Abstract
Polymorphisms in the genes encoding enzymes in the folate metabolism pathway have been associated with male infertility and chromosome abnormalities. The aim of this study was to analyze the distribution of the methylenetetrahydrofolate reductase (MTHFR), methionine synthase (MTR), and methionine synthase reductase (MTRR) polymorphisms in fertile men and infertile men with non-obstructive azoospermia (NOA). A case-control study comprising 85 infertile men with NOA and 246 fertile men as controls was carried out. MTHFR c.677C > T (rs1801133), MTHFR c.1298A > C (rs1801131), MTR c.2756A > G (rs1805087), and MTRR c.66A > G (rs1801394) polymorphisms were determined using the polymerase chain reaction restriction fragment length polymorphism technique. There were significant differences in AC + CC genotype (OR = 1.9, 95% CI = 1.1–3.2) and C allele frequencies (OR = 1.8, 95% CI = 1.2–2.8) of MTHFR c.1298A > C polymorphism between NOA patients and controls after applying the Bonferroni correction. Moreover, the 1298AC genotype, 1298AC + CC genotype, and 1298C allele frequencies were statistically significant in NOA with chromosomal abnormalities and/or a Y chromosome deletion compared to the controls (AC genotype: OR = 3.0; AC + CC genotype: OR = 3.0; C allele: OR = 2.3). Considering the other polymorphisms, no differences were found between cases and controls. Our findings suggest the MTHFR c.1298A > C polymorphism is associated with an increased risk of male infertility, i.e., NOA.
Introduction
Infertility is estimated to affect 10–15% of couples, and roughly half of these cases are due to the male factor [De Kretser and Baker Citation1999]. Spermatogenic failure is the most common form of male infertility; however, the etiology remains unknown in most cases. About 15% of infertile men may carry genetic abnormalities, including chromosomal aberrations and single-gene detects [Ferlin et al. Citation2006; O'Flynn O'Brien et al. Citation2010]. Deleterious gene polymorphisms in key genes involved in testicular function, combined with environmental factors, may be responsible for the reduced number of sperm and poor sperm quality that is observed in many infertile men.
Folate metabolism is important for maintaining the stability and integrity of the genome because of its role in DNA synthesis, repair, and methylation [Donnelly Citation2001]. Methylenetetrahydrofolate reductase (MTHFR) is a key regulatory enzyme involved in folate metabolism. Methionine, the precursor for the universal methyl donor (S-adenosylmethionine) is produced through the irreversible transfer of a methyl group from 5-methyltetrahydrofolate. This reaction is regulated by two enzymes, methionine synthase (MTR) and methionine synthase reductase (MTRR) [Chen et al. Citation2001]. Disturbances in the catalytic activity of MTRR could lead to higher levels of homocysteine (Hcy). MTHFR, MTR, and MTRR play an important role in folate metabolism, and Hcy levels could affect DNA synthesis and methylation, leading to increased oxidative stress [Agarwal et al. Citation2006] and disrupted methylation [Frosst et al. Citation1995].
MTHFR, located on the short arm of chromosome 1 (1p36.3), presents two common polymorphisms involving nucleotides c.677C > T and c.1298A > C. The c.677C > T results in the substitution of alanine by valine, and the enzyme has only 30% residual activity in a homozygous condition and about 70% residual activity in a heterozygous condition [Frosst et al. Citation1995]. The c.1298A > C results in substitution of glutamine by alanine, reducing the enzymatic activity, but to a lesser extent than c.677C > T [Frosst et al. Citation1995; van der Put et al. Citation1998]. Individuals (particularly with low folate status) carrying these variants can present with mild hyperhomocysteinemia [Frosst et al. Citation1995]. MTR is polymorphic at nucleotide c.2756A > G and has been associated with decreased plasma Hcy levels [Fowler Citation2005]. The most common polymorphism reported in the MTRR gene is at nucleotide position 22 (c.66A > G), an isoleucine to methionine change. Although the MTRR c.66A > G polymorphism does not appear to alter the catalytic activity of the protein, the 66GG genotype is associated with a modest but significant decrease in plasma total Hcy levels [Gaughan et al. Citation2001].
