investigated if hESC lines can be further differentiated in general into germ cells when stimulated with ATRA.
Results
Three different hESC lines derived in two different laboratories were used to evaluate whether hFFs could support germ cell differentiation in general during long term culture without supplementing the culture media with FGF2. These cells were also examined to determine whether they could be further stimulated with ATRA towards germ cell lineage and if the stimulation was dosage dependent.
Gene expression comparison of three undifferentiated hESC lines before and after spontaneous differentiation in adherent culture on hFFs
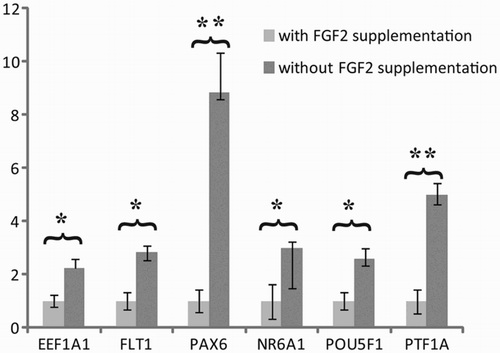
A panel of 96 genes was used to compare the gene signature for three undifferentiated hESC lines (LRB010, LRB017, and CLS1). Each cell line showed a unique signature of both stem cell related genes and genes linked to differentiation. However, to get an overview about a more general effect of the absence of exogenous FGF2, we compared the gene expression of 96 genes pooled for all three hESC lines (LRB010, LRB017, and CLS1) treated with FGF2 against untreated. Hereby, we revealed significant differences between six genes eukaryotic elongation factor 1alpha1 (EEF1A1), fms-related tyrosine kinase 1 (FLT1), paired box-6 (PAX6), nuclear receptor subfamily 6, group A, member 1 (NR6A1), POU class 5 homeobox 1 (POU5F1), and pancreas specific transcription factor, 1A (PTF1A) (). All six genes were up-regulated by the absence of exogenous FGF2, whereas the other 90 genes showed no significant difference between treated and untreated cultures (Supplementary Table 1).
Figure 1. Effect of long-term hESC culture without exogenous FGF2. The pooled analysis revealed a significant up-regulation of six genes related to endothelial (FLT-1 and PTF1A-p48), neuronal cells (PAX6 and NR6A1), as well as pluripotency (POU5F1) and cancer related genes when up-regulated (EEF1A1). Relative quantity was calculated by ddCt method for each sample, from the mean of four replicates. Error bars: SEM: standard deviation of the mean, Significance: *P < 0.05, **P < 0.01
Effect of ATRA stimulation on DDX4, DAZL, POU5F1, and NANOG gene expression in hESCs cultured in the absence of FGF2
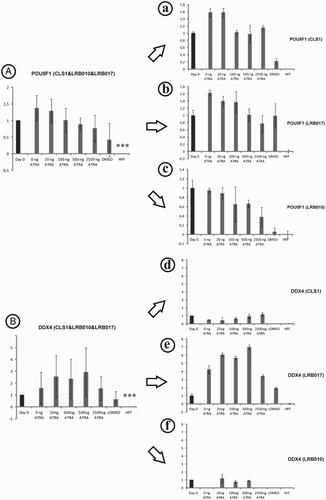
Expression of germ cell marker (DDX4 and DAZL) as well as markers for pluripotency (POU5F1 and NANOG) was evaluated with quantitative polymerase chain reaction (Q-PCR) for three hESC lines (LRB010, LRB017, and CLS1) after differentiating for 7 days with ATRA stimulation, but without exogenous stimulation of FGF2 for 12 weeks. This revealed no significant elevated gene expression of any of the tested genes, when all cell lines were analyzed together.
Whereas, the analysis of these genes showed a quite homogenous expression pattern for POU5F1 (A), NANOG and DAZL (Supplementary ) for each cell line (a-c), the expression of DDX4 instead varies between the different cell lines (B). While CLS1 and LRB010 showed no increased DDX4 expression levels after stimulation with different ATRA concentrations (d and f), when compared to the starting cell population (cultured for 12 weeks without exogenous FGF2), LRB017 showed a strong response (highest after stimulation with 500 ng) after stimulation for 7 days when compared to the situation at day 0 (e). Normal hFFs were used as negative control for all genes. hFFs showed a significant lower expression of all analyzed genes when compared to the hESCs at the start of the culture (day 0; ).
