Abstract
Embryo screening for aneuploidy (AS) is part of preimplantation genetic diagnostics (PGD) and is aimed at improving the efficiency of assisted reproduction. Currently, several technologies, including the well-established fluorescence in situ hybridization (FISH) technique, cover the screening of all chromosomes in a single cell. This study evaluates a novel 24-chromosome FISH technique protocol (FISH-24). A total of 337 embryos were analyzed using the traditional 9-chromosome FISH technique (FISH-9) while 251 embryos were evaluated using the new FISH-24 technique. Embryos deemed nontransferable on Day 3 were cultured in vitro to Day 5 of development, then fixed and reanalyzed according to the technique allocated to each treatment cycle (107 embryos analyzed by FISH-9 and 111 by FISH-24). The global error rate (discrepancy between Day 3 and Day 5 results for a single embryo) was 2.8% after FISH-9 and 3.6% after FISH-24, with a p value of 0.95. Thus, we have established and validated a 24-chromosome FISH-based single cell aneuploidy screening technique, showing that the error rate obtained for FISH-24 is independent of the number of chromosomes analyzed and equivalent to the error rate observed for FISH-9, as a useful tool for chromosome segregation studies and clinical use.
Introduction
A certain percentage of embryos produced by in vitro fertilization (IVF) have chromosomal anomalies [Magli et al. Citation2001; Munne et al. Citation1995, Citation2007], the great majority of which are incompatible with life. An embryo with chromosomal anomalies will not give rise to a pregnancy at all, or will result in an early miscarriage or a child with a genetic syndrome. Aneuploidy, closely related to fertility problems, accounts for many of the chromosomal anomalies observed and occurs either as monosomies or trisomies of the 22 autosomes and the sex chromosomes [Fragouli et al. Citation2008; Treff et al. Citation2010].
Preimplantation genetic diagnosis (PGD) has been used since the 1990s as a tool to detect genetically abnormal embryos so that they can be discarded to achieve, with some certainty, only genetically normal embryos transferred. Aneuploidy screening (PGD-AS) has been shown to improve the efficiency of assisted reproduction, increasing implantation rates while lowering pregnancy loss rates [Forman et al. Citation2012; Gianaroli et al. Citation1999; Munne et al. Citation2005, Citation1999, Citation2003; Scott et al. Citation2012; Yang et al. Citation2012].
However, the predictive value of PGD-AS has several limitations, some deriving from technical procedures, such as the reliability of the genetic test used or the adverse impact of embryo biopsy on implantation, and others arising from intrinsic issues, such as genuine mosaicism and self-correction of aneuploidy within the embryo [Frumkin et al. Citation2008]. Embryo mosaicism occurs naturally during mitosis and is therefore unavoidable, but over the past years PGD-AS techniques have been optimized to overcome many of the former technical limitations. These include fluorescence in situ hybridization (FISH) on the fixed nuclei of biopsied cells which has been the technique of choice for some time. However, the classic PGD-AS for nine chromosomes using FISH technique (FISH-9) can analyze only a restricted number of chromosomal loci [Munne et al. 1998]. Currently, most of the available techniques, including the FISH technique [Ioannou et al. Citation2011, Citation2012], have been developed to enable the analysis of all the chromosomes contained in a single cell. Recent years have seen a rise in the number of molecular techniques in clinical practice that has superseded FISH. Despite this, their efficiency has been assessed in most cases by comparison to the original FISH technique (FISH-9) which analyzes a limited number of chromosomes. As a result, these new methods have shown excellent results, unfairly positioning FISH as an outdated technique. Thus, further improvement of this technique, which is easily applicable in most clinics, has been largely abandoned by some in favor of expensive molecular protocols and equipment. Our group is involved in the implementation of a novel 24-chromosome FISH protocol (FISH-24), consisting of a fine-tuning of the hybridization protocol and validation of its accuracy. The aim of this study is to establish the error rate of FISH-24 by comparing it with the error rate of FISH-9.
Results
PGD-AS by FISH-9 was performed in a total of 58 cycles and PGD-AS by FISH-24 in 47 cycles. shows the study flow chart. On Day 3, single blastomeres from 337 embryos were biopsied and analysed by FISH-9 and 251 were biopsied and analyzed by FISH-24. On Day 5, 107 and 111 embryos from the FISH-9 group and FISH-24 group, respectively, were fixed. A total of 5,635 nuclei were obtained; 2,717 were analyzed using the FISH-9 technique, and 2,918 using the FISH-24 method ().