Folate deficiency occurs frequently and the related hyperhomocysteinaemia is considered a risk factor for various diseases including infertility. Several association studies have suggested that polymorphic variants in the MTHFR gene may be associated with reduced sperm counts in humans, leading to male infertility in some populations [Botezatu et al. Citation2014; Lee et al. Citation2006; Singh et al. Citation2005; Shen et al. Citation2012]. Turner syndrome may provide a model for MTHFR gene polymorphisms associated somatic chromosomal non-disjunction. These patients typically present with a high frequency of chromosome mosaicism [Santos et al. Citation2006]. Similarly, O'Leary et al. [Citation2002] showed that MTR polymorphisms may be a modest risk factor for Down syndrome [O'Leary et al. Citation2002].
Thus, the objective of the present study was to assess the distribution of the polymorphisms in folate-related enzyme genes MTHFR c.677C > T and c.1298A > C, MTR c.2756A > G, and MTRR c.66A > G in infertile men with non-obstructive azoospermia (NOA). These were compared to controls to define the association of these polymorphisms to non-obstructive male infertility.
Results
We analyzed the four most frequent polymorphisms of the MTHFR, MTR, and MTRR genes in 85 infertile men with NOA and 246 fertile men by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) using the primers outlined in . Of the 85 infertile men, 59 men had a normal karyotype (46,XY) without any genetic abnormalities and 26 men had abnormal chromosomes and/or microdeletions in Y chromosome azoospermia factors (). Electrophoretic analysis of a multiplex PCR with Y chromosome microdeletions in three infertile men is shown in . The mean age was significantly different between NOA and the control groups (33.9 ± 4.9 years vs. 42.5 ± 8.5 years, p < 0.001). In addition, the mean ultrasonic testicular volume of the right and left testis in NOA was 10.0 ± 5.2 mL and 10.3 ± 5.4 mL, respectively. Testicular histology revealed seminiferous tubules containing sertoli cells with an absence of germ cells in 67.1% (57/85) of NOA patients, whereas maturation arrest was found in 32.9% (28/85) of the NOA patients.
Figure 1. Multiplex PCR analysis with STS markers for microdeletion of the azoospermia factor (AZF) regions in the Y chromosome. M: low molecular weight DNA ladder; Lane 1: blank; Lane 2: male control; Lane 3: female control; Lane 4: AZFa deleted patient; Lane 5: AZFc deleted patient; Lane 6: AZFb + c deleted patient. PCR: polymerase chain reaction; STS: sequence tagged site; ZFX: zinc finger X chromosomal protein.
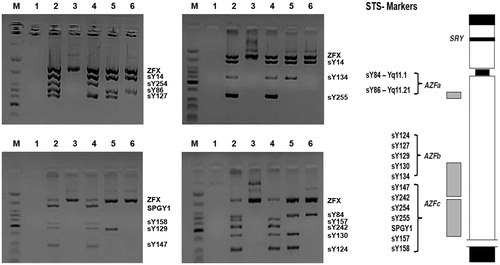
Table 1. SNPs analyzed along with PCR primers and detection methods used for genotyping.
Table 2. Genetic abnormalities (chromosomal aberrations, variants, and Y chromosomal microdeletions) in infertile men with non-obstructive azoospermia (NOA).
The PCR-RFLP results of MTHFR c.677C > T, MTHFR c.1298A > C, MTR c.2756A > G, and MTRR c.66A > G polymorphisms are shown in . The genotype and allele distributions of MTHFR c.677C > T, MTHFR c.1298A > C, MTR c.2756A > G, and MTRR c.66A > G polymorphisms in NOA patients and controls are shown in . The genotype frequencies of these polymorphisms in both NOA and control groups were in accordance with Hardy-Weinberg equilibrium (HWE) (data not shown). The frequency of MTHFR 1298AC + CC genotype was significantly higher in the NOA group than in controls (38.8% vs. 25.2%, odds ratio (OR) = 1.9, 95% confidence interval (CI) = 1.1–3.2, p = 0.019). Moreover, the mutant 1298C allele frequency was significantly higher in the NOA group compared with controls (22.4% vs. 13.8%, OR = 1.8, 95% CI = 1.2–2.8, p = 0.011). When the Bonferroni correction was applied, the frequency of 1298AC + CC genotype borderlined statistical significance and 1298C allele remained significant. The MTHFR c.677C > T, MTR c.2756A > G, and MTRR c.66A > G polymorphism data showed no significant differences between the NOA and control groups before and after the Bonferroni correction.