Figure 2. Stem (POU5F1, A) and germ cell (DDX4, B) markers pooled (A and B) and single evaluation (a-f) in three hESC lines after stimulation with ATRA for 7 days in vitro. No up- or down-regulation could be observed by Q-PCR gene expression analysis after supplementation with ATRA when all cell lines were pooled (A and B). Gene expression of germ cell markers in LRB017 showed a positive response to the supplementation with ATRA when compared to day 0. Day 0 is control hESCs that have been spontaneously differentiated on hFFs for 12 weeks and showed a significant higher expression of POU5F1 and DDX4 genes compared to hFF alone. Relative quantity was calculated by ddCt method for each sample, from the mean of three replicates. Error bars: SD: standard deviation, Significance: ***P < 0.001
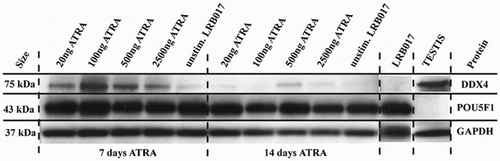
Focusing the effect of ATRA stimulation on DDX4 expression in LRB017, we further investigated protein levels of DDX4, POU5F1, and PAX6 in LRB017 after stimulation with different concentrations of ATRA (). While the gene expression analysis revealed in the strongest up-regulation with 500 ng/ml, the strongest protein expression was observed after 7 days stimulation with 100 ng/ml ATRA in vitro (). After 14 days of ATRA stimulation, the highest protein expression was observed after stimulation with 500 ng/ml (). Evaluating POU5F1 expression on protein level, POU5F1 revealed a strong expression when stimulated for 7 day or 14 days with ATRA (). However, PAX6 expression was not observed after ATRA stimulation for 7 or 14 days (data not shown).
Figure 3. Western blot analysis of DDX4 and POU5F1 in LRB017 after stimulation with ATRA for 7 and 14 days. ATRA is all-trans retinoic acid dissolved in DMSO. Positive control for DDX4 is human adult testis and for POU5F1 is undifferentiated hESCs (LRB017). All markers analyzed DDX4, POU5F1, and GAPDH showed bands of expected size (75kDa, 43kDa, and 35kDa, respectively).
DDX4 expression in undifferentiated hESCs
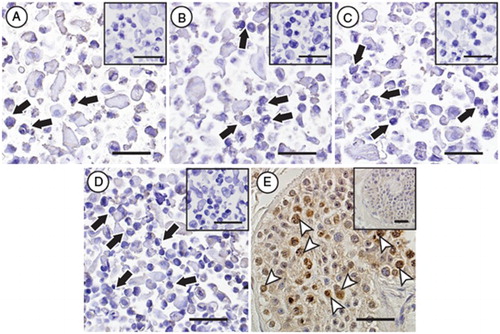
In addition to the three cell lines used in the experiments mentioned above, we detected DDX4 RNA in 8 additional undifferentiated hESC lines and 3 human induced pluripotent stem cell (hiPSC) lines when defining the status of DDX4 in those stem cell lines. Our experiment showed expression of DDX4 RNA in all undifferentiated cell lines analyzed (in total 14 different undifferentiated pluripotent cell lines), albeit at varying levels (). DDX4 protein expression was not detected when analyzing four undifferentiated hESC lines (H9, HS207, HS360, and HS401) by immunocytochemical analysis ().
Figure 4. DDX4 expression at RNA level in 8 undifferentiated hESC lines and 3 hiPSC lines established in 5 different laboratories. DDX4 expression was detected at RNA level in all cell lines analyzed. Relative quantity was calculated by ddCt method for each sample from mean of three replicates.
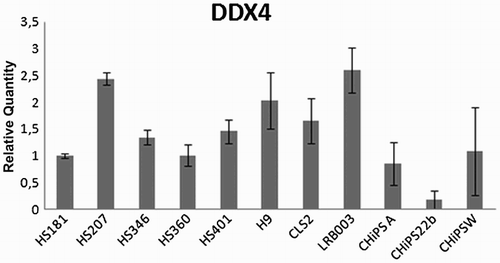
Figure 5. Immunocytochemical evaluation of DDX4 expression in hESC lines. No specific protein expression of DDX4 could be observed in any of the evaluated stem cells lines (black arrows, H9 (A), HS207 (B), HS360 (C) and HS401 (D)), whereas positive protein expression of DDX4 (white arrow heads) could be observed in male germ cells in cross-sections of a human testicular biopsy (E). Negative controls for the primary antibody for all samples are shown as small Figures (A-E). Scale bars: 50µm.
