Abstract
We explored the application of single nucleotide polymorphism microarray (SNP array) in molecular karyotype analysis for early spontaneous abortion detection in assisted reproductive technology (ART). SNP array was performed in 81 cases. Of the 81 cases, 16 experienced natural conception (NC) and 65 were pregnant by ART. Of the 65 cases, 4 underwent artificial insemination (AI), 32 fresh in vitro fertilization-embryo transfer (IVF-ET), 9 fresh intracytoplasmic sperm injection (ICSI), and 20 thawed embryo transfer. In the 81 cases examined 69.1% displayed an abnormal molecular karyotype. In the subjects greater than 35 years of age, the abnormal molecular karyotype rate was 87.5% higher compared to 61.4% in younger individuals (P < 0.05). There was no significant difference in the abnormal molecular karyotype rate or type between ART (64.6%) and NC (87.5%). Compared with traditional cytogenetic diagnosis, the SNP array can identify a greater number of abnormal karyotypes.
Introduction
Assisted reproductive technology (ART) has enabled many more infertile couples to have children. Over the years the ART pregnancy rate has greatly increased yet the take-home baby rate is still 20-30%. In comparison fetal loss in the general population is close to 14% and varies with age [Nybo Andersen et al. 2000].The spontaneous abortion of natural conception is strongly associated with embryonic chromosomal abnormalities which occur in more than 50% of spontaneous abortions after natural conception. The pregnancy loss rate is 22-63% in ART, and 58% occur before 6-weeks gestation and 10-45% occur in the 6-weeks gestation [Farr et al. 2007]. The causes of spontaneous abortion in ART are complex, and are not well-understood. Karyotyping of chorionic villi suggests that embryonic chromosomal abnormalities are still the main cause of spontaneous abortion in ART.
Traditional cytogenetic karyotype analyses are generally limited to 5 – 10 Mb resolution [Emanuel and Saitta 2007] and are subject to cell culture failure (10-40%). Array comparative genomic hybridization (array CGH), provides an effective alternative affording a resolution of approximately 3 kb. This technology has been used in the analysis of spontaneous miscarriages [Schaeffer et al. 2004]. Compared with array aCGH, single nucleotide polymorphism microarrays (SNP arrays) can provide an even higher resolution and are capable of discerning polyploidy and chromosomal imbalance [Lathi et al. 2012]. In order to explore appropriateness and etiology the SNP array was used in molecular karyotype analysis of ART of early fetal demise.
Results and Discussion
SNP array analysis was performed for 81 tissue samples. The average age of the subject was 32.9 ± 5.1 years in the 81 cases. The call rate for each probe was greater than 99%. A normal chromosomal pattern was observed in 25 cases (30.9%). In comparison chromosomal abnormalities were observed in 56 cases (69.1%). The abnormal molecular karyotype rate for women 35 years of age and less was 61.4% (35/57). This was substantially lower than the rate of 87.5% (21/24) in the cases of women greater than 35 years (P < 0.05, ).
Table 1. Abnormal molecular karyotype in different age groups.
There were no significant differences in maternal age, paternal age, maternal body mass index (BMI), previous live birth history, and previous history of miscarriage between ART and natural conception (NC) groups (). The abnormal molecular karyotype rate was lower in ART (64.6%) than in NC (87.5%), but the difference was not statistically significant ().
Table 2. General status in ART and NC groups.
Table 3. Abnormal molecular karyotype in NC and ART groups.
Of the 56 samples presenting an abnormal karyotype, trisomy was the most frequent (62.5%), followed by chromosome deletion (16.1%), duplication (5.4%), triploidy (5.4%), and mosaicism (5.4%). There was no significant difference in the type of abnormal molecular karyotype between ART and NC () . As expected trisomy chromosome 16 (28.6%) was the most common followed by trisomy chromosome 22 (20.4%), trisomy chromosome 14 (6.1%), trisomy chromosome 12 (6.1%), and trisomy chromosome 8 (6.1%) (examples shown in and ). All provide useful information for the choice of probe in prenatal diagnosis of fluorescence in situ hybridization.
