Abstract
The genus Langeronia parasitizing the intestine of several species of anurans is distributed from North to Central America. We identified Langeronia macrocirra and Langeronia cf. parva from the same host and localities, and present here new data not applicable about their tegumental surface by scanning electron microscopy. We compared sequences of the rDNA ITS2 region and mtDNA cox1 gene for the two morphotypes. ITS2 exhibited a high degree of conservation. Phylogenetic reconstruction using cox1 revealed three clades (I, II, and III), which did not correspond to a previous identification or host. Little divergence was found within clades: sequences were identical in clade I, whereas clade II had 0.27% and clade III had 1.08%. Inter-clade divergence reached 8.69% (I vs. III). This pattern of genetic divergence indicated that both taxa probably belong to the same species, so we posit that the morphological changes could be correlated with development. Increasing sample size and geographical coverage will contribute to the taxonomy of the genus based on morphological and molecular evidence, and will open tracks toward the use of DNA barcodes to the genus in Mexico.
Introduction
The genus Langeronia includes six species distributed from North America to Central America, and occurs in the intestine of several anurans (Lithobates, Rhinella, and Incilius): Langeronia macrocirra, Langeronia provitellaria, Langeronia parva, Langeronia jimenezi, Langeronia brenesi, and Langeronia burseyi. Two species have been recorded in México: L. macrocirra parasitizing several species of Lithobates spp., Rhinella marina, and Incilius valliceps (Caballero and Bravo-Hollis Citation1949; Guillén-Hernandez et al. Citation2000; Bursey and Goldberg Citation2001; Paredes-Calderón et al. Citation2004; Espínola-Novelo and Guillén-Hernández Citation2008; Cabrera-Guzmán et al. Citation2010; Yáñez-Arenas and Guillén-Hernández Citation2010), and L. jimenezi parasitizing Lithobates berlandieri (Iruegas-Buentello and Salinas-López Citation1989; León-Règagnon et al. Citation2005). Little is known about the biology of this genus, and only the life cycle of L. brenesi is known (Goodman Citation1989). Morphological differentiation among species is based on a combination of characters such as body shape, pharynx, intestinal ceca, genital pore, and the distribution of vitellaria (Dailey and Goldberg Citation2000). The identification is difficult due to variability and a lack of distinctive characters to discriminate species (Ubelaker Citation1965). For this reason, some species have been misidentified, e.g. specimens originally determined to be L. macrocirra that were later transferred to L. provitellaria by Christian (Citation1970). Morphology alone is not enough to distinguish species in several groups; therefore, new sources of characters are being explored for digeneans, such as the body surface using scanning electron microscopy (SEM) or DNA (León-Règagnon et al. Citation2001; Razo-Mendivil et al. Citation2008).
Several molecular studies have been conducted using sequences of nuclear rDNA (internal transcribed spacer 2, ITS2) or mitochondrial DNA (cytochrome c oxidase subunit I, cox1) to determine relationships among species on parasites (Bell et al. Citation2001), species identification (Dallas et al. Citation2000; Bell and Sommerville Citation2002), discovery of genetically distinct but morphologically very similar species or species complex (McManus and Bowles Citation1996; Razo-Mendivil et al. Citation2010), as well as to complement taxonomic descriptions or re-descriptions (León-Règagnon et al. Citation2001; Razo-Mendivil et al. Citation2008).
A molecular study might be pertinent in the search for evidence if one is dealing with a single species and is detecting genetic variation levels. This is one of the objectives of a biodiversity research program that is currently extending around the world and is being developed in Mexico: The International Barcode of Life Initiative (http://www.barcodeoflife.org; http://www.mexbol.org/). This initiative consists of developing a DNA-based taxonomic system-based DNA barcode using partial sequences of the mitochondrial cox1 gene, which provides strong species-level resolution (Hebert et al. Citation2003; Tautz et al. Citation2003).