Discussion
Spontaneous differentiation of hESCs towards early germ cell lineage on mEFs has previously been shown [Bucay et al. Citation2009; Laursen et al. Citation2007; West et al. Citation2008]. In order to answer the question if hESCs in general could be spontaneously differentiated towards the germ cell lineage on hFFs we cultured three hESCs in absence of exogenous FGF2 and combined the results of all three cell lines, in order to reduce cell line specific effects which are well-known for hESC lines. The expression analysis showed a significant up-regulation of 6 out of 96 genes when three hESC lines (LRB010, LRB017, and CLS1) were cultured without supplementation of FGF2 for 12 weeks and analyzed together. A significant up-regulation was observed in EEF1A1, which is ubiquitously expressed in mammalian cells and takes part in various important cellular mechanisms (e.g., cytoskeletal remodeling) as well as control mechanisms of cell cycle, growth, and death [Kobayashi and Yonehara Citation2009; Koiwai et al. Citation2008; Scaggiante et al. Citation2008] and the modulation of cell signaling [Yan et al. Citation2008]. Recently, the over expression of EEF1A1 has been connected to the formation of cancer (e.g., prostate cancer) and cell proliferation activities [Scaggiante et al. Citation2008; Scaggiante et al. Citation2011]. When focusing on pluripotent markers, known to be expressed by stem as well as pluripotent cancer cells, significant up-regulation of POU5F1 (also known as OCT4) and NR6A1 (also known as GCNF) has been observed. NR6A1 is known to repress POU5F1 [Chung et al. Citation2001] . Therefore the up-regulation of these genes might be the result of the interaction between both genes. Additionally, NR6A1 has been mentioned to be involved in the transition from primitive neuronal stem cells (pNSC) to definitive neural stem cells (dNSC) [Akamatsu et al. Citation2009]. Consistent with this finding Greber et al. [2011] demonstrated very recently, that FGF2 inhibits neuronal induction of hESCs when cultured on the extracellular matrix Matrigel. Focusing on genes expressed in neuronal cells, our combined analysis of all three different hESCs showed a significant increase of NR6A1 and PAX6 supporting the published data in a more general way. Another marker, up-regulated after 12 weeks of culture without FGF2 was FLT-1 (also known as VEGFR-1), which is known to play a role in tumor growth in different organs (e.g., brain, lung, prostate) [Gille et al. Citation2001; Shibuya Citation1995], and is expressed in the vascular endothelium. Consistent with this we found PTF1A up-regulated. PTF1A is a basic helix-loop-helix (bHLH) transcription factor which is activated by vascular endothelium [Krapp et al. Citation1996; Krapp et al. Citation1998]. In summary, the combined analysis of all three hESCs revealed that culturing without exogenous FGF2 initiates the up-regulation of genes expressed in neuronal as well as endothelial cells, which is consistent with previous studies using single cell lines. However, a spontaneous up-regulation of markers crucial for germ cell differentiation as well as a down-regulation of pluripotency markers could not be demonstrated. Therefore, the lack of exogenous FGF2 does not ensure the absence of the FGF pathway.
Additionally to spontaneous differentiation of hESCs without exogenous FGF2, we investigated the general effect of different concentrations of ATRA on the direction towards germ cell differentiation. ATRA is the active form of vitamin A and has been shown to be essential for spermatogenesis [Thompson et al. Citation1964; Wolbach and Howe Citation1925]. Various concentrations of ATRA have been used in previously published studies for inducing germ cell differentiation from ESCs but an optimal concentration has not yet been published [Aflatoonian et al. Citation2009; Eckhoff and Nau Citation1990; Geijsen et al. Citation2004; Nayernia et al. Citation2006]. When analyzing all three cell lines together after culture for 7 days with different concentrations of ATRA, no overall significant up-regulation of the germ cell specific markers DDX4 or DAZL could be shown for all 3 cell lines. In addition, no significant effect of down-regulation of the pluripotent markers POU5F1 and NANOG could be observed. However, it has been shown in previous studies that hESC gene profiles differ even if they have common features. To show this effect on the reported germ cell specific marker DDX4, we checked additionally to our three hESC lines, eight hESC, as well as three hiPSC lines for their ‘natural’ expression of DDX4. As shown by other studies [Aflatoonian et al. Citation2009; Kee et al. Citation2006; West et al. Citation2008; West et al. Citation2010] and as expected, all cell lines showed an expression of DDX4 on RNA level, but not on protein level when analyzed with immunocytochemistry (ICC). Therefore, we checked our three cell lines on a single cell line basis for the expression of DDX4 on RNA as well as protein after stimulation with ATRA. By this analysis we could demonstrate that only one cell line (LRB017) was able to show augmented DDX4 expression of RNA as well as protein after stimulation with ATRA for 7 and 14 days. DDX4 shows the highest expression after 7 day stimuli with 100 ng/ml ATRA on protein level while on RNA level the highest DDX4 expression is observed after stimuli with 500 ng/ml ATRA. However, when evaluating POU5F1 expression on protein level, we observed no difference between the different stimuli with ATRA or the unstimulated LRB017 sample. The difference observed between DDX4 expression of RNA and protein can be explained by post transcriptional regulation and/or variation in mRNA and protein turnover rates [Cox et al. Citation2005; Hack Citation2004]. This result demonstrates that hESCs have different tendencies to differentiate into different cell types and that choosing the ‘optimal’ hESC line is crucial for differentiation into specific cell types.
In summary, our study showed for the first time by analyzing hESCs in a combined way that culturing these cell lines on hFFs without exogenous supplementation of FGF2 results in a significant up-regulation of genes specific for endothelial as well as neuronal cells, but not for genes related to germ cell differentiation. We could also demonstrate that differentiation towards the germ lineage by stimulation with ATRA is cell line dependent and that DDX4 is already expressed on RNA level in different levels in hESCs as well as hiPSCs.
Materials and Methods
Ethics
All issues concerning the experimental setups and evaluation techniques have been approved by the local Scientific Ethical Committee, Region Midtjylland, Denmark and the Ethics Board of Karolinska Institutet, Stockholm, Sweden.