Figure 2. PCR-RFLP analysis of MTHFR c.677C > T (A), MTHFR c.1298A > C (B), MTR c.2756A > G (C), and MTRR c.66A > G (D) polymorphisms. PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; B: blank; M: low molecular weight DNA ladder.
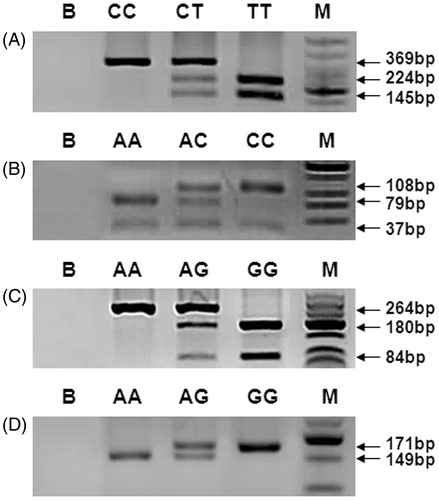
Table 3. Genotype and allele distribution of MTHFR c.677C > T, MTHFR c.1298A > C, MTR c.2756A > G, and MTRR c.66A > G polymorphisms in NOA and control groups.
We compared the genotype and allele frequencies of MTHFR c.1298A > C polymorphism among the NOA group with chromosome abnormalities and/or Yq microdeletions, to the unexplained NOA group with normal karyotype and no Yq microdeletion, and controls. showed that the frequencies of MTHFR 1298AC genotype, 1298AC + CC genotype, and 1298C allele were significantly higher in the NOA group with chromosome abnormalities and/or Yq microdeletion than the controls (AC: OR = 3.0, 95% CI = 1.2–7.6, p = 0.014, AC + CC: OR = 3.0, 95% CI = 1.2–7.3, p = 0.011; C: OR = 2.3, 95% CI = 1.1–4.7, p = 0.023, respectively). However, the unexplained NOA group showed no significant difference in the genotype and allele frequencies of MTHFR c.1298A > C polymorphism in comparison to controls. There was also no statistically significant difference in the frequencies of MTHFR 1298AC, 1298CC, 1298AC + CC, and 1298C between unexplained and explained NOA groups (p = 0.077, 1.000, 0.094, 0.217).
Table 4. Genotype and allele distribution of MTHFR c.1298A > C polymorphisms in NOA subgroups.
Discussion
In the present study, we evaluated the association of polymorphisms in MTHFR (c.677C > T and c.1298A > C), MTR (c.2756A > G), and MTRR (c.66A > G) genes involved in folate metabolism and their possible risk of non-obstructive male infertility. We observed that the functional polymorphism of the MTHFR c.1298A > C was strongly associated with the risk of NOA. The 1298AC genotype and 1298AC + CC genotype had a 1.8 - and 1.9-fold increased risk of NOA, respectively. Furthermore, the 1298AC genotype, 1298AC + CC genotype, and 1298C allele had a 3.0-fold, 3.0-fold, and 2.3-fold increased risk of NOA with chromosome abnormalities and/or Yq microdeletion. We did not find any association between the MTHFR c.677C > T, MTR c.2756A > G, and MTRR c.66A > G and infertile men with NOA. Our findings demonstrate the relevance of the MTHFR c.1298A > C polymorphism of folate metabolism in susceptibility to male infertility, especially with NOA including genetic abnormalities.
