Abstract
Follicle stimulating hormone receptor (FSHR) genetic variation at position 2039 A > G (rs6166, p.Asn680Ser) was repetitively shown to correspond to the measures of ovarian sensitivity and the outcomes of gonadotropin stimulation. However, to date, there has been no study revealing the mechanisms behind the associations observed. The aim of the present research was to investigate the relationship between rs6166 and mRNA expression of FSHR-dependent genes such as LHCGR, CYP19A1, and FSHR itself with particular reference to the FSHR transcript variants (deletions of exon 2, 6, and 9 and insertion of a novel exon between exons 8 and 9) in the cell culture model. Steroid production and its dependency on FSHR genotype were also assessed. A total of 22 normoovulatory patients undergoing IVF treatment were recruited. Granulosa cells were obtained by ovary puncture and cultured for 7 days to regain responsiveness to FSH in a gonadotropin-free medium. Stimulation was carried out for 24 hours in a serum-depleted environment using 0.5 UI/l rhFSH. Gene expression was assessed by real-time PCR and genotype was determined by allele-specific PCR. The distribution of p.Asn680Ser genotypes was as follows: 10 homozygotes Asn/Asn, 8 heterozygotes Asn/Ser, and 4 homozygotes Ser/Ser. Expression of total FSHR in all samples studied increased by a mean factor of 1.9 (95% CI: 0.39–11.53, p < 0.001) upon stimulation. All of the analyzed FSHR transcript variants were detectable in non-stimulated and stimulated cells. The only distinct transcript that followed up-regulation was deletion of exon 2. Homozygotes Asn/Asn tended to have higher rhFSH-induced expression of FSHR as compared to the carriers of Ser/Ser genotype. Relative expression of LHCGR and CYP19A1 although up-regulated showed no significant difference with respect to the FSHR genotype. Variable modulation of FSHR expression by its own ligand is likely to explain different clinical behavior of patients with FSHR genetic variants. The putative contribution of rs6166 requires further investigation.
Introduction
The controlled ovarian hyperstimulation with gonadotropins is an integral step of assisted reproductive technologies. Treatment outcomes measured by the number and quality of oocytes retrieved show high inter-individual variability. It is believed that this is determined by individual ovarian sensitivity to gonadotropins, with the size of the ovarian reserve and the effectiveness of the follicular recruitment being of utmost relevance. According to many authors, the type of the gonadotropic preparation as well as the initial and total dose are of secondary importance [Dickey et al. Citation2002; Klinkert et al. Citation2005]. However, there is some scope for drug-induced modulation of multifollicular recruitment and growth, and it seems reasonable to search for predictive factors enabling optimal personalized stimulation.
Recently, attention has been directed to pharmacogenetic aspects of the treatment with gonadotropins. Numerous reports confirm the importance of follicle stimulating hormone receptor (FSHR) genetic variation in exon 10 at position 2039 (A > G) in the prediction of response to hyperstimulation [Binder et al. Citation2008; Jun et al. Citation2006; Perez Mayorga et al. Citation2000]. This single nucleotide substitution results in amino acid change in the encoded protein sequence and is commonly referred to as p. Asn680Ser. It is tightly linked to p. Thr307Ala [Aittomäki et al. Citation1995; Simoni et al. Citation1999] and together with much less prevalent p. Ile160Thr they are the main substitutions responsible for sequence variations of the FSHR protein. Moreover, the promoter region of the FSHR gene harbors frequent polymorphic sites located at positions -29 (G > A) and -114 (T > C) [Wunsch et al. Citation2005].
Given the critical role of FSHR-dependent signaling in reproduction, the non-synonymous nature of the coding polymorphisms and its location in a sequence encoding important intracellular domain a direct effect of rs6166 on protein receptor functions could be expected. Current functional studies investigating this and other single nucleotide polymorphisms (SNPs) of the FSHR did not reveal the biological basis of the SNP-phenotype associations [Simoni et al. Citation1999; Sudo et al. Citation2002]. Recently, one study showed an association of the promoter -29 (G > A) polymorphism with ovarian response and FSHR expression both at the mRNA and protein level in an Indian population [Desai et al. Citation2011]. Functional experiments performed by this group suggested different binding affinities of proteins for both alleles. These results need confirmation as other experimental approaches failed to show an influence of -29 (G > A) substitution on the transcriptional activity. Additionally, in a mixed German-Indonesian cohort, no associations with clinical parameters were observed [Wunsch et al. Citation2005].