Stem cell culture
All hESC lines used in this study were fully characterized and showed teratoma formation in vivo. The three hESC lines used for spontaneous differentiation and ATRA stimulation were: LRB017 (46,XY) and LRB010 (46,XY) both derived at the University Hospital in Copenhagen, Denmark, and CLS1 (46,XY) derived at Ciconia Private Hospital, Højbjerg, Denmark. All three cell lines had been derived using immunosurgery. The eight hESC lines used for detection of the reported germ cell specific marker DDX4 were: HS181 (46,XX), HS207 (46,XX), HS346 (46,XX), HS360 (46,XY), HS401 (46,XY) (all derived after isolating the inner cell mass mechanically at Karolinska Institutet, Karolinska University Hospital Huddinge, Sweden), H9 (46,XX, derived using immunosurgery at University of Wisconsin, USA), CLS2 (46,XY, derived using immunosurgery at Ciconia Private Hospital, Højbjerg, Denmark), and LRB003 (46,XY, derived using immunosurgery at the University Hospital in Copenhagen, Denmark). The three hiPSC lines used for detection of DDX4 were reprogrammed by transfecting hFFs using a lentiviral system. CHiPSW and CHiPS22b were transfected with POU5F1, SOX2, LIN28, and NANOG while CHiPSA was transfected with NANOG, POU5F1, SOX2, KLF4, and MYC (University of Geneva, Geneva, Switzerland [Chicha et al. Citation2011]. The undifferentiated stem cell lines were cultured in two different laboratories on inactivated (irradiation 40 Gy) hFFs (CRL-2429; ATCC, Manassas, VA, USA) at 37°C in 5% CO2. Culture media used for the undifferentiated hESCs consisted of Knockout Dulbeccós Modified Eaglés medium (P/N 10829018, Invitrogen, Paisley, UK) supplemented with 20% Knockout serum replacement (P/N 10828028, Invitrogen), 0.5% penicillin-streptomycin (P/N 15140122, Invitrogen), 2mM L-Glutamine (P/N 25030024, Invitrogen), 1% non-essential amino acids (P/N 11140035, Invitrogen) and 0.5mM 2-mercaptoethanol (P/N M3148-25ml, Sigma Aldrich, Stockholm, Sweden). The media used for culturing undifferentiated stem cells was supplemented with either 4ng/ml (CLS1, CLS2, LRB010, LRB017, LRB003) or 8ng/ml FGF2 (the remaining cell lines) (P/N PHG-0021, Biosource, Camarillo, CA, USA). Undifferentiated stem cells were passaged enzymatically using either trypsin (TRYPSIN 0.05%, EDTA, P/N 25300062, Invitrogen) every 9-14 d (CLS1, CLS2, LRB010, LRB017, LRB003) or TrypLE select (P/N 12563011, Invitrogen) every 5-7 d (the remaining cell lines). Under these culture conditions all the hESC lines showed typical morphology of undifferentiated hESCs and expressed the six consensus markers for undifferentiated hESCs (NANOG, POU5F1, GDF3, DNMT3B, GABRB3, and TDGF1) [Adewumi et al. Citation2007].
Differentiation of hESCs
To induce spontaneous differentiation the CLS1, LRB017, and LRB010 hESC lines were cultured in an adherent culture on hFFs for 12 w without supplementing the culture media with FGF2. Cells were passaged every 9-14 d during the 12 w. After the 12 w of spontaneous differentiation, the cells were stimulated with different concentrations of ATRA dissolved in dimethyl sulfoxide ((DMSO) 20, 100, 500, 2500ng/ml, P/N R2625, Sigma-Aldrich) and cells were harvested for analysis at day 7 and 14 (Supplementary ). The media was changed every other day and the cells were not passaged during the stimulation time.
RNA isolation and cDNA amplification
hESCs were harvested enzymatically using either trypsin (CLS1, CLS2, LRB010, LRB017, LRB003) or TrypLE select (remaining cell lines) and stored in RNAprotect Cell Reagent until time of RNA extraction. The RNA was isolated with RNeasy Mini Kit (P/N 74104, Qiagen, Hilden, Germany) according to manufactures protocol. The cells were homogenized by using QIAshredder (P/N 79654, Qiagen, Hilden, Germany). The RNA was treated with DNase 1 (P/N 048K6043, Sigma Aldrich). RNA concentration and quality was measured by Nanovue plus (GE Healthcare, Little Chalfont, UK). cDNA amplification was performed from 100 ng of RNA with High capacity cDNA RT Kit (P/N 4368814, Applied Biosystems (ABI), Carlsbad, CA, USA). The cDNA was pre-amplified with TaqMan PreAmplification Pool for human stem cell pluripotency array according to manufactureŕs protocol (P/N 4405625, ABI) before loading on TaqMan low density array cards (TLDA, P/N 438544, ABI).