Figure 1. Distribution of autosomal trisomies. The percentage of total for each trisomy is shown. Note that trisomies were evident for chromosomes 2, 3, 5, 6, 7, 8, 9, 10, 12, 13, 14, 15, 16, 20, 21, and 22. Chromosomes 16 and 22 display the highest rate of trisomy.
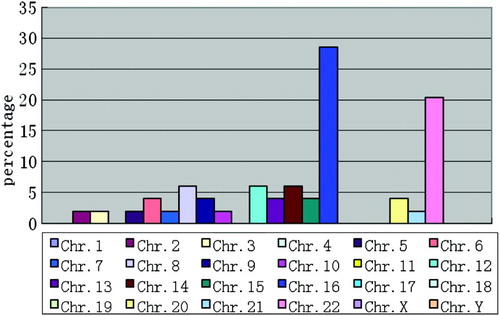
Figure 2. Molecular karyocyte analysis of single nucleotide polymorphism microarray from spontaneous abortion. A) shows the normal diploid chromosome 1 diagnostic reading obtained from POC, Normal AA, AB, and BB alleles and a 0 reading for the smooth log R ratio is observed. B) shows the trisomy chromosome 16 diagnostic reading obtained from POC, AAA, AAB, ABB, and BBB alleles are observed and a significant shift in the smooth log R ratio is observed, consistent with the trisomy karyotype. C) shows the monosomy chromosome 21 diagnostic reading obtained from POC, AA, BB alleles are observed without AB and A significant shift in the smooth log R ratio is observed, consistent with the monosomy karyotype. D) demonstrates the duplication of q24.32 → q24.33 reading of chromosome 12. AAA, AAB, ABB, and BBB alleles are observed from q24.32 to q24.33. A significant shift in the smooth log R ratio is observed from q24.32 to q24.33 of chromosome 12. E) demonstrates the deletion of p11.12 → p11.23 reading of chromosome 11. AA,AB, and BB alleles are observed from p11.2 to pter and from q10 to qter of chromosome 11, however AA and BB alleles are observed without AB from p11.12 to p11.23 of chromosome 11 represented. A significant shift in the smooth log R ratio is observed from p11.12 to p11.23 of chromosome 11.
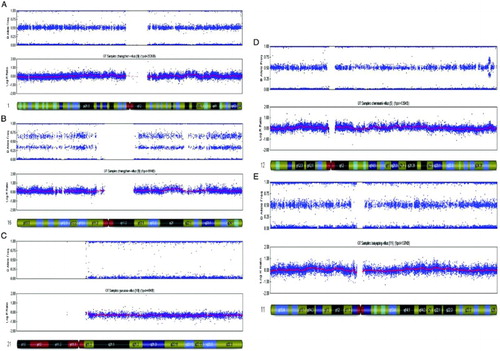
Table 4. Type of abnormal molecular karyotype in abortion tissue.
Advantages and limitations of SNP microarrays in the molecular karyotyping and diagnosis of spontaneous abortion
The causes of spontaneous abortion are complex and are associated with many factors such as chromosome abnormality (early pregnancy loss) and single-gene disorders (late pregnancy loss) [Laurino et al. 2005], infectious diseases, uterine malformation, endocrine abnormality, and immune-related diseases [Toth et al. 2010]. Chromosomal abnormalities are associated with more than 60% of the spontaneous abortions. Therefore, genetic analysis provides an important approach for determining the cause of spontaneous abortion and for providing appropriate counseling for couples undergoing ART. The use of cell culture followed by G-banding karyotype analysis has traditionally been applied affording a 5 – 10 Mb view of the genome. The failure rate of chromosome analysis caused by cell culture is 5-23.5% [Gu et al. 2009]. New analytical methods such as aCGH and SNP array have shown their advantages. Chromosome microarray analysis (CGH and SNP) is becoming a first-tier clinical diagnostic test of prenatal diagnosis for individuals with developmental disabilities or congenital anomalies [Miller et al. 2010]. As reported the SNP array was used in molecular karyotype analysis of 81 samples. A total of 69.1% of the samples displayed an abnormal karyotype. This was significantly higher than that of traditional method. The chromosome abnormalities that occurred in 56 cases included trisomies, triploidies, mosaics, deletions, and duplications. The rate of chromosome deletions and duplications were significantly higher (21.5%) as compared to that observed for traditional cytogenetic diagnosis (4.8%) [Bettio et al. 2008]. However, SNP arrays still have some limitations. They cannot identify chromosomal balanced structural aberrations such as balanced translocation, inversion, or tetraploid chromosomes. However, chromosomal balanced translocations and inversions are rarely observed in these cases [Zuffardi et al. 2011].