Apart from genetic polymorphism, another potential mechanism creating protein diversity stems from posttranscriptional processing of pre-mRNA, which includes alternative splicing. It results in a generation of different mature mRNAs from the same gene. Some of the alternative mRNA transcripts may be used as templates for variant protein species. Interestingly the occurrence and regulation of alternative splicing may be allele-specific [Nembaware et al. Citation2004]. Allelic variants rather than altering an encoded protein may affect the mechanisms governing gene expression and posttranscriptional modifications. A number of FSHR transcriptional isoforms were identified in human female and male gonads [Gerasimova et al. Citation2010; Song et al. Citation2002]. At least 3 truncated variants exist, which differ from the full-length transcript (composed of exons 1-10) by the absence of exons 2, 6, or 9. One variant is associated with insertion of an extra exon (corresponding to an inverted 102-bp sequence of intron 2) between exons 8 and 9. The variant proteins generated as a consequence of alternative splicing, may potentially function as receptors since they contain the transmembrane region encoded by exon 10. It was suggested that allelic variation of the FSHR gene may determine the presence of splice variants in addition to the full-length isoform [Gerasimova et al. Citation2010].
Finding a biological link between FSHR genetic polymorphism and reproductive potential may provide an important argument for the incorporation of FSHR genotyping in the standard panel of tests in patients being prepared for IVF. Relatively easy access to human luteinized granulosa cells (LGCs) and their responsiveness to stimulatory factors renders them a natural target for studies addressing ovarian reproductive function. Hence, the present study aimed to explore the association between p. Asn680Ser and the expression of FSHR transcripts in primary, long-term cultures of human luteinized granulosa cells. Additionally steady-state mRNA levels of the other critical genes regulated by FSHR signaling, namely luteinizing hormone/chorionic gonadotropin receptor (LHCGR) and aromatase (CYP19A1) as well as steroid biosynthesis were analyzed.
Results
Steady-state mRNA levels of genes of interest in cultured LGCs and its dependence on the FSHR rs6166 genotype
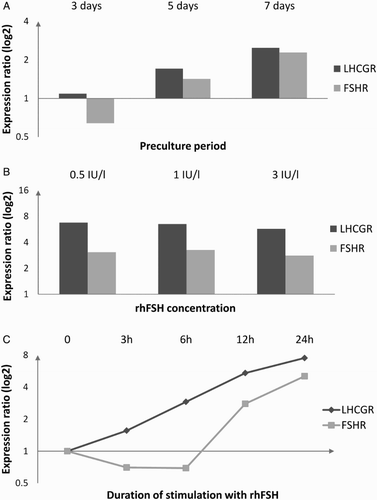
In the initial experiments performed to establish an experimental model we observed a tendency toward higher levels of steady-state mRNA with longer culturing periods preceding rhFSH treatment (A). Decreasing the dose of rhFSH down to 0.5 IU/l did not seem to influence the response (B). The FSH-induced change in the relative mRNA levels of FSHR and LHCGR was dependent on the length of incubation (C). In case of LHCGR the steady increase in transcript level was observed at the selected time intervals. The FSHR time-response curve suggested a period of transrepression/decreased mRNA stability followed by transactivation/increased mRNA stability (C). Consequently, 7-day preculture followed by 24 hours of stimulation with 0.5 IU/l of rhFSH were chosen for the subsequent experiments.