Figure 1. Study flowchart of the validation process. After a Day-3 FISH analysis of a one-cell biopsy, following a 9-chromosome or a 24-chromosome protocol, non-transferable and non vitrifiable embryos were fixated and their nuclei re-analyzed on Day 5. PGD-AS: preimplantation genetic diagnostics - aneuploidy screening; FISH: fluorescence in situ hybridization.
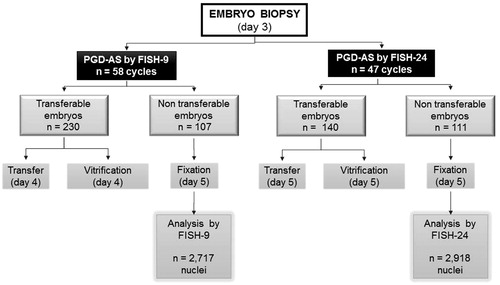
Table 1. Error rate obtained for FISH-9 and FISH-24.
The results obtained on Day 3 by FISH-9 were not confirmed on Day 5 in 3 of 107 reanalyzed embryos (average of 25.4 nuclei per embryo) giving a global error rate of 2.8%. The global error rate was equivalent to the pure error rate in that group (). The discordances for these embryos are described in .
Table 2. Discordance results in FISH-9 and in FISH-24.
The results obtained on Day 3 by FISH-24 were not confirmed on Day 5 in 4 of 111 reanalyzed embryos (average of 26.3 nuclei per embryo), giving a global error rate of 3.6%. Otherwise, the exact results obtained on Day 3 were not confirmed on Day 5 in 6 of 111 reanalyzed embryos, giving a pure error rate of 5.4% (). The discordances for these embryos are described in .
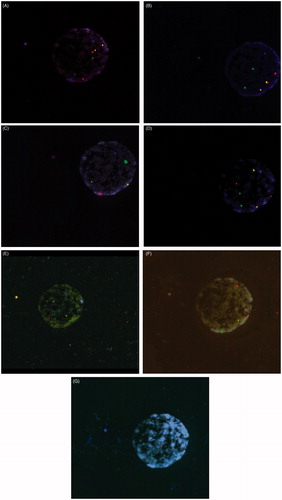
Global error rates of FISH-9 and FISH-24 were statistically equivalent (p = 0.95). In order to determine whether or not the discordant results from Day 3 to Day 5 were due to mosaicism or technical error, the analyzed discordant nuclei from Day 3 (Day 3 vs. Day 5) were re-analyzed, using an extra round of subtelomeric probe hybridization. In each case, the Day 3 diagnosis was confirmed. shows a nucleus in which six rounds of hybridization and a subsequent round of subtelomeric probes have been performed.
Figure 2. Fluorescence in situ hybridization (FISH) of a blastomere nucleus. (A) Round 1- chromosome 1 in spectrum aqua located at 1q12, chromosome 2 in spectrum green, located at D2Z1, chromosome 8 in spectrum gold located at D8Z2 and chromosome 12 in spectrum red located at D12Z1. (B) Round 2- chromosome 3 in spectrum gold located at D3Z1, chromosome 6 in spectrum aqua located at D6Z1, chromosome 7 in spectrum green located at D7Z1 and chromosome 20 in spectrum red located at D20Z1. (C) Round 3- X chromosome in spectrum red located at DXZ1, Y chromosome in spectrum green located at Yq12, chromosome 15 in spectrum gold, located at D15Z1 and chromosome 17 in spectrum aqua located at D17Z1. (D) Round 4- chromosome 4 in spectrum red located at D4Z1, chromosome 9 in spectrum aqua located at 9q12, chromosome 10 in spectrum gold located at D10Z1, and chromosome 11 in spectrum green located at D11Z1. (E) Round 5- chromosome 13 in spectrum red located at 13q14, chromosome 16 in spectrum aqua located at 16q11.2 satellite II DNA, chromosome 18 in spectrum blue located at 18p11.1-q11.1 Alpha satellite DNA, chromosome 21 in spectrum green located at 21q22 and chromosome 22 in spectrum gold located at 22q11.2. (F) Round 6- chromosome 5 in spectrum green located at 5q11.2, chromosome 14 in spectrum red located at 14p11.1-q11.1 and chromosome 19 in spectrum gold located at 19p13.11. (G) Round 7- subtelomeric probes 18p spectrum orange and 18q spectrum green.