Factors affecting the abnormal molecular karyotype associated with loss
The age of the mother is a primary factor in the presentation of abnormal molecular karyotype. In women over 35 years of age the frequency of an abnormal karyotype associated with fetal demise was 87.5%, significantly higher than 61.4% in the cases aged 35 years or younger. In older infertile couples seeking ART, preimplantation genetic diagnosis (PGD) can be used to identify abnormal embryos, so only the most fit are selected for embryo transfer and thus improving ART outcomes. SNP microarray has been used in PGD, and the results suggest SNP-PGD can increase pregnancy rate and reduce early pregnancy loss in older couples [Treff et al. 2011; Benner et al. 2010].
Our data indicated that there were no statistical differences in the mean age and abnormal karyotype rate between NC and ART, which is consistent with the results reported by Bettio et al. [2008] and Kim et al. [2010]. The causes of ART early spontaneous abortion are complex, and abnormal molecular karyotype is the main cause. Compared with traditional cytogenetic method, SNP microarray can identify more and smaller abnormal karyotypes such as deletion and duplication. The data presented above using SNP microarrays suggest that ART does not increase the frequency of abnormal molecular karyotypes observed in fetal demise. Additional confirmation studies will be required.
Materials and Methods
All study methods were approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University. All subjects enrolled in the study gave written formal consent to participate.
Materials
Between June 2011 and February 2012, products of conception (POC) were collected from 81 couples with normal chromosomes. Of the 81 cases, 16 experienced NC and 65 were pregnant by ART. Of the 65 cases, 4 underwent AI, 32 fresh IVF-embryo transfer, 9 fresh ICSI-embryo transfer, and 20 thawed embryo transfer. All chorionic villi were obtained from the 81 cases. Under anatomic microscope, decidua was carefully removed from chorionic villi. The chorionic villi were washed with phosphate buffer solution (PBS) to remove coagulated blood and decidua, and then stored at -80°C for DNA extraction.
DNA extraction
Sample (25 mg) was ground in a mortar, and then DNA was extracted with QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturers' instructions. The extracted DNA was quantitated with Nanovue Plus (GE, Fairfield, Connection, USA), and then stored at −20°.
SNP microarray analysis
DNA (200ng) was used as an input for a single array. DNA amplification, tagging, and hybridization were performed according to the manufacturer's protocols. The arrays were scanned on a HiScanSQ (Illumina,USA). Data analysis was performed using GenomeStudio (Illumina, standard settings). The HapMap control set provided by the manufacturer was used as a control. Standard settings for SNP arrays were adjusted for a cutoff value of 0.15, homozygous frequency threshold of 0.95. Plot of two parameters, the B allele frequency (BAF) and the log2R ratio, providing information about genotype and copy number, respectively, were examined by visual inspection using GenomeStudio software. The BAF is a value between 0 and 1, and represents the proportion contributed by one SNP allele (B) to the total copy number. A BAF value of 0.5 indicates a heterozygous genotype (AB), whereas 0 and 1 indicate homozygous genotypes (AA and BB, respectively). For example, a region with a deletion in all cells will show homozygosity – bands at 1 and 0. In other words, a loss should always be accompanied by a LOH visible in the BAF plot. A region of single-copy-number gain in all cells, in addition to the two bands of homozygous SNPs at BAF¼0 (AAA) and BAF¼1 (BBB), also showed two bands: one at BAF¼0.33 with SNPs having genotype AAB and one at BAF¼0.67 with SNPs having genotype ABB.
Statistical analysis
Data were analyzed with SPSS10.0 software. Chi square test was used in the comparison of rates. The t test was used in comparison of mean. Statistical significance was established at P < 0.05.