Figure 1. The effect of the length of preculture before 24-hour-long stimulation with 3 IU/l rhFSH (A), rhFSH concentration in the culture medium during 24-hour-long stimulation after 7 days of preculture (B), and duration of stimulation with 0.5 IU/l rhFSH after 7 days of preculture (C) on changes in relative quantity of FSHR and LHCGR mRNA as compared to non-stimulated cells (based on pooled cells from 3 random patients, different for each analysis). rhFSH: recombinant human follicle stimulating hormone; FSHR: follicle stimulating hormone receptor; LHCGR: luteinizing hormone/chorionic gonadotropin receptor
The distribution of Asn680Ser genotypes in 22 patients recruited for the study was as follows: 10 homozygotes Asn/Asn, 8 heterozygotes Asn/Ser, and 4 homozygotes Ser/Ser. presents an example of SNP genotyping results. In this set of patients, carriers of different genotypes presented with similar clinical characteristics (). Total RNA isolated from cultured LGCs was of very good quality with RNA integrity number (RIN) exceeding 9 in all tested samples. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was chosen to serve as a reference gene (normalizer) and was validated to be a good internal control as its expression was constant with mean fold change in the stimulated vs. non-stimulated LGCs cultures equal to 1.05 (95% CI: 0.061-20.393, p = 0.89).
Figure 2. Allele-specific PCR of FSHR gene in three different samples visualized on agarose gel. Genotypes of each individual are shown beneath the figure: band of 61 bp present or absent. Number one and two above each line stand for A and G allele, respectively. FSHR: follicle stimulating hormone receptor; M: molecular size marker (pBR322 DNA/BsuRI Digest, Fermentas)
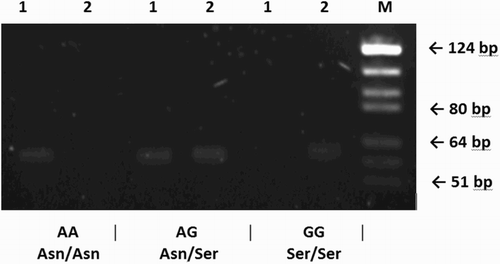
Table 1. Characteristics of the patients (means [95% CI]) recruited to the study with respective p values (ANOVA).
Normalized relative expression data as retrieved from the Relative Expression Software Tool software are presented in . All tested FSHR transcript variants, as well as other transcripts, were detectable in each analyzed sample. Sequences of primers and probes used for FSHR variant detection are presented in . In the whole analyzed group of 22 patients, levels of total transcripts of FSHR, LHCGR, and CYP19A1 were elevated after 24 hours of stimulation with rhFSH as compared to non-stimulated cells (fold change: 1.9 [0.39-11.63), p = 0.001; 3.44 [0.43-22.43], p < 0.001; 6.07 [1.05-57.4], p < 0.001, respectively for FSHR, LHCGR and CYP19A1). FSHR splicing variant del ex2 was similarly up-regulated and p-value approached statistical significance (1.58 [0.25-18.85], p = 0.053). The other tested FSHR variants remained unaltered with normalized relative quantity ranging from 1.04 to 1.14 and p > 0.05. When stratifying the subjects by genotype, the trend was held in homozygotes Asn/Asn and heterozygotes Asn/Ser, although in the former group increase of the FSHR del ex2 variant did not reach statistical significance (1.69 [0.23-25.9], p = 0.23). In Ser/Ser homozygotes the only significantly elevated transcript was CYP19A1 (6.76 [1.62-17.41], p = 0.001) and LHCGR which turned out to be overexpressed at borderline significance (2.8 [0.98-13.13], p = 0.050).
Table 2. Normalized relative quantity (fold change [95% CI]) with levels of significance (p) of studied genes of interst (GOI) and follicle stimulating hormone receptor (FSHR) splicing variants.
Table 3. Sequences of primers and probes used for detection of analyzed transcripts of FSHR gene.
Pairwise comparisons of normalized expression (2−ΔCt) in non-stimulated cultures of LGCs did not reveal any statistically significant differences between the genotypes (data not shown). Analyses comparing normalized relative quantities (2−ΔΔCt) showed that the homozygotes SerSer were associated with lower estimates of fold change of FSHR ins ivs8 and FSHR del ex2 variants (p = 0.018 and p = 0.05, respectively). Total FSHR transcript, representing all variants and remaining distinct FSHR splicing variants followed similar rhFSH-induced changes in steady-state mRNA. Normalized relative expression did not reach statistical significance (p > 0.05).