Discussion
The root cause of infertility in a large proportion of patients attending reproductive medicine centers is the formation of embryos containing chromosome imbalances, such as aneuploidies. In recent years, PGD-AS for all the chromosomes in a single cell has been used with increasing frequency, and various research groups have described an improvement in clinical outcomes [Scott, et al. Citation2012; Schoolcraft et al. Citation2010; Wells et al. Citation2008].
To date, different strategies have been developed to assess PGD [Handyside Citation2013]: the use of molecular biology techniques based on Comparative Genomic Hybridization (CGH) [Fragouli, et al. Citation2008; Wells, et al. Citation2008], and/or arraySNP technologies [Treff et al. Citation2010, Citation2011], real-time quantitative PCR (qPCR) [Treff et al. Citation2012], next generation sequencing [Fiorentino et al. Citation2014], karyomapping [Natesan et al. Citation2014], or FISH-24 [Ioannou et al. Citation2011, Citation2012].
So far, FISH-24 has resolved the main limitations of the standard FISH technique: (a) the limited number of chromosomes that could be evaluated due to the low number of fluorochromes available and (b) the impact of denaturation/hybridization measures. These limitations were circumvented with the development of new probes and our optimized protocol. FISH-24 involves several rounds of denaturation with formamide treatment. This treatment, if not used properly, can severely damage the DNA structure. However, after denaturation times have been adjusted, a sufficient number of rounds of hybridization can be completed, and all chromosomes can be analyzed. In fact, up to seven rounds of hybridization can be performed with our protocol without breaking or losing the DNA.
The main differences between the FISH-24 protocol presented here and the previously published methods are: (1) there is no need to mix fluorochromes, (2) all spectra are visible to the human eye and can be evaluated under the microscope, and (3) ‘non-result rescue’ (NRR) can be performed. NRR is performed in order to diagnose non-result signals or to confirm monosomies by the use of different extra rounds of subtelomeric probes, in a timeframe compatible with the Day 5 embryo transfer, thus avoiding the need to cryopreserve the embryos.
Nevertheless, validation of new technology is a fundamental part of introducing these procedures into clinical practice. This particular technique is especially difficult as it entails analysis of a single cell. This study validates the analysis of all chromosomes using FISH technology with OligoFISH® probes. Some studies have validated the efficiency of the different techniques applied using the original FISH technique for a limited number of chromosomes, to make comparisons with the abnormalities detected using newly introduced techniques, so in fact they were performing only partial validations [Gutierrez-Mateo et al. Citation2011; Mir et al. Citation2013; Rius et al. Citation2010; Treff et al. Citation2010]. For instance, one of these studies involved the validation of array-CGH (aCGH) [Gutierrez-Mateo et al. Citation2011], in which results obtained following aCGH on Day 3 of embryo development were compared with those obtained from a 12-chromosome FISH study of the embryos on Day 5. This resulted in a 1.9% error rate when the SurePlex amplification technique was used (BlueGnome, Cambridge, UK), and a 9.1% error rate when the GenomePlex was used (Sigma-Aldrich, Dorset, UK). This group defined error rate as: “…a FISH-reanalyzed embryo being erroneously classified by aCGH when the embryo was initially classified as normal or abnormal, but found by FISH to be abnormal or normal, respectively” [Gutierrez-Mateo et al. Citation2011]. This error rate is equivalent to the global error rate defined in our study. However, to further validate the technique it is also necessary to describe the pure error rate defined as the discordance between the exact results found on Day 5 compared to Day 3. Mir et al. [Citation2013] presented another study with a preclinical phase comparing FISH-9 performed on Day 3 with PGD-AS of 24 chromosomes performed by aCGH on Day 4 in all embryos that showed an abnormal result on Day 3. The number of discordant results observed comparing both techniques was 1.3% (2 of 37). One of the misdiagnosed embryos was due to a global error rate and the other due to a pure error rate.