A recent study on men with idiopathic infertility and healthy fertile controls of Indian origin concluded that the MTHFR 1298CC genotype is a genetic risk factor for idiopathic male infertility in an Indian population [Singh et al. Citation2010]. In a Brazilian study Gava et al. [Citation2011] suggested that MTHFR c.1298A > C polymorphism could be an important genetic factor predisposing NOA men to infertility. A study of 1,633 cases and 1,735 controls from seven case control studies identified the MTHFR 1298C allele as a genetic risk factor for infertility, whereas a more comprehensive meta-analyses of 3,850 cases and 4,085 controls from twelve published case-control studies did not observe an association of the 1298C genotype with male infertility, although an association with the MTHFR 677T allele was detected [Shen et al. Citation2012; Wei et al. Citation2012]. These meta-analyses of the published data concerning MTHFR polymorphisms and infertility are inconclusive. A possible role of ethnic differences in genetic backgrounds and the environment they live in may affect these results. Other factors such as selection bias, different matching criteria, and methodological differences may also play a role. Lee et al. [Citation2006] evaluated polymorphisms of 3 folate metabolism enzymes (MTHFR c.677C > T and c.1298A > C, MTR c.2756A > G, and MTRR c.66A > G) and the association of non-obstructive male infertility. The results showed genetic evidence that MTHFR c.677C > T, MTR c.2756A > G, and MTRR c.66A > G genotypes were independently associated with male infertility. In these studies, the genetic association was observed in specific phenotypic subgroups [Lee et al. Citation2006; Shen et al. Citation2012; Wei et al. Citation2012]. Genetic association following phenotype stratification was also reported by Wu et al. [Citation2011]. Moreover, the interactions of genetic and nutrient/environmental factors (such as levels of dietary folate uptake) have been shown to affect the impact of these genetic variants [Toffoli and De Mattia Citation2008].
Changes in folate status could negatively impact spermatogenesis by causing DNA hypomethylation and thereby disrupting gene expression or by inducing uracil misincorporation during DNA synthesis leading to errors in DNA repair, strand breakage, and chromosomal anomalies. Experimentally induced hypomethylation of premeiotic germ cells in mice has been shown to inhibit their differentiation into spermatocytes [Raman and Narayan Citation1995]; therefore, it is possible that the MTHFR mutations in men cause infertility via the same mechanism. MTHFR encodes a key regulatory enzyme involved in folate metabolism, and genetic polymorphisms of this gene may predispose some men to reduced sperm counts [Ebisch et al. Citation2007; Paracchini et al. Citation2006]. Another obvious effect of the MTHFR mutation on cell physiology is auto-oxidation, leading to the production of toxic reactive oxygen metabolites, such as hydrogen peroxide [Loscalzo Citation1996]. An increased production of reactive oxygen species results in homocysteine-mediated DNA damage [Micheal et al. Citation2009]. Human spermatozoa are particularly susceptible to peroxidative damage, and antioxidants such as folate can overcome oxidative stress and maintain the integrity of sperm cells by preventing oxidative damage to sperm DNA. Thus, in addition to hypomethylation, homocysteine-mediated DNA damage caused by oxidative stress may be another plausible mechanism of infertility in men with the MTHFR polymorphisms [Huang et al. Citation2000; Micheal et al. Citation2009].
An abnormal level of folate while impairing methylation can lead to chromosome breakage, defective chromosome recombination, and abnormal chromosome segregation [Fenech Citation2001]. Previous studies showed that impaired folate metabolism due to genetic polymorphisms of metabolic enzymes could increase the risk for having an infant with Down syndrome [James et al. Citation1999; Rai et al. Citation2014]. Oliveira et al. [Citation2008] reported that Turner syndrome may be an investigative model for MTHFR gene polymorphisms for chromosomal non-disjunction. These authors studied 140 Turner syndrome patients and 209 controls and found a high frequency of 1298CC genotype in Turner syndrome patients. They concluded that MTHFR c.1298A > C polymorphism can have an effect on chromosomal non-disjunction through a decrease in MTHFR activity in Turner syndrome patients. In addition, Ozbek et al. [Citation2008] found that the c.1298A > C MTHFR polymorphism, mainly genotype 1298CC, was more frequent in the patients with Klinefelter syndrome, the most common sex chromosome abnormality related with male infertility and azoospermia, suggesting its involvement in the origin of chromosomal imbalance.