Steroid production by cultured LGCs and its association with the FSHR rs6166 genotype
Steroid concentrations in culture media were normalized to total RNA content in cell lysates. Total RNA isolated from the cell cultures was highly correlated with GAPDH arbitrary values assigned from the standard curve with 10-fold serial dilutions (Pearson r = 0.82 and r = 0.85, respectively for non-stimulated and stimulated cells). Estradiol and progesterone levels assessed in the non-stimulated LGCs tended to be higher in the carriers of at least one Asn allele, whereas the opposite trend was observed for the relative increase in hormone production (). The differences were not statistically significant (p > 0.05).
Table 4. Normalized to total RNA steroid concentrations and relative increase in steroid concentration (median [interquartile range]) in the culture media of the luteinized granulosa cells.
The relative increase of estradiol and progesterone in culture media correlated with the relative expression of CYP19A1 mRNA (Pearson's r = 0.6, p = 0.003 and r = 0.49, p = 0.02, for estradiol and progesterone, respectively). Correlation between progesterone biosynthesis and LHCGR relative expression approached statistical significance (Pearson's r = 0.41, p = 0.0558). No associations between steroid production and FSHR mRNA expression were observed ().
Table 5. Pearson correlation coefficients (r) with respective levels of significance (p) between normalized increase in steroid concentration and normalized relative quantity of studied genes of interest (GOI).
Discussion
The rs6166 (p. Asn680Ser) polymorphism of the FSHR gene has emerged as a marker of ovarian sensitivity to FSH. Female carriers of different genotypes differ by biological and clinical characteristics related to reproductive function. The differences are minor and impossible to reproduce in some populations studied. The observations at the clinical level may reflect subtle alterations in receptor activity which, in certain conditions, alters the effectiveness of folliculogenesis.
While there are no studies providing mechanistic support for the role of rs6166 polymorphism, it is still not generally recommended to include FSHR genotyping in a routine assessment of infertile women. Studies addressing the issue of functionality of different genetic variants assumed a direct change in downstream signaling of encoded variant proteins. The studies failed to find differences in the production of a major transducer of the FSHR cascade, namely cAMP in the in vitro systems [Simoniet al. 1999; Sudo et al. Citation2002]. However, although FSHR is linked to a G-protein it apparently couples to multiple signaling pathways [Ulloa-Aguirre et al. Citation2007]. It should be recognized that rs6166 could be a marker with no direct impact on FSHR biology. Functional polymorphisms may reside in vast, only partially explored non-coding sequences of the FSHR gene region. It is suffice to mention that a search of Entez dbSNP database for intronic polymorphisms of FSHR with a minor allele frequency exceeding 10% gives 292 results. It becomes widely accepted that polymorphisms located in introns can alter the expression of genetic information in multiple ways [Johnson et al. Citation2005]. Interestingly phenotypic changes evoked by coding or non-coding DNA sequence variations may result from changes in gene regulation, including mRNA stability and transcription as well as mRNA processing, posttranslational modification, and translation itself [Johnson et al. Citation2005].
In many species analyzed posttranscriptional processing seems to be important at different stages of follicle development and may account for pleiotropic effects of FSHR activation in target cells. Data from animal models suggests that the expression of FSHR proteins may be even more dependent on splicing than changes in mRNA levels [Findlay and Drummond Citation1999]. In some species FSHR splice variants with different properties were consistently documented [Sairam and Babu Citation2007]. Results of the current study indicate that the FSHR gene undergoes alternative splicing in human luteinized granulosa cells cultured in gonadotropin-free conditions. In the in vitro model adopted, distinct alternative variants were co-expressed constitutively in non-stimulated cells and we found no detectable dependency on FSHR genotype. The dominant variant co-amplified by the assay designed for capturing a maximum number of transcripts (total FSHR) appears to be different from the four alternative variants tested and most probably represents a full-length isoform. Expression of all distinct variants correlated with total FSHR and the only one which was significantly up-regulated was that lacking exon 2 (del ex2).