The most common errors in the FISH technique are the interpretation of a false trisomy (when a signal is split) or a false monosomy (when two hybridization signals are very close). The discrepancies between results obtained on Day 3 and Day 5 of diagnosis using both techniques could either be due to embryo mosaicism or to a technical error in the Day 3 analyses. For this reason we reanalyzed slides from Day 3 using subtelomeric probes to determine whether or not the error was technical. In each case Day 3 diagnosis was confirmed, so we can hypothesize that in these particular embryos, discrepancies occurred due to the existence of various cell lines.
Our results show that the diagnostic error obtained after FISH-24 is statistically equivalent to the error rate obtained with FISH-9 (3.6% vs. 2.8%, respectively). The error rate for FISH-24 is also equivalent to the error rate observed for alternative techniques: 1.9% in aCGH BACs [Gutierrez-Mateo et al. Citation2011], 1% (consistency 99%) in aCGH SNPs [Treff et al. Citation2010], and 2% (consistency 98%) in qPCR [Treff and Scott Citation2013; Treff et al. Citation2012].
Moreover, the observed rate of misdiagnosis with FISH-24 is lower than the rate stated as acceptable in the European Society for Human Reproduction and Embryology (ESHRE) PGD Consortium guidelines [Harton et al. Citation2011]. In summary, this study describes and validates an optimized FISH-24 protocol. PGD-AS by aCGH, next generation sequencing or karyotyping seem to have more applications than FISH-24, but FISH-24 stands as a robust and cost-effective test (in comparison to any other 24-chromosome techniques) and can be useful in the clinical setting for patients with economic limitations or in laboratories in which aCGH or other molecular biology methods are not available.
Furthermore, FISH-24 is the only technique that allows cell by cell reanalysis of embryos at an affordable cost. It can also be of great utility in investigating chromosomal segregation, embryo mosaicism, or as part of internal quality control programs in PGD laboratories. In this study we have shown that the error rate of the FISH-24 protocol presented is equivalent to the error rates of other techniques already in clinical use, so it may be acceptable for routine clinical practice and may also constitute an important tool for research purposes.
Materials and Methods
Patients
One hundred and one couples undergoing IVF-PGD cycles between August 2011 and October 2012 were included in the study. PGD-AS was indicated for advanced maternal age, recurrent miscarriage, and genetic male factor. The experimental design is shown in . All women included in the program gave written informed consent for the procedures and for digital recording of the data related to their medical history. Data were accessed and retrieved only by authorized healthcare professionals and anonymity was ensured for subsequent analysis, according to the Spanish Personal Data Protection Act 15/1999 (LOPD) and the principles of the Declaration of Helsinki.
Embryo biopsy and blastomere fixation
On Day 3 of development, embryos were biopsied to perform PGD-AS by FISH-9 or FISH-24. Embryo biopsy was performed with Tyrode’s acid and a single blastomere was removed. Blastomeres were fixed according to the methods described by Tarkowsky and modified as previously described [Munne et al. 1998].
FISH-9 technique: 2 rounds
PGD-AS was performed in nine chromosomes with two rounds of FISH with centromeric, locus specific, and subtelomeric probes [Munne et al. 1998]. The first round was performed using an in-house hybridization mixture with probes for chromosomes X11.1-q11.1 (spectrum green), Yq12 (spectrum orange and aqua), 15p11.1-q11.1 (spectrum orange), 17p11.1-q11.1 (spectrum aqua) (Abbott Molecular, Abbott Park, IL, USA). The second round contained probes for chromosomes 13q14 (spectrum red), 16q11.2 (spectrum aqua), 18p11.1-q11.1 (spectrum blue), 21q22.13-q22.2 (spectrum green), 22q11.2 (spectrum gold) (Abbot Molecular). In order to avoid false monosomies and to rescue ‘non result’ chromosomes, a subsequent round of subtelomeric probes (Abbot Molecular) was routinely included [Colls et al. Citation2007].
The scoring criterion described by Munne et al. [1998] was used.
FISH-24 technique: 6 rounds
The 24-chromosome FISH technique involved 6 rounds of hybridization.