The presence of deletions in the distal euchromatic region of the long arm of chromosome Y in patients with NOA has suggested the existence of a locus responsible for spermatogenesis, the azoospermia factor (AZF) [Tiepolo and Zuffardi Citation1976]. Generally, Y chromosome microdeletions are essentially focused on screening the AZF region using sequence tagged site (STS) markers that have been mapped at high density along of the Y chromosome. At least three distinct non-overlapping regions, each associated with variable degrees of spermatogenic impairment, have been defined by numerous deletion mapping studies in the AZF region. These regions named as AZFa, AZFb, and AZFc for AZF a, b, and c indicate that at least three different loci on the long arm of the Y chromosome are critical for germ cell differentiation. In this study, we detected three NOA patients with STS deleted within the AZFa, AZFc, or AZFb + c region. Microdeletions in AZFa are associated with azoospermia disorders, such as Sertoli-cell only syndrome [Hopps et al. Citation2003; Kamp et al. Citation2001]. AZFb microdeletions are frequently associated with spermatogenic arrest, while men with AZFc microdeletions may suffer from severe oligozoospermia to NOA [O'Flynn O'Brien et al. Citation2010]. The AZF region is deleted in about 13% of men with NOA and in 7% to 10% of men with oligozoospermia [Girardi et al. Citation1997]. In infertile patients with Y chromosome microdeletions, the frequency of deletions in AZFa (5%) and AZFb (10% to 16%) is lower compared with the frequency of AZFc deletions (60%) [Hopps et al. Citation2003; Foresta et al. Citation2001; Vogt Citation1998]. Deletions bearing two or three AZF regions are detected at a frequency of 14% [Foresta et al. Citation2001].
There are several limitations in this study. First, in the subgroup analysis, the number of subjects in each subgroup was relatively small, not having enough statistical power to explore the real association. Second, our results were based on unadjusted estimates, while a more precise analysis should be conducted if all individual data were available, which would allow for the adjustment by other co-variants including age, body mass index, smoking status, drinking status, and other lifestyle factors. Third, we only included the Korean population in this case-control study. The association of the MTHFR c.1298A > C polymorphism with male infertility does require confirmation.
In conclusion, our study found that the MTHFR c.1298A > C polymorphism was associated with an increased risk for male infertility, especially with NOA including genetic abnormalities. However, because our study is relatively small, larger cohort studies are necessary to confirm these findings. Since male infertility has a multifactorial etiology, more studies or complete case-control studies, especially stratified by different ethnic background, environmental exposure, or other risk factors, should be performed to clarify possible roles of the MTHFR c.1298A > C polymorphism in the pathogenesis of male infertility in the future.
Materials and Methods
Subjects
Among the patients of the Department of Urology at Cheil General Hospital and Women's Healthcare Center in Seoul, Korea, we recruited a total of 85 infertile men with NOA, consisting of 59 unexplained NOA with normal karyotype and no Y chromosome deletion and 26 NOA with chromosomal abnormalities and/or Y chromosome deletion. NOA patients had an infertility history of at least two years. From January 2009 and August 2011, eighty-five infertile men with NOA diagnosed by physical examination, semen analysis, and hormone estimation underwent cytogenetic and Y chromosomal microdeletion analysis. Two hundred and forty-six fertile men who have at least one child by direct survey and who lacked any history of requiring assisted reproduction technology were included as the control group. Infertile men who were found to have cryptorchidism and varicocele via the physical examination and clinical tests were excluded from the study. Appropriate institutional review board approval was obtained from the Ethics Committee at Cheil General Hospital and Women's Healthcare Center for this study (#CGH-IRB-2011-61). Written informed consent was obtained from each participant before the collection of samples and subsequent analysis.
Semen analysis was performed strictly according to the World Health Organization [WHO, Citation1999] guidelines. The diagnosis of azoospermia (no spermatozoa in the ejaculate) was made on the basis of two semen analyses performed according to the WHO-recommended procedure. The diagnosis of azoospermia was made on the basis of two semen analyses. NOA was determined after physical examination, sperm analysis (including assessment of sperm volume, pH, and evaluation of fructose concentration), endocrine profile (follicle-stimulating hormone, luteinizing hormone, testosterone, estradiol, prolactin, and sex hormone-binding globulin), ultrasound testicular volume measurement, and seminal vesicle evaluation.