Our observation of the existence of the splice variants is in line with the previous studies evaluating FSHR transcription profile in human cumulus cells and testicular tissue. These studies documented the presence of one out of four splicing variants in addition to the full-length transcript in a fraction of the samples and attempted to correlate individual transcripts with reproductive potential [Gerasimova et al. Citation2010; Song et al. Citation2002]. In contrast to the results presented above, these studies did not show a concurrent presence of multiple transcripts in any sample analyzed. This may be attributed to different cell types (parietal granulosa vs. cumulus granulosa vs. Sertoli) and experimental settings, namely long-term cell culturing or measures on cells or tissues taken directly from their natural environment. As opposed to isolated cells grown in culture, analysis of tissues processed immediately after acquisition by biopsy allows one to consider the influence of the follicular milieu and modulating local factors inter-connecting cellular components. Additionally, the technical factors could also contribute to the differences observed. Previous studies utilized semi-quantitative RT-PCR with gel detection of amplification products which has limited sensitivity. Even more importantly, PCR conditions, including the initial concentration of target and PCR substrates and co-amplification of the more abundant variants must be taken into account. Predominance of one isoform, particularly when coupled with a small quantity of initial template may significantly reduce the detection level of less abundant transcripts [Vandenbroucke et al. Citation2001]. For the same reasons we did not attempt to perform accurate quantitative analysis of transcriptional variants studied.
There was a tendency to lessen the relative increase in expression of the total and distinct alternative transcripts of FSHR after rhFSH stimulation in Ser/Ser homozygotes. The inability to show significant associations is most likely due to the lack of power caused by limited sample size and overall low levels of variants (high Ct values). However allele-specific imbalance in FSHR expression could be an intriguing explanation of different receptor activity. One can speculate that rs6166 itself or alternatively a linked, intronic polymorphism or a set of individual genetic variants in a haplotype block can influence pre-mRNA stability or transcription efficiency. Thus, down-regulation and desensitization might be genotype-dependent. We decided to measure mRNA expression after 24 hours of stimulation because we believed relatively small changes in steady-state mRNA would be easiest to record at this time point. Our assumptions were based on the initial experiments which indicated a period of down-regulation followed by up-regulation clearly apparent after 24 hours of incubation in the presence of saturating doses of rhFSH. Therfore it can be presumed that FSHR autoregulation by its ligand may be allele-specific with carriers quantitatively differing in terms of mRNA expression. The potential predictive role of FSHR expression was demonstrated by a previous study that showed lower mRNA and protein levels in granulosa cells of females with poor ovarian response to stimulation [Cai et al. Citation2007].
FSH-induced expression of LHCGR and CYP19A1 was significantly increased and showed no differences between genotypic groups. The tendency toward lower levels of steroid hormones in Ser/Ser homozygotes was observed but it was not statistically significant. Basal as well as rhFSH-induced increase in steroid concentrations showed very high variability even within genotype groups. Using a similar cell culture model, Nordhoff et al. [2011] were unable to record major differences in steroid biosynthesis. This may indicate that activity of steroidogenic machinery was predetermined before oocyte retrieval.
As rs6166 polymorphic variants occur in the Caucasian population at almost equal frequency, it suggests that none are associated with considerable advantage in terms of reproductive potential and natural selection. Nevertheless, in certain clinical settings, including multifollicular ovarian stimulation with exogenous gonadotropins, FSHR polymorphism may considerably influence follicular growth. Quantitative variability in FSHR activity attributable to differences in mRNA stability or regulation of the transcription is one of the tempting hypotheses that requires verification in future studies. Differences in FSHR expression of even 20-50% may be responsible for noticeable variability in granulosa cell and entire follicle biology. Our study indicates the need for further functional studies in this area. Lessons from animal models allow one to presume that alternative transcripts may also add to phenotypic variability at least at some stages of folliculogenesis. Significance and allelic dependence of alternative posttranscriptional processing, although not documented in our study cannot be excluded.
Materials and Methods
Subjects
The initial experiments to establish the final experimental model were performed on pooled cells from 3 random, consecutive patients. For the final study, 22 women undergoing IVF treatment were recruited. Inclusion criteria comprised: i) age < 40 y, ii) normoovulatory cycles, iii) presence of both ovaries in USG, iv) male, tubal, or idiopathic factor infertility, v) absence of polycystic ovary syndrome, endometriosis, chemo- or radiotherapy, and vi) Polish nationality. Informed consent was obtained and the study was approved by the Ethics Committee of the Medical Univeristy of Białystok.