The first four rounds were carried out using oligonucleotide OligoFISH® probes from Cellay (Cellay Inc, Cambridge, MA, USA; 100% specificity). First round: 12p11.1-q11.1 (spectrum red), 2p11.1-q11.1 (spectrum green), 1q12 (spectrum aqua), 8p11.1q11.1 (spectrum gold); Second round: 20p11.1-q11.1 (spectrum red); 7p11.1-q11.1 (spectrum green), 6p11.1-q11.1 (spectrum aqua), 3p11.1-q11.1 (spectrum gold); Third round: Xp11.1q11.1 (spectrum red), Yq12 (spectrum green), 15p11.1-q11.1 (spectrum gold), 17p11.1-q11.1 (spectrum aqua); and Fourth round: 4p11.1-q11.1 (spectrum red), 11p11.1q11.1 (spectrum green); 9q12 (spectrum aqua), 10p11.1-q11.1 (spectrum gold).
The fifth round contained chromosome enumeration probes (CEP) and locus-specific identifier (LSI) probes for chromosomes 13q14 (spectrum red), 16q11.2 (spectrum aqua), 18p11.1-q11.1 (spectrum blue), 21q22.13-q22.2 (spectrum green), 22q11.2 (spectrum gold) (Abbott Molecular; 100% specificity). The sixth round consisted of BAC probes from Cellay: 5q11.2 (spectrum green; 99.5% sensitivity and 100% specificity), 14p11.1q11.1 (spectrum red; 98.6% sensitivity and 100% specificity); 19p13.11 (spectrum gold; 99.5% sensitivity and 100% specificity).
In order to avoid false monosomies and to rescue ‘non result’ chromosomes, two supplementary rounds of subtelomeric probes (Abbott Molecular; 100% specificity) were added when discrepancies arose [Colls, et al. Citation2007].
Although the probes do not have 100% specificity/sensitivity for all chromosomes, working with rounds of subtelomeric probes provides clear results and diagnosis can be obtained in all cells. Using the protocol described here, two extra rounds of subtelomeric probes can be used. Based on our results, two extra rounds are sufficient because the number of signals to be confirmed is six or less.
Hybridization protocol of FISH-24
Denaturation/hybridization of panel 1-4: Slides were denatured for 2 min in the denaturation solution (Formamide 70% at 72°C) for the first hybridization round and 1 min for subsequent hybridizations. Slides were dehydrated in 70%, 80%, 90%, and 100% ethanol for 2 min each. When slides were dried the FISH mixture was dropped onto them. They were then covered with a cover slip and incubated at 37°C for 5 min. gives a chronological description of the denaturation and hybridization processes.
Denaturation/hybridization of panel 5: The process was the same but slides were incubated overnight at 37°C.
Denaturation/hybridization of panel 6: slides were incubated 4 h at 37°C; otherwise the process was the same.
Panel 1-5 washing: Slides were placed in 2 × SSC until the coverslip came off. They were washed at 50°C for 2 min in the wash solution (0.2 × SSC, 0.1% SDS). Afterwards they were placed in 2 × SSC at room temperature and counterstained with 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (panel 1-4) or Antifade II (panel 5).
Panel 6 washing: Slides were placed in 2 × SSC until the coverslip came off. They were then washed at 72°C for 2 min in the wash solution (0.4 × SSC, 0.3% NP-40). Afterwards, they were placed in 2 × SSC, 0.1% NP-40 for 2 min at room temperature and counterstained with DAPI.
Cleaning slides before next hybridization round: Slides were placed in 2xSSC to remove the coverslip and they were washed twice in the wash solution (2xSSC, 0.1% SDS) for 5 min each. They were then dehydrated in 70%, 80%, 90%, and 100% ethanol for 2 min each.
Table 3. Timetable description of the denaturation and hybridization processes of FISH-24.
Fluorescence analysis
Analysis was carried out with an Olympus BX61 microscope equipped with specific filters for FITC (fluorescein isothiocyanate), Texas Red, Aqua, Blue, Orange, Gold, and a triple band-pass filter for DAPI/Green/Red.