Cytogenetic and FISH analyses
Cytogenetic analysis was performed on metaphase spreads of cultured peripheral lymphocytes. Cells were cultured in RPMI 1640 culture medium (Welgene, Daegu, Korea). Karyotypes were analyzed using GTL, CBG, and DA-DAPI staining techniques. For each case, at least 20 metaphases were analyzed at a high resolution level of 700-band per haploid chromosome set. Fluorescent in situ hybridization (FISH) probes specific for SRY/DXZ1 loci (LSI SRY, Spectrum Orange/CEP X, Spectrum Green), Y centromeric DYZ3 (CEP Y alpha satellite, Spectrum Orange), and Y heterochromatin specific DYZ1 locus (Satellite III, Spectrum Green) were used to analyze X- and Y-chromosome (Vysis, Downers Grove, IL, USA).
Y chromosome microdeletion analysis
Peripheral blood was collected in EDTA vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ, USA), and genomic DNA was extracted from whole blood using a QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s recommendations. Genomic DNA extracted from peripheral blood was amplified by a multi-PCR method with primers for 15 Y-specific STS markers (SRY, sY84, sY86, sY124, sY127, sY129, sY130, sY134, sY147, sY242, sY254, sY255, SPGY1, sY157, s Y158) [Ghorbian Citation2012] and X-linked gene encoding a zinc-finger protein marker ZFX. PCR reactions were performed in a final volume of 10 uL containing 50 ng of genomic DNA, 1 × PCR buffer, 1.5 mM MgCl2, 1 mM of each dNTP, 10 pmol of each specific primer, and 1 unit of AmpliTaq Gold DNA polymerase (Thermo Fisher, Foster City, CA, USA). The PCR conditions consisted of an initial denaturation at 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 62°C for 1 min 30 s, extension at 65°C for 1 min 30 s, and a final extension at 65°C for 10 min in an ABI PRISM 2700 thermal cycler (Thermo Fisher). The reaction products were analyzed by electrophoresis on 3% NuSieve gels containing ethidium bromide (0.1 mg/mL) and visualized under ultraviolet light. A positive control (sample from a normal fertile male) and one negative control (normal female sample) were included in every PCR analysis.
Genotype analysis
The MTHFR c.677C > T, MTHFR c.1298A > C, MTR c.2756A > G, and MTRR c.66A > G polymorphisms were analyzed by the PCR-RFLP method as described previously () [Kim et al. Citation2011]. To ensure reliability of all assays, genotyping was repeated for 10% of the samples, and two samples of each genotype were randomly selected and subjected to direct DNA sequencing to verify genotype; concordance was found to be 100%.
Statistical analysis
Data are expressed as a mean ± standard deviation (SD) or number (%). Genotype frequencies for each of the four polymorphisms were tested for deviation from HWE using the χ2 test (www.fourmilab.ch/rpkp/experiments/analysis/chiCalc.html). Comparison of genotype and allele frequencies between groups was analyzed using the Fisher’s exact test. OR and 95% CI were calculated by the χ2 test. For all statistical analyses, p < 0.05 was considered statistically significant. Considering the potential false positive rate incurred by multiple comparisons of single nucleotide polymorphisms, we used the Bonferroni correction method to adjust the pc value. Statistical analysis was performed using the Statistical Package for Social Sciences version 12.0 (SPSS Inc, Chicago, IL, USA).
| Abbreviations | ||
| MTHFR | = | methylenetetrahydrofolate reductase |
| MTR | = | methionine synthase |
| MTRR | = | methionine synthase reductase |
| Hcy | = | homocysteine |
| NOA | = | non-obstructive azoospermia |
| FISH | = | fluorescent in situ hybridization |
| STS | = | sequence tagged site |
| PCR-RFLP | = | polymerase chain reaction-restriction fragment length polymorphism |
Declaration of interest
The authors report no conflicts of interest.
Author contributions
Conceived and designed the experiments: SYK, SYP; Performed the experiments: SYK, JWL, JWK; Analyzed the data: HJK; Contributed reagents/materials/analysis tools: SYP, JTS; Wrote the manuscript: SYK, SYP, JTS.