Gene expression analysis
TaqMan low density array cards (TLDA, P/N438544, ABI), designed by the International Stem Cell Initiative [Adewumi et al. Citation2007], were used for comparative analysis of CLS1, LRB017, and LRB010 in undifferentiated state as well as after spontaneous differentiation for 12 w on hFFs. The analysis was done according to manufactures protocol. Briefly, the TLDA cards are based on TaqMan chemistry where gene expression of a panel of 96 genes was analysed in one run. The TLDA cards are pre-loaded with 96 TaqMan gene expression assays of importance for hESC pluripotency, self-renewal, and differentiation, together with six endogenous controls assigned for normalization. Mean of endogenous controls 18S, ACTB, and GAPDH was used for normalization. Gene expression was calculated by taking mean of the quadruplicates of each sample, then normalized to mean of 18S, ACTB, and GAPDH of the same sample (dCt). Undifferentiated CLS1 was used as calibrator (ddCT) and gene expression was finally presented as relative quantity (fold change (2−ddCT)). Q-PCR was performed on 7500 Fast Real-time PCR System (ABI) for ATRA stimulated hESCs as well as the 11 additional lines analyzed for DDX4 expression. TaqMan Gene expression Master Mix (P/N 4369510, ABI) was used for TaqMan Gene expression assays from ABI: DDX4 (VASA, Assay ID Hs00251859_m1), DAZL (Assay ID Hs00154706_m1), and 18S (Assay ID Hs99999901_s1) was used for normalization. SYBR Green PCR Master Mix (P/N 4309155, ABI) was used for analysis with SYBR Green primers: NANOG fw 3′-CAA AGG CAA ACA ACC CAC TT–5′ rev 3′-CTG GAT GTT CTG GGT CTG G0T–5′. POU5F1 fw 3′-GAC AAC AAT GAA AAT CTT CAG GAG A–5′ rev 3′-TTC TGG CGC CGG TTA CAG AAC CA – 5′. GAPDH fw 3′- GAA GGT GAA GGT CGG AGT CAA C–5′ rev 3′- CAG AGT TAA AAG CAG CCC TGG T– 5′. Gene expression was calculated by taking mean of the triplicates ran for each sample, and then normalized to mean of 18S or GAPDH (depending on chemistry used) of the same sample (dCt). Cells from Day 0 of ATRA differentiation was used as calibrator of each cell line (ddCT) and gene expression was finally presented as relative quantity (fold change (2−ddCT)).
Protein isolation and quantification
The LRB017 hESCs were harvested enzymatically with trypsin and lysed in lysis buffer (P/N 9803, Cell Signalling Technology, Danvers, MA, USA) and supplemented with 1mM phenylmethanesulphonylfluoride (PMSF), 5mM sodium fluoride (NaF), and 15mM Complete Mini Protease Inhibitor (P/N 11836153001, Roche Diagnostics, Indianapolis, IN, USA). The samples were incubated on ice for 30 min followed by 10 min centrifugation at 20,000g at 4°C. The lysate was harvested and protein concentration measured by Bio-Rad protein assay (Bradford method, P/N 500-0006, Bio-Rad).
Western blot analysis
14 µg of each protein lysate were mixed with sample buffer (62.5 mM Tris base, 2% sodium dodecyl sulphate (SDS) and 10% glycerol, pH 6.9), 0.5% dithiothreitol (DTT), and a small quantity of bromophenol blue. The samples were heated for 5 min at 95°C. The proteins were separated on Criterion TGX Precast gel, 4-15% (P/N 567-1084, Bio-Rad, Hercules, CA, USA) with 1 × running buffer (P/N SC-24949, Bio-Rad) for 1 h at 140V and then electro-transferred to a AmershamTM Hybond-P membrane (P/N RPN303F, GE Healthcare) at 130mA in transfer buffer (P/N 161-0734, Bio-Rad) for 1 h. The membranes were blocked with AmershamTM ECL Prime Blocking Agent (50 g/L, P/N RPN418, GE Healthcare) in TBS (P/N SC-24951, Santa Cruz Biotechnology, Santa Cruz, CA, USA) with 0.1% Tween 20 for 1 h at room temperature (RT). The membranes were incubated with primary antibodies overnight and secondary antibodies for 2 h. Primary antibodies used for the western blotting were: goat polyclonal anti-GAPDH (diluted 1:200 (1 µg/ml), P/N SC-20357, Santa Cruz Biotechnology), rabbit polyclonal anti-DDX4 (diluted 1:1000 (1µg/ml), P/N ab13840, AbCam, Cambridge, UK), rabbit polyclonal anti-POU5F1 (diluted 1:400, P/N ab19857 Abcam) and rabbit polyclonal anti-PAX-6 (diluted 1:500 (0.4 µg/ml), P/N SC-11357, Santa Cruz Biotechnology). Secondary antibodies used for the western blotting were: horseradish peroxidase-conjugated (HRP) donkey anti-goat (diluted 1:5000 (0.08 µg/ml), P/N SC-2033, Santa Cruz Biotechnology) and HRP donkey anti-rabbit (diluted 1:5000 (0.08 µg/ml), P/N NA934V, GE Healthcare). The antibodies were all diluted in the blocking solution. As positive control for DDX4, POU5F1, and PAX-6, human protein lysates from adult testicular biopsies, hESCs (LRB017) and human fetal brain (P/N P1244035, BioChain, Hayward, CA, USA) were used. The membranes were developed with AmershamTM ECL Prime Western Blotting Detection reagent (P/N RPN2232, GE Healthcare).