Controlled ovarian stimulation
Patients were stimulated in the long protocol in which the administration of gonadotropins starts after the pituitary desensibilization with GnRH agonist (Diphereline SR 3.75; Ipsen Pharma, Paris, France). Stimulation was performed with the use of rhFSH (Gonal F; Merck Serono, Darmstadt, Germany). Development of follicles was monitored by a transvaginal ultrasonography and hormonal tests. Gonadotropin dose was corrected according to clinical judgment. When at least 2 follicles reached 17-18 mm in diameter and estradiol concentration was at least 200pg/ml/follicle > 14 mm patients were given 10,000 IU rhCG (Ovitrelle; Merck Serono) to induce ovulation. Collection of oocytes was performed after 36h from rhCG injection. Developed follicles were punctured under transvaginal ultrasound guidance and the contents were aspirated. Follicular fluids from each patient were collected after retrieval of oocytes, pooled, and stored at 4°C.
Granulosa cell cultures
Luteinized granulosa cells were separated from follicular fluid by hypotonic erythrocyte lysis and centrifugation as described previously [Lobb and Younglai Citation2006]. Next, 1.5 - 2 x 105 of the viable cells per well were seeded in at least 4 wells of 12-well microplates (TPP) and cultured for 7 d at 37°C in a humidified, 5% CO2 atmosphere in 1 ml of medium. The culture medium consisted of DMEM/HAM's-F12 (1:1 mixture) (PAA; Cölbe, Germany) supplemented with 5% defined FBS ‘GOLD’ (PAA), testosterone (Sigma-Aldrich; Poznań, Poland) 10ng/ml and antibiotics (1% Streptomycin-penicillin, PAA). The cells were washed every 24 h with PBS. The isolation and establishing cell cultures were the same for initial and final experiments. The initial experiments were undertaken to evaluate the responsiveness to rhFSH stimulation under different experimental conditions. Additionally, kinetics of FSHR and LHCGR mRNA expression was assessed (). In the final experiment following the 7-d-long preculturing period half of the cell cultures from each patient were incubated in serum-depleted environment in the presence of 0.5 IU/l rhFSH (Gonal F; Merck Serono, Darmstadt, Germany). The remaining non-stimulated cells were cultured in the medium devoid of serum and rhFSH and used as calibrators. After 24 h of incubation, the medium was collected and guanidinium-based lysis solution was added to washed LGCs. Lysates and culture media were stored at -80°C until subsequent analyses.
Quantitative real-time PCR (qPCR)
Total RNA was isolated with RNAqueous-Micro Kit supplied by Ambion, Life Technologies, Warsaw, Poland. The Agilent 2100 Bioanalyzer and RNA 6000 Pico Kit was used to control the quality and assess quantity of total RNA. Gene expression was measured after reverse transcription (High Capacity RNA-to-cDNA Master Mix, Applied Biosystems, Warsaw, Poland) by real-time PCR with predesigned TaqMan Gene Expression Assays (LHCGR, CYP19A1) or designed to our specifications for custom TaqMan assays (Applied Biosystems). Manufacturer's instructions were followed during the analyses. The twenty-two cDNA samples, inter-run calibrators and controls were run in triplicates on an ABI7900HT and analyzed by sequence detection systems (SDSs) v2.3. Expression levels were normalized to GAPDH (TaqManRNase P Control Reagents; Applied Biosystems).
Genotyping
Genotypes were determined by allele-specific PCR (AS-PCR) in the DNA samples extracted directly from LGCs remaining after the separation and seeding (Genomic mini, A&A Biotechnology, Gdynia, Poland). Allele specific primers were designed to differ from each other by a single base pair at 3' end: 5' - CAGCTCCCAGAGTCACCAA and 5' - CAGCTCCCAGAGTCACCAG. The common reverse primer sequence was 5' - TGGGCTAAATGACTTAGAGGGACA. Forward and reverse primer pairs flanked a 61 bp sequence. To increase specificity of the reaction SNPase (Bioron, Ludwigshafen, Germany), which has a decreased mutation rate as compared to standard Taq polymerase with 5'-3' activity, was utilized. The reaction mixture, prepared in duplicate with different allele-specific forward primers contained 1 x buffer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 2 pmol of each primer, and 20 – 70 ng of template DNA in a volume of 10 µl. Thermal cycling conditions were as follows: 5 min at 94°C, followed by 30 cycles of 94°C for 30s, 64°C for 45s, and 72°C for 30s, with a final extension at 72°C for 5 min. The amplification products were visualized using 3.5% agarose electrophoresis with ethidium bromide staining. During optimization of the AS-based discrimination of the alleles the accuracy and reliability of the reactions with different template concentrations were confirmed by sequencing. For this purpose, a 152 bp fragment was amplified using forward primer: 5' - TGTGGCTGCTATGAAATGCAA and reverse primer described above.