References
- Agarwal, A., Prabakaran, S. and Allamaneni, S.S. (2006) Relationship between oxidative stress, varicocele and infertility: a meta-analysis. Reprod Biomed Online 12:630–633
- Botezatu, A., Socolov, R., Socolov, D., Iancu, I.V. and Anton, G. (2014) Methylation pattern of methylene tetrahydrofolate reductase and small nuclear ribonucleoprotein polypeptide N promoters in oligoasthenospermia: a case-control study. Reprod Biomed Online 28:225–231
- Chen, Z., Karaplis, A.C., Ackerman, S.L., Pogribny, I.P., Melnyk, S., Lussier-Cacan, S., et al. (2001) Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet 10:433–443
- De Kretser, D.M. and Baker, H.W. (1999) Infertility in men: recent advances and continuing controversies. J Clin Endocrinol Metab 84:3443–3450
- Donnelly, J.G. (2001) Folic acid. Crit Rev Clin Lab Sci 38:183–223
- Ebisch, I.M., Thomas, C.M., Peters, W.H., Braat, D.D. and Steegers-Theunissen, R.P. (2007) The importance of folate, zinc and antioxidants in the pathogenesis and prevention of subfertility. Hum Reprod Update 13:163–174
- Fenech, M. (2001) The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutat Res 475:57–67
- Ferlin, A., Arredi, B. and Foresta, C. (2006) Genetic causes of male infertility. Reprod Toxicol 22:133–141
- Foresta, C., Moro, E. and Ferlin, A. (2001) Prognostic value of Y deletion analysis. The role of current methods. Hum Reprod 16:1543–1547
- Fowler, B. (2005) Homocysteine: overview of biochemistry, molecular biology, and role in disease processes. Semin Vasc Med 5:77–86
- Frosst, P., Blom, H.J., Milos, R., Goyette, P., Sheppard, C.A., Matthews, R.G., et al. (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10:111–113
- Gaughan, D.J., Kluijtmans, L.A., Barbaux, S., McMaster, D., Young, I.S., Yarnell, J.W., et al. (2001) The methionine synthase reductase (MTRR) A66G polymorphism is a novel genetic determinant of plasma homocysteine concentrations. Atherosclerosis 157:451–456
- Gava, M.M., Chagas Ede, O., Bianco, B., Christofolini, D.M., Pompeo, A.C., Glina, S., et al. (2011) Methylenetetrahydrofolate reductase polymorphisms are related to male infertility in Brazilian men. Genet Test Mol Biomarkers 15:153–157
- Ghorbian, S. (2012) Routine diagnostic testing of Y chromosome deletions in male infertile and subfertile. Gene 503:160–164
- Girardi, S.K., Mielnik, A. and Schlegel, P.N. (1997) Submicroscopic deletions in the Y chromosome of infertile men. Hum Reprod 12:1635–1641
- Hopps, C.V., Mielnik, A., Goldstein, M., Palermo, G.D., Rosenwaks, Z. and Schlegel, P.N. (2003) Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod 18:1660–1665
- Huang, C., Li, J., Zheng, R. and Cui, K. (2000) Hydrogen peroxide-induced apoptosis in human hepatoma cells is mediated by CD95(APO-1/Fas) receptor/ligand system and may involve activation of wild–type p53. Mol Biol Rep 27:1–11
- James, S.J., Pogribna, M., Pogribny, I.P., Melnyk, S., Hine, R.J., Gibson, J.B., et al. (1999) Abnormal folate metabolism and mutation in the methylenetetrahydrofolate reductase gene may be maternal risk factors for Down syndrome. Am J Clin Nutr 70:495–501
- Kamp, C., Huellen, K., Fernandes, S., Sousa, M., Schlegel, P.N., Mielnik, A., et al. (2001) High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. Mol Hum Reprod 7:987–994
- Kim, S.Y., Park, S.Y., Choi, J.W., Kim, D.J., Lee, S.Y., Lim, J.H., et al. (2011) Association between MTHFR 1298A > C polymorphism and spontaneous abortion with fetal chromosomal aneuploidy. Am J Reprod Immunol 66:252–258
- Lee, H.C., Jeong, Y.M., Lee, S.H., Cha, K.Y., Song, S.H., Kim, N.K., et al. (2006) Association study of four polymorphisms in three folate-related enzyme genes with non-obstructive male infertility. Hum Reprod 21:3162–3170