Although several studies have employed partial sequences of cox1 in digeneans (McManus and Bowles Citation1996; Bell et al. Citation2001; Razo-Mendivil et al. Citation2010), this fragment does not correspond to the specific region proposed for DNA barcodes (Folmer et al. Citation1994; Casiraghi et al. Citation2004); however, DNA barcodes have recently been explored systematically with the design of specific primers for digeneans (Moszczynska et al. Citation2009). For other parasite groups, successful amplification of the barcode region has been achieved (Casiraghi et al. Citation2004; Oceguera-Figueroa et al. Citation2005; Junker et al. Citation2010). No universal primers have been developed able to successfully amplify this region across every phylum and new efforts should focus on testing these new primers proposed by Moszczynska et al. (Citation2009). In spite of this impediment, the antecedents regarding the performance of the other cox1 fragment are valuable information (e.g. McManus and Bowles Citation1996), because this could represent an initial and complementary source for progress in biodiversity inventories through DNA barcodes of helminths.
As part of a research program to establish an inventory of the helminth fauna of amphibians and reptiles in Mexico, we analyzed specimens of Langeronia from several localities for morphological and molecular studies in order to contribute to the taxonomy of the genus.
Materials and methods
Samples and morphological study
From November 1998 to May 2006, Lithobates vaillanti, L. Berlandieri, and R. marina were collected by hand or using nets in several localities in Mexico (). Hosts were killed with an overdose of anesthetic, and all organs were examined under a stereomicroscope. Digeneans were removed from the intestine of their hosts and placed in 0.65% w/v saline solution, and afterwards were killed by sudden immersion in hot 70% ethanol to preserve morphological traits for further identification. These were stained with Meyer's paracarmine and or Gomori's trichrome, cleared in methyl salicylate, and mounted in Canada balsam. Measurements are given in millimeters unless otherwise stated. Minimum and maximum values are given followed by the arithmetic mean and SD in parentheses (data not shown). For SEM, specimens were stored in 4% formalin, dehydrated in series of gradual alcohol baths, and critical-point dried. Specimens were coated with a gold–palladium mixture and examined in a Hitachi S-2460N, Tokyo, Japan at 15 kV, scanning electron microscope. For taxonomic determination at the species level, we used original descriptions and specialized literature.
Table I. Host, locality, geographical coordinates, and GenBank accession numbers for cox1 and ITS2 sequences of analyzed samples.
Voucher specimens were deposited at the Coleccion Nacional de Helmintos, Instituto de Biología, UNAM, Mexico City (CNHE), with the following accession numbers: L. macrocirra from L. vaillanti: Laguna Escondida, Veracruz (4874, 4891), San José Independencia, Oaxaca (4876), Chapultepec (4877), Río Pizote, Guanacaste, Costa Rica (7226); from L. berlandieri: La Selva Biological Station, Costa Rica (7227), El Petén (7228); and from R. marina: Lago de Catemaco, Veracruz (4875, 4892), Salto de Eyipantla, Veracruz (7229); Coquimatlán (4883, 4884, 4895, 4896); and Pseudosonsinotrema chabaudi Caballero and Caballero Citation1969 from L. vaillanti: Río Pizote, Guanacaste, Costa Rica (7230). Specimens of genus Langeronia from Colección de Helmintos de Costa Rica, Universidad de Costa Rica (CHCR), US National Parasite Collection (USNPC, Beltsville, MD, USA) and CNHE were also examined during this study for comparison: L. macrocirra (CNHE 1127, 1525, 1526, 1527, 3307, 3631, 4092, 4093, 4891, 13885; CHCR 200-19, 200-19bis, USNPC 89185), L. provitellaria (USNPC 47569, 47570), L. parva (USNPC 70557, 70558), L. brenesi (USNPC 76941), and L. burseyi (USNPC 89628).
DNA extraction, PCR amplification, and sequencing
For molecular work, specimens were washed with saline solution and preserved in 100% ethanol. Genomic DNA was extracted individually from the adult worms following a standard phenol/chloroform extraction (Hillis et al. Citation1996; Palumbi Citation1996) and the DNeasy Blood and Tissue Kit (Qiagen, Valencia, California, USA) according to the manufacturer's instructions.