Embryo fixation
After PGD-AS on Day 3, chromosomally normal embryos were transferred on Day 4 if they had undergone FISH-9 and on Day 5 if they had undergone FISH-24. Surplus chromosomally normal embryos that reached the blastocyst stage were vitrified for later use by the patient. Non-transferable and non vitrifiable embryos (chromosomally abnormal embryos or normal embryos that did not reach good quality blastocyst stage) were fixed on Day 5 of development. Fifty percent of those were blastocysts, 49% were at the morula stage, and 1% were at the cell stage. In order to avoid diagnostic errors, only fixed nuclei of high quality were considered for inclusion in the analysis, whereas those nuclei presenting overlaps were discarded. As a result, embryos included in the analysis had six or more good quality nuclei (range from 6 to 89). A mean of 25.8 nuclei per embryo was analyzed. The technique of whole embryo fixation was performed in three steps with: (1) ultrapure water, (2) methanol: acetic acid (3:1), and (3) ultrapure water: methanol: acetic acid (4:3:1) [Sandalinas et al. Citation2001].
Error rate estimation
All embryos that were neither transferable nor vitrifiable were incubated until Day 5 and the same number of chromosomes that had been studied on Day 3 were reanalyzed in order to establish the error rate from both techniques. Two different error rates were calculated from the results: (1) global error rate, that was defined as the confirmation of the normal or abnormal diagnosis on Day 5 vs. the one found on Day 3; (2) pure error rate, that was defined as the discordance between the exact type of aneuploidy result found on Day 5 vs. the one found on Day 3.
Statistical analysis
To analyze the incidence of discordant results between Day 3 and Day 5, error rate estimates were compared with the Chi-squared test with a 0.05 confidence interval. All statistical analyses were performed using the software package SPSS 15.0 and R 2.8.0 (IBM Corp., Armonk, New York).
Acknowledgment
Leonardo Marques Foundation for financial and technical support.
Declaration of interest
The authors declare that they have no conflict of interest.
Authors contributions
Took part in project conception, design, coordination and follow up, analysis, and interpretation of the data: MLT, SF, EV; Took part in IVF and PGD-AS procedures and data acquisition: SF, ET, AC; Took part in manuscript writing: EV, MLT, SF. All authors contributed to the critical review and final approval of the manuscript.
References
- Colls, P., Escudero, T., Cekleniak, N., Sadowy, S., Cohen, J. and Munne, S. (2007) Increased efficiency of preimplantation genetic diagnosis for infertility using “no result rescue". Fertil Steril 88:53–61
- Fiorentino, F., Biricik, A., Bono, S., Spizzichino, L., Cotroneo, E., Cottone, G., et al. (2014) Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril 101:1375–82
- Forman, E.J., Tao, X., Ferry, K.M., Taylor, D., Treff, N.R. and Scott, R.T., Jr. (2012) Single embryo transfer with comprehensive chromosome screening results in improved ongoing pregnancy rates and decreased miscarriage rates. Hum Reprod 27:1217–22
- Fragouli, E., Lenzi, M., Ross, R., Katz-Jaffe, M., Schoolcraft, W.B. and Wells, D. (2008) Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod 23:2596–608
- Frumkin, T., Malcov, M., Yaron, Y. and Ben-Yosef, D. (2008) Elucidating the origin of chromosomal aberrations in IVF embryos by preimplantation genetic analysis. Mol Cell Endocrinol 282:112–19
- Gianaroli, L., Magli, M.C., Ferraretti, A.P. and Munne, S. (1999) Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: Identification of the categories for which it should be proposed. Fertil Steril 72:837–44
- Gutierrez-Mateo, C., Colls, P., Sanchez-Garcia, J., Escudero, T., Prates, R., Ketterson, K., et al. (2011) Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril 95:953–8
- Handyside, A.H. (2013) 24-chromosome copy number analysis: A comparison of available technologies. Fertil Steril 100:595–602
- Harton, G.L., Harper, J.C., Coonen, E., Pehlivan, T., Vesela, K. and Wilton, L. (2011) ESHRE PGD consortium best practice guidelines for fluorescence in situ hybridization-based PGD. Hum Reprod 26:25–32