Immunocytochemistry (ICC)
hESCs (HS207, HS360, HS401, and H9) were harvested from culture media and fixed in 4% paraformaldehyde (PFA) overnight at 4°C and routinely embedded in paraffin (P/N P3808, Sigma Aldrich) after dehydration via gradually increasing ethanol concentration. Afterwards, samples were cut into 4 µm sections. 4% PFA fixed and paraffin embedded human testicular tissue was used as positive control for DDX4 detection of germ cells. The samples were rehydrated using xylene and gradually decreasing EtOH series. Briefly, the samples were blocked with 5% goat serum (P/N S-1000, Vector Laboratories, Peterborough, UK) in 0.1% bovine serum albumin (BSA) in phosphate buffered saline (PBS) for 20 min. Primary antibody, rabbit polyclonal anti-DDX4 (P/N ab13840, AbCam) was diluted (1:100 (10 µg/ml)) in 0.1% BSA in PBS and incubated overnight at 4°C. For the negative control for the primary antibody, samples were incubated with rabbit polyclonal IgGs (P/N ab27478, AbCam) with the same concentration of the primary antibody overnight at 4°C. The samples were washed three times for 5 min in PBS and afterwards directly incubated with the secondary antibody, biotinylated goat anti-rabbit IgG (ready to use, P/N ab64256, AbCam) for 1 h at 37°C. Samples were washed three times for 5 min in PBS and incubated with ABC reagent from Vectastain ABC-kit (P/N PK6100, Vector Laboratories) for 30 min at 37°C. The samples were washed three times and stained with DAB for 45 s at room temperature, washed again twice for 2 min in H2O, incubated for 7 s in haemalaun (P/N 1.09249.1000 Merck, Germany) and rinsed for 5 min in tap water. Samples were dehydrated in a gradually increasing EtOH series and xylene and mounted with Entellan® new (P/N 1.07961.0100 Merck) and analyzed under the microscope (Eclipse E800; Nikon; Japan) and pictures were taken with a 12.5 million-pixel cooled digital color camera system (Olympus DP70, Japan).
Statistical evaluation
The gene expression levels were calculated as mean ± standard deviation (SD) or standard deviation of the mean (SEM) as stated in the figure legends. Student test (SigmaPlot 11.0; Systat Software Inc. CA, USA) was used to compare the difference between two experimental settings. A difference was considered statistically significant when the p value was ≤ 0.05.
Abbreviations
| ESCs: | = | embryonic stem cells |
| ABI: | = | Applied Biosystems |
| RNA: | = | ribonucleic acid |
| cDNA: | = | complementary deoxyribonucleic acid |
| TLDA: | = | taqman low density array |
| Q-PCR: | = | quantitative polymerase chain reaction |
| PMSF: | = | phenylmethanesulphonylfluoride |
| NaF: | = | sodium fluoride |
| SDS: | = | sodium dodecyl sulphate |
| DTT: | = | dithiothreitol |
| PFA: | = | paraformaldehyde |
| PBS: | = | phosphate buffered saline |
| hESCs: | = | human embryonic stem cells |
| mESCs: | = | mouse embryonic stem cells |
| ATRA: | = | all-trans retinoic acid |
| PGCs: | = | primordial germ cells |
| hFFs: | = | human foreskin fibroblasts |
| mEFs: | = | mouse embryonic fibroblasts |
| hiPSCs: | = | human induced pluripotent stem cells |
| FGF2: | = | fibroblast growth factor |
| SSCs: | = | spermatogonial stem cells |
| DMSO: | = | dimethyl sulfoxide |
| BSA: | = | bovine serum albumin |
| pNSC: | = | primitive neuronal stem cells |
| dNSC: | = | definitive neural stem cells |
| bHLH: | = | basic helix-loop-helix |
| ICC: | = | immunocytochemistry. |
Supplementary Material
Download TIFF Image (4.3 MB)Supplementary Material
Download TIFF Image (4 MB)Supplementary Material
Download MS Word (617.5 KB)Acknowledgments
We would like to thank Drs. Svechnikov, Colón, Lim, and Høllsberg as well as Bettina Bundgaard, Maja Kaspersen for technical assistance and fruitful discussions of different aspects of the manuscript. Special thanks to Ciconia Private Hospital for supporting this project.
Declaration of interests: Financial support for this project was received from Aase and Ejnar Danielsen foundation, Frimurare Barnhuset in Stockholm, Academy of Finland, Paediatric Research Foundation, Sällskåpet Barnavård in Stockholm as well as from Region Midtjylland. J.B. Stukenborg is supported by the “Deutsche Forschungsgemeinschaft” (DFG-Grant No.: STU 506/3-1) and was not involved in handling any hESC lines. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Author contribution Conceived and designed the experiments: KRK, OH, JBS, JF. Performed the experiments: KRK, AG, AR, JBS, RBT. Analyzed the data: KRK, AR, CYA, RBT, JBS. Contributed reagents/materials, analysis tools: OS, CYA, OH, JF. Wrote the manuscript: KRK, OS, CYA, OH, AR, JBS, JF.