Hormone measurements
Concentrations of 17-β-estradiol and progesterone in the culture media were measured by an enzyme-linked fluorescent assay with a Vidas instrument (bioMérieux, Warsaw, Poland) using the Vidas estradiol II and a Vidas progesterone kits (bioMérieux). Manufacturer's protocols were followed during the analyses.
Statistical analysis
Relative expression was assessed by ∆∆Ct method with normalization to GADPH utilizing REST 2009 software [Pfaffl et al. Citation2002]. As the efficiency of the qPCR reactions for FSHR, LHCGR, and CYP19A1, assessed by serial dilution standard curve was between 102 and 110 % no correction in this regard was applied. Normalized relative quantity (NRQ) data and non – normally distributed steroid concentrations were log-transformed before comparisons between genotypic groups. Differences between three genotypes with regard to the clinical parameters and the outcomes (mean log NRQ, log steroid concentrations) were compared by one-way ANOVA followed by pair-wise comparisons using two-tailed unpaired t-tests. For correlations between the parameters Pearson's correlation was used. A p-value less than 0.05 was considered significant. Data were presented as mean (95% CI) or median (interquartile range) unless otherwise indicated.
Abbreviations
| FSHR: | = | follicle stimulating hormone receptor |
| LHCGR: | = | luteinizing hormone/chorionic gonadotropin receptor |
| CYP19A1: | = | aromatase FSHR del ex2, FSHR sel ex6, FSHR del ex9: FSHR transcript variants with deletion of exon 2,6 and 9 |
| FSHR ins ivs8: | = | FSHR transcript variant with insertion of intron 2 sequence between exons 8 and 9 |
| LGCs: | = | luteinized granulosa cells |
| SNP: | = | single nucleotide polymorphism |
| AS-PCR: | = | allele-specific PCR |
| qPCR: | = | quantitative real-time PCR |
| GnRH: | = | gonadotropin-releasing hormone |
| rhFSH: | = | recombinant human follicle stimulating hormone |
| GAPDH: | = | glyceraldehyde 3-phosphate dehydrogenase. |
Declaration of interests: The research was financed by the Polish National Science Centre (grant No. NN401062535). The authors report no conflicts of interest.
Author contributions: Conceived and designed the experiments: GZ, SW, LC. Performed the experiments: GZ, SW. Analyzed the data: GZ. Contributed materials: SW. Wrote the manuscript: GZ, SW, LC.
References
- Aittomäki, K., Lucena, J.L., Pakarinen, P., Sistonen, P., Tapanainen, J., Gromoll, J. (1995) Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell 82:959–968.
- Binder, H., Dittrich, R., Hager, I., Müller, A., Oeser, S., Beckmann, M.W. (2008) Association of FSH receptor and CYP19A1 gene variations with sterility and ovarian hyperstimulation syndrome. Reproduction 135:107–116.
- Cai, J., Lou, H.Y., Dong, M.Y., Lu, X.E., Zhu, Y.M., Gao, H.J. (2007) Poor ovarian response to gonadotropin stimulation is associated with low expression of follicle-stimulating hormone receptor in granulosa cells. Fertil Steril 87:1350–1356.
- Desai, S.S., Achrekar, S.K., Pathak, B.R., Desai, S.K., Mangoli, V.S., Mangoli, R.V. (2011) Follicle-stimulating hormone receptor polymorphism (G-29A) is associated with altered level of receptor expression in Granulosa cells. J Clin Endocrinol Metab 96:2805–2812.
- Dickey, R.P., Thornton, M., Nichols, J., Marshall, D.C., Fein, S.H., Nardi, R.V. (2002) Comparison of the efficacy and safety of a highly purified human follicle-stimulating hormone (Bravelle) and recombinant follitropin-beta for in vitro fertilization: a prospective, randomized study. Fertil Steril 77:1202–1208.