- Ioannou, D., Fonseka, K.G., Meershoek, E.J., Thornhill, A.R., Abogrein, A., Ellis, M., et al. (2012) Twenty-four chromosome FISH in human IVF embryos reveals patterns of post-zygotic chromosome segregation and nuclear organisation. Chromosome Res 20:447–60
- Ioannou, D., Meershoek, E.J., Thornhill, A.R., Ellis, M. and Griffin, D.K. (2011) Multicolour interphase cytogenetics: 24 chromosome probes, 6 colours, 4 layers. Mol Cell Probes 25:199–205
- Magli, M.C., Gianaroli, L. and Ferraretti, A.P. (2001) Chromosomal abnormalities in embryos. Mol Cell Endocrinol 183:S29–34
- Mir, P., Rodrigo, L., Mercader, A., Buendia, P., Mateu, E., Milan-Sanchez, M., et al. (2013) False positive rate of an arrayCGH platform for single-cell preimplantation genetic screening and subsequent clinical application on day-3. J Assist Reprod Genet 30:143–9
- Munne, S., Alikani, M., Tomkin, G., Grifo, J. and Cohen, J. (1995) Embryo morphology, developmental rates, and maternal age are correlated with chromosome abnormalities. Fertil Steril 64:382–91
- Munne, S., Chen, S., Colls, P., Garrisi, J., Zheng, X., Cekleniak, N., et al. (2007) Maternal age, morphology, development and chromosome abnormalities in over 6000 cleavage-stage embryos. Reprod Biomed Online 14:628–34
- Munne, S., Chen, S., Fischer, J., Colls, P., Zheng, X., Stevens, J., et al. (2005) Preimplantation genetic diagnosis reduces pregnancy loss in women aged 35 years and older with a history of recurrent miscarriages. Fertil Steril 84:331–5
- Munne, S., Magli, C., Cohen, J., Morton, P., Sadowy, S., Gianaroli, L., et al. (1999) Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod 14:2191–9
- Munne, S., Marquez, C., Magli, C., Morton, P. and Morrison, L. (1998) Scoring criteria for preimplantation genetic diagnosis of numerical abnormalities for chromosomes X, Y, 13, 16, 18 and 21. Mol Hum Reprod 4:863–70
- Munne, S., Sandalinas, M., Escudero, T., Velilla, E., Walmsley, R., Sadowy, S., et al. (2003) Improved implantation after preimplantation genetic diagnosis of aneuploidy. Reprod Biomed Online 7:91–7
- Natesan, S.A., Bladon, A.J., Coskun, S., Qubbaj, W., Prates, R., Munne, S., et al. (2014) Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med 16:838--45
- Rius, M., Obradors, A., Daina, G., Cuzzi, J., Marques, L., Calderon, G., et al. (2010) Reliability of short comparative genomic hybridization in fibroblasts and blastomeres for a comprehensive aneuploidy screening: First clinical application. Hum Reprod 25:1824–35
- Sandalinas, M., Sadowy, S., Alikani, M., Calderon, G., Cohen, J. and Munne, S. (2001) Developmental ability of chromosomally abnormal human embryos to develop to the blastocyst stage. Hum Reprod 16:1954–8
- Scott, R.T., Jr. Ferry, K., Su, J., Tao, X., Scott, K. and Treff, N.R. (2012) Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: A prospective, blinded, nonselection study. Fertil Steril 97:870–5
- Schoolcraft, W.B., Fragouli, E., Stevens, J., Munne, S., Katz-Jaffe, M.G. and Wells, D. (2010) Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil Steril 94:1700–6
- Treff, N.R., Northrop, L.E., Kasabwala, K., Su, J., Levy, B. and Scott, R.T., Jr. (2011) Single nucleotide polymorphism microarray-based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil Steril 95:1606–12 e1601–1602
- Treff, N.R. and Scott, R.T., Jr. (2013) Four-hour quantitative real-time polymerase chain reaction-based comprehensive chromosome screening and accumulating evidence of accuracy, safety, predictive value, and clinical efficacy. Fertil Steril 99:1049–53
- Treff, N.R., Su, J., Tao, X., Levy, B. and Scott, R.T., Jr. (2010) Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril 94:2017–21
- Treff, N.R., Tao, X., Ferry, K.M., Su, J., Taylor, D. and Scott, R.T., Jr. (2012) Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil Steril 97:819–24
- Wells, D., Alfarawati, S. and Fragouli, E. (2008) Use of comprehensive chromosomal screening for embryo assessment: Microarrays and CGH. Mol Hum Reprod 14:703–10
- Yang, Z., Liu, J., Collins, G.S., Salem, S.A., Liu, X., Lyle, S.S., et al. (2012) Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: Results from a randomized pilot study. Mol Cytogenet 5:24. doi:10.1186/1755-8166-5-24