References
- Abeyta, M.J., Clark, A.T., Rodriguez, R.T., Bodnar, M.S., Pera, R.A. and Firpo, M.T. (2004) Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet 13:601–608.
- Adewumi, O., Aflatoonian, B., Ahrlund-Richter, L., Amit, M., Andrews, P.W., Beighton, G., (2007) Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 25:803–816.
- Aflatoonian, B., Ruban, L., Jones, M., Aflatoonian, R., Fazeli, A. and Moore, H.D. (2009) In vitro post-meiotic germ cell development from human embryonic stem cells. Hum Reprod 24:3150–3159.
- Akamatsu, W., DeVeale, B., Okano, H., Cooney, A.J. and van der Kooy, D. (2009) Suppression of Oct4 by germ cell nuclear factor restricts pluripotency and promotes neural stem cell development in the early neural lineage. J Neurosci 29:2113–2124.
- Akmal, K.M., Dufour, J.M. and Kim, K.H. (1997) Retinoic acid receptor alpha gene expression in the rat testis: potential role during the prophase of meiosis and in the transition from round to elongating spermatids. Biol Reprod 56:549–556.
- Bongso, A., Fong, C.Y., Ng, S.C. and Ratnam, S. (1994) Isolation and culture of inner cell mass cells from human blastocysts. Hum Reprod 9:2110–2117.
- Bucay, N., Yebra, M., Cirulli, V., Afrikanova, I., Kaido, T., Hayek, A., (2009) A novel approach for the derivation of putative primordial germ cells and sertoli cells from human embryonic stem cells. Stem Cells 27:68–77.
- Chicha, L., Feki, A., Boni, A., Irion, O., Hovatta, O. and Jaconi, M. (2011) Human pluripotent stem cells differentiated in fully defined medium generate hematopoietic CD34– and CD34+ progenitors with distinct characteristics. PLoS One 6:e14733.
- Chung, A.C., Katz, D., Pereira, F.A., Jackson, K.J., DeMayo, F.J., Cooney, A.J., (2001) Loss of orphan receptor germ cell nuclear factor function results in ectopic development of the tail bud and a novel posterior truncation. Mol Cell Biol 21:663–677.
- Clark, A.T., Bodnar, M.S., Fox, M., Rodriquez, R.T., Abeyta, M.J., Firpo, M.T., (2004) Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet 13:727–739.
- Cox, B., Kislinger, T. and Emili, A. (2005) Integrating gene and protein expression data: pattern analysis and profile mining. Methods 35:303–314.
- de Kretser, D.M. (1997) Male infertility. Lancet 349:787–790.
- Eckhoff, C. and Nau, H. (1990) Identification and quantitation of all-trans- and 13-cis-retinoic acid and 13-cis-4-oxoretinoic acid in human plasma. J Lipid Res 31:1445–1454.
- Eguizabal, C., Shovlin, T.C., Durcova-Hills, G., Surani, A. and McLaren, A. (2009) Generation of primordial germ cells from pluripotent stem cells. Differentiation 78:116–123.
- Geijsen, N., Horoschak, M., Kim, K., Gribnau, J., Eggan, K. and Daley, G.Q. (2004) Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 427:148–154.
- Gille, H., Kowalski, J., Li, B., LeCouter, J., Moffat, B., Zioncheck, T.F., (2001) Analysis of biological effects and signaling properties of Flt-1 (VEGFR-1) and KDR (VEGFR-2). A reassessment using novel receptor-specific vascular endothelial growth factor mutants. J Biol Chem 276:3222–3230.
- Greber, B., Coulon, P., Zhang, M., Moritz, S., Frank, S., Muller-Molina, A.J., (2011) FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J 30:4874–4884.
- Hack, C.J. (2004) Integrated transcriptome and proteome data: the challenges ahead. Brief Funct Genomic Proteomic 3:212–219.
- Hongisto, H., Vuoristo, S., Mikhailova, A., Suuronen, R., Virtanen, I., Otonkoski, T., (2012) Laminin-511 expression is associated with the functionality of feeder cells in human embryonic stem cell culture. Stem Cell Res 8:97–108.
- Hovatta, O., Mikkola, M., Gertow, K., Strömberg, A.M., Inzunza, J., Hreinsson, J., (2003) A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum Reprod 18:1404–1409.
- Hubner, K., Fuhrmann, G., Christenson, L.K., Kehler, J., Reinbold, R., De La Fuente, R., (2003) Derivation of oocytes from mouse embryonic stem cells. Science 300:1251–1256.
- Kee, K., Gonsalves, J.M., Clark, A.T. and Pera, R.A. (2006) Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev 15:831–837.
- Kobayashi, Y. and Yonehara, S. (2009) Novel cell death by downregulation of eEF1A1 expression in tetraploids. Cell Death Differ 16:139–150.