- Findlay, J.K. and Drummond, A.E. (1999) Regulation of the FSH Receptor in the Ovary. Trends Endocrinol Metab 10:183–188.
- Gerasimova, T., Thanasoula, M.N., Zattas, D., Seli, E., Sakkas, D. and Lalioti, M.D. (2010) Identification and in vitro characterization of follicle stimulating hormone (FSH) receptor variants associated with abnormal ovarian response to FSH. J Clin Endocrinol Metab 95:529–536.
- Johnson, A.D., Wang, D. and Sadee, W. (2005) Polymorphisms affecting gene regulation and mRNA processing: broad implications for pharmacogenetics. Pharmacol Ther 106:19–38.
- Jun, J.K., Yoon, J.S., Ku, S.Y., Choi, Y.M., Hwang, K.R., Park, S.Y. (2006) Follicle-stimulating hormone receptor gene polymorphism and ovarian responses to controlled ovarian hyperstimulation for IVF-ET. J Hum Genet 51:665–670.
- Klinkert, E.R., Broekmans, F.J., Looman, C.W., Habbema, J.D. and te Velde, E.R. (2005) Expected poor responders on the basis of an antral follicle count do not benefit from a higher starting dose of gonadotrophins in IVF treatment: a randomized controlled trial. Hum Reprod 20:611–615.
- Lobb, D.K. and Younglai, E.V. (2006) A simplified method for preparing IVF granulosa cells for culture. J Assist Reprod Genet 23:93–95.
- Nembaware, V., Wolfe, K.H., Bettoni, F., Kelso, J. and Seoighe, C. (2004) Allele-specific transcript isoforms in human. FEBS Lett 577:233–238.
- Nordhoff, V., Sonntag, B., von Tils, D., Götte, M., Schüring, A.N., Gromoll, J. (2011) Effects of the FSH receptor gene polymorphism p.N680S on cAMP and steroid production in cultured primary human granulosa cells. Reprod Biomed Online 23:196–203.
- Perez Mayorga, M., Gromoll, J., Behre, H.M., Gassner, C., Nieschlag, E. and Simoni, M. (2000) Ovarian response to follicle-stimulating hormone (FSH) stimulation depends on the FSH receptor genotype. J Clin Endocrinol Metab 85:3365–3369.
- Pfaffl, M.W., Horgan, G.W. and Dempfle, L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36.
- Sairam, M.R. and Babu, P.S. (2007) The tale of follitropin receptor diversity: a recipe for fine tuning gonadal responses? Mol Cell Endocrinol 260-262:163–171.
- Simoni, M., Gromoll, J., Höppner, W., Kamischke, A., Krafft, T., Stähle, D. (1999) Mutational analysis of the follicle-stimulating hormone (FSH) receptor in normal and infertile men: identification and characterization of two discrete FSH receptor isoforms. J Clin Endocrinol Metab 84:751–755.
- Song, G.J., Park, Y.S., Lee, Y.S., Lee, C.C. and Kang, I.S. (2002) Alternatively spliced variants of the follicle-stimulating hormone receptor gene in the testis of infertile men. Fertil Steril 77:499–504.
- Sudo, S., Kudo, M., Wada, S., Sato, O., Hsueh, A.J. and Fujimoto, S. (2002) Genetic and functional analyses of polymorphisms in the human FSH receptor gene. Mol Hum Reprod 8:893–899.
- Ulloa-Aguirre, A., Zariñán, T., Pasapera, A.M., Casas-González, P. and Dias, J.A. (2007) Multiple facets of follicle-stimulating hormone receptor function. Endocrine 32:251–263.
- Vandenbroucke, I.I., Vandesompele, J., Paepe, A.D. and Messiaen, L. (2001) Quantification of splice variants using real-time PCR. Nucleic Acids Res 29:E68–68.
- Wunsch, A., Ahda, Y., Banaz-Yaşar, F., Sonntag, B., Nieschlag, E., Simoni, M. (2005) Single-nucleotide polymorphisms in the promoter region influence the expression of the human follicle-stimulating hormone receptor. Fertil Steril 84:446–453.