- Loscalzo, J. (1996) The oxidant stress of hyperhomocyst(e)inemia. J Clin Invest 98:5–7
- Micheal, S., Qamar, R., Akhtar, F., Khan, M.I., Khan, W.A. and Ahmed, A. (2009) MTHFR gene C677T and A1298C polymorphisms and homocysteine levels in primary open angle and primary closed angle glaucoma. Mol Vis 15:2268–2278
- O'Flynn O'Brien, K.L., Varghese, A.C. and Agarwal, A. (2010) The genetic causes of male factor infertility: a review. Fertil Steril 93:1–12
- O'Leary, V.B., Parle-McDermott, A., Molloy, A.M., Kirke, P.N., Johnson, Z., Conley, M., et al. (2002) MTRR and MTHFR polymorphism: link to Down syndrome? Am J Med Genet 107:151–155
- Oliveira, K.C., Bianco, B., Verreschi, I.T., Guedes, A.D., Galera, B.B., Galera, M.F., et al. (2008) Prevalence of the polymorphism MTHFR A1298C and not MTHFR C677T is related to chromosomal aneuploidy in Brazilian Turner Syndrome patients. Arq Bras Endocrinol Metabol 52:1374–1381
- Ozbek, M., Oztürk, M.A., Ureten, K., Ceneli, O., Erdogan, M. and Haznedaroglu, I.C. (2008) Severe arterial thrombophilia associated with a homozygous MTHFR gene mutation (A1298C) in a young man with Klinefelter syndrome. Clin Appl Thromb Hemost 14:369–371
- Paracchini, V., Garte, S. and Taioli, E. (2006) MTHFR C677T polymorphism, GSTM1 deletion and male infertility: a possible suggestion of a gene-gene interaction? Biomarkers 11:53–60
- Rai, V., Yadav, U., Kumar, P., Yadav, S.K. and Mishra, O.P. (2014) Maternal methylenetetrahydrofolate reductase C677T polymorphism and down syndrome risk: a meta-analysis from 34 studies. PLoS One 9:e108552
- Raman, R. and Narayan, G. (1995) 5-Aza deoxyCytidine-induced inhibition of differentiation of spermatogonia into spermatocytes in the mouse. Mol Reprod Dev 42:284–290
- Santos, K., Lemos-Marini, S.H.V., Baptista, M.T.M., Bonadia, L.C., Pinto W. Jr and Bertuzzo, C.S. (2006) Frequency of 677(C → T) and 1298(A → C) polymorphisms in the 5,10-methylenetetrahydrofolate reductase (MTHFR) gene in Turner syndrome individuals. Genet Mol Biol 29:41–44
- Shen, O., Liu, R., Wu, W., Yu, L. and Wang, X. (2012) Association of the methylenetetrahydrofolate reductase gene A1298C polymorphism with male infertility: a meta-analysis. Ann Hum Genet 76:25–32
- Singh, K., Singh, S.K. and Raman, R. (2010) MTHFR A1298C polymorphism and idiopathic male infertility. J Postgrad Med 56:267–269
- Singh, K., Singh, S.K., Sah, R., Singh, I. and Raman, R. (2005) Mutation C677T in the methylenetetrahydrofolate reductase gene is associated with male infertility in an Indian population. Int J Androl 28:115–119
- Tiepolo, L. and Zuffardi, O. (1976) Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet 34:119–124
- Toffoli, G. and De Mattia, E. (2008) Pharmacogenetic relevance of MTHFR polymorphisms. Pharmacogenomics 9:1195–1206
- van der Put, N.M., Gabreëls, F., Stevens, E.M., Smeitink, J.A., Trijbels, F.J., Eskes, T.K., et al. (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 62:1044–1051
- Vogt, P.H. (1998) Human chromosome deletions in Yq11, AZF candidate genes and male infertility: history and update. Mol Hum Reprod 4:739–744
- Wei, B., Xu, Z., Ruan, J., Zhu, M., Jin, K., Zhou, D., et al. (2012) MTHFR 677C > T and 1298A > C polymorphisms and male infertility risk: a meta-analysis. Mol Biol Rep 39:1997–2002
- World Health Organization (WHO). (1999) Laboratory manual for the examination of human semen and sperm-cervical mucus interaction. 4th ed. Cambridge, UK: Cambridge University Press
- Wu, W., Shen, O., Qin, Y., Lu, J., Niu, X., Zhou, Z., et al. (2011) Methylenetetrahydrofolate reductase C677T polymorphism and the risk of male infertility: a meta-analysis. Int J Androl 35:18–24