- Koiwai, K., Maezawa, S., Hayano, T., Iitsuka, M. and Koiwai, O. (2008) BPOZ-2 directly binds to eEF1A1 to promote eEF1A1 ubiquitylation and degradation and prevent translation. Genes Cells 13:593–607.
- Koshimizu, U., Watanabe, M. and Nakatsuji, N. (1995) Retinoic acid is a potent growth activator of mouse primordial germ cells in vitro. Dev Biol 168:683–685.
- Krapp, A., Knofler, M., Frutiger, S., Hughes, G.J., Hagenbuchle, O. and Wellauer, P.K. (1996) The pp48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J 15:4317–4329.
- Krapp, A., Knofler, M., Ledermann, B., Burki, K., Berney, C., Zoerkler, N., (1998) The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev 12:3752–3763.
- Laursen, S.B., Mollgard, K., Olesen, C., Oliveri, R.S., Brochner, C.B., Byskov, A.G., (2007) Regional differences in expression of specific markers for human embryonic stem cells. Reprod Biomed Online 15:89–98.
- Levenstein, M.E., Ludwig, T.E., Xu, R.H., Llanas, R.A., VanDenHeuvel-Kramer, K., Manning, D., (2006) Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells 24:568–574.
- Melichar, H., Li, O., Ross, J., Haber, H., Cado, D., Nolla, H., (2011) Comparative Study of Hematopoietic Differentiation between Human Embryonic Stem Cell Lines. PLoS One 6:e19854.
- Nayernia, K., Nolte, J., Michelmann, H.W., Lee, J.H., Rathsack, K., Drusenheimer, N., (2006) In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring mice. Dev Cell 11:125–132.
- Newman, A.M. and Cooper, J.B. (2010) Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 7:258–262.
- Panula, S., Medrano, J.V., Kee, K., Bergstrom, R., Nguyen, H.N., Byers, B., (2011) Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum Mol Genet 20:752–762.
- Richards, M., Fong, C.Y. and Bongso, A. (2010) Comparative evaluation of different in vitro systems that stimulate germ cell differentiation in human embryonic stem cells. Fertil Steril 93:986–994.
- Rosler, E.S., Fisk, G.J., Ares, X., Irving, J., Miura, T., Rao, M.S., (2004) Long-term culture of human embryonic stem cells in feeder-free conditions. Dev Dyn 229:259–274.
- Scaggiante, B., Bonin, S., Cristiano, L., Siracusano, S., Stanta, G., Dapas, B., (2008) Prostate-tumor-inducing gene-1 analysis in human prostate cancer cells and tissue in relation to Mycoplasma infection. Cancer Invest 26:800–808.
- Scaggiante, B., Dapas, B., Bonin, S., Grassi, M., Zennaro, C., Farra, R., (2011) Dissecting the expression of EEF1A1/2 genes in human prostate cancer cells: the potential of EEF1A2 as a hallmark for prostate transformation and progression. Br J Cancer Epub 2011 Nov 17.
- Shibuya, M. (1995) Role of VEGF-flt receptor system in normal and tumor angiogenesis. Adv Cancer Res 67:281–316.
- Stukenborg, J.B., Colón, E. and Söder, O. (2010) Ontogenesis of testis development and function in humans. Sex Dev 4:199–212.
- Thompson, J.N., Howell, J.M. and Pitt, G.A. (1964) Vitamin A and Reproduction in Rats. Proc R Soc Lond B Biol Sci 159:510–535.
- Thomson, J.A., Itskovitz-Eldor, J., Shapiro, S.S., Waknitz, M.A., Swiergiel, J.J., Marshall, V.S., (1998) Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147.
- Toyooka, Y., Tsunekawa, N., Akasu, R. and Noce, T. (2003) Embryonic stem cells can form germ cells in vitro. Proc Natl Acad Sci USA 100:11457–11462.
- West, F.D., Machacek, D.W., Boyd, N.L., Pandiyan, K., Robbins, K.R. and Stice, S.L. (2008) Enrichment and differentiation of human germ-like cells mediated by feeder cells and basic fibroblast growth factor signaling. Stem Cells 26:2768–2776.
- West, F.D., Mumaw, J.L., Gallegos-Cardenas, A., Young, A. and Stice, S.L. (2011) Human haploid cells differentiated from meiotic competent clonal germ cell lines that originated from embryonic stem cells. Stem Cells Dev 20:1079–1088.
- West, F.D., Roche-Rios, M.I., Abraham, S., Rao, R.R., Natrajan, M.S., Bacanamwo, M., (2010) KIT ligand and bone morphogenetic protein signaling enhances human embryonic stem cell to germ-like cell differentiation. Hum Reprod 25:168–178.
- Wolbach, S.B. and Howe, P.R. (1925) Tissue Changes Following Deprivation of Fat-Soluble A Vitamin. J Exp Med 42:753–777.
- Yan, G., You, B., Chen, S.P., Liao, J.K. and Sun, J. (2008) Tumor necrosis factor-alpha downregulates endothelial nitric oxide synthase mRNA stability via translation elongation factor 1-alpha 1. Circ Res 103:591–597.