Abbreviations
| SNP array: | = | single nucleotide polymorphism microarray |
| ART: | = | assisted reproductive technology |
| aCGH: | = | array comparative genomic hybridization |
| NC: | = | natural conception |
| AI: | = | artificial insemination |
| IVF-ET: | = | in vitro fertilization-embryo transfer |
| ICSI: | = | intracytoplasmic sperm injection |
| CGH: | = | comparative genomic hybridization |
| POC: | = | products of conception |
| PBS: | = | phosphate buffer solution |
| BAF: | = | B allele frequency |
| PGD: | = | preimplantation genetic diagnosis. |
Declaration of interests: This study was supported by the grants from key project of Henan Province (112102310103), medical tackle key problems in science and technology of Henan Province (2011020011, 2011020045), and Youth Innovation Project of the First Affiliated Hospital of Zhengzhou University. The authors have no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
Author contributions: Conceived and designed the study: GL,Y-pS; Performed the study: GL, YL, N-nH, L-lH, Y-lZ, YW, F-lD,Y-cS; Analyzed the data: Y-hG; Wrote the manuscript: GL.
References
- Benner, A., Chipko, C., Pen, R. and Kearns, W.G. (2010) Clinical results on single cells from 470 embyros using 23-chromosome single nucleotide polymorphism (SNP) microarray preimplantation genetic screening (PGS) from 45 patients. Fertil Steril 94:S123.
- Bettio, D., Venci, A. and Levi Setti, P.E. (2008) Chromosomal abnormalities in miscarriages after different assisted reproduction procedures. Placenta 29: pSB126–S128.
- Emanuel, B.S and Saitta, S.C. (2007) From microscopes to microarrays: dissecting recurrent chromosomal rearrangements. Nat Rev Genet 8:p869-883.
- Farr, S.L., Schieve, L.A. and Jamieson, D.J. (2007) Pregnancy loss among pregnancies conceived through assisted reproductive technology, United States, 1999-2002. Am J Epidemiol 165:1380-1388.
- Kim, J.W., Lee,W.S., Yoon, T.K., Seok, H.H., Cho, J.H., Kim, Y.S. , (2010) Chromosomal abnormalities in spontaneous abortion after assisted reproductive treatment. BMC Medical Genetics 11:153.
- Lathi, R.B., Massie, J.A., Loring, M., Demko, Z.P., Johnson, D., Sigurjonsson, S., (2012) Informatics enhanced SNP microarray analysis of 30 miscarriage samples compared to routine cytogenetics. PLoS One 7:e31282.
- Laurino, M.Y., Bennett, R.L., Saraiya, D.S., Baumeister, L., Doyle, D.L., Leppig, K., (2005) Genetic evaluation and counseling of couples with recurrent miscarriage: recommendations of the National Society of Genetic Counselors. J Genet Couns 14:165–181.
- Miller, D.T, Adam, M.P., Aradhya, S., Biesecker, L.G., Brothman, A.R., Carter, N.P., , (2010) Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am J Hum Genet 86:749–764.
- Nybo Andersen, A.M., Wohlfahrt, J., Christens, P., Olsen, J. and Melbye, M. (2000) Maternal age and fetal loss: population based register linkage study. BMJ 320:p1708-1712.
- Schaeffer, A.J., Chung, J., Heretis, K., Wong, A., Ledbetter, D.H. and Lese Martin, C. (2004) Comparative Genomic Hybridization–Array Analysis Enhances the Detection of Aneuploidies and Submicroscopic Imbalances in Spontaneous Miscarriages. Am. J. Hum. Genet 74:1168–1174.
- Toth, B., Jeschke, U., Rogenhofer, N., Scholz, C., Würfel, W., Thaler, C.J., (2010) Recurrent miscarriage: current concepts in diagnosis and treatment. J Reprod Immunol 85:25–32.
- Treff, N.R., Northrop, L.E., Kasabwala, K., Su, J., Levy, B. and Scott, R.T. (2011) Single nucleotide polymorphism microarray-based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil Steril 95:1606-12.e1-2.
- Zuffardi, O., Vetro, A., Brady, P. and Vermeesch, J. (2011) Array technology in prenatal diagnosis. Semin Fetal Neonatal Med 16:94-98.
- Gu, Y., Xie, J. S., Luo, F.-W., Geng, X., Zhang, H.-K., Shen, H.-N., (2009) Chromosome karyotyping of 220 cases of early spontaneous abortion. Chinese Journal of Birth Health and Heredity 17:38-39.