ITS2 and cox1 (partial sequence) were amplified by PCR. All PCRs were performed in a final volume of 25 μl (2.5 μl of 10x PCR buffer, 2.0 μl of 10 mM dNTP mixture [200 mM each], 1.25 mM MgCl2 [50 mM], 1.0 μl each primer [10 pmol/μl], 1–2 μl template DNA, 0.125 μl Taq DNA polymerase [5 units; Biogenica, Mexico city, Mexico], and the remaining volume of sterilized distilled water). Primers ITS-F (5′-TGTGTCGATGAAGAACGC AG-3′) and ITS2-RIXO (5′-TTCTATGCTTAAATTCAGGG-3′) were used to amplify ITS2 region (Gasser and Hoste Citation1995). Primers JB3 (5′-TTTTTTGGGCATCCTGAG GTTTAT-3′) and JB4.5 (5′-TAAAGAAAGAACATAATGAAAATG-3′) were used to amplify partial fragments of cox1 (Morgan and Blair Citation1998). ITS2 PCR conditions were: 1 min at 94°C, 35 cycles of 30 s at 92°C, 30 s at 50–55°C, 1 min 30 s at 72°C, and a final 5-min elongation period at 72°C. cox1 PCR conditions were: 1 min at 94°C, 30 cycles of 1 min at 92°C, 1 min 30 s or 2 min at 47–50°C, 3 min 30 s at 72°C, and a final 4-min elongation period at 72°C. PCR products were purified using the QIAquick (Qiagen) purification kit and were sequenced with a Dye Terminator Cycle Sequencing kit (Applied Biosystems, Inc., Foster City, CA, USA) on an ABI PRISM 3100 Genetic Analyzer.
Sequence alignment, genetic distances, and phylogenetic reconstruction
Sequences were aligned in the program Clustal W (Thompson et al. Citation1994) and corrected manually with BioEdit v5.0.6 (Hall Citation1999). Uncorrected pairwise distance measures were calculated in PAUP* v4b10 (Swofford Citation2002). The phylogenetic relationships among cox1 sequences were analyzed using the maximum parsimony optimality criterion in PAUP*. Heuristic searches were performed with the tree bisection–reconnection branch-swapping option, and all characters were unweighted and unordered. Node support was evaluated using the non-parametric bootstrap method and 1000 pseudo-replicates (Felsenstein Citation1985). Sequences were deposited in GenBank ().
Results
Morphological analysis
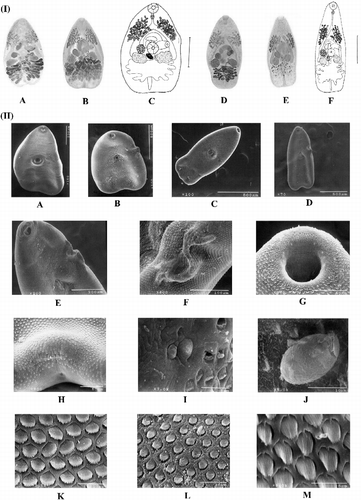
We found two species (L. macrocirra and L. cf. parva; ) differing in body size dimensions (data not shown), e.g. distinct pre-pharynx, esophagus length, and the position of acetabulum. By standardizing the technique of fixation (i.e. not flattened), the position of the genital pore and presence of pre-pharynx do not remain constant (personal observation). The body shape dimensions as well as the shape and distribution of vitellaria could be related to the degree of development of the fluke, rather than the setting process of fixation. A “tuck” was observed in the morphotypes close to the genital pore (), and is not consistently present; it seems to be a product of the ventral adhesion suckers between individuals during cross-fertilization (in vivo observation). By examining the surface morphology of adults (), we found that the body tegument surface has spines that are more abundant in the middle body, and the quantity and size of the spines markedly diminish toward the posterior end; spines have a see-saw shape; two types of papillae exist around the oral sucker and the acetabulum (type I, ciliated dome-shaped papillae and type II, non-ciliated dome-shaped papillae), and small ciliated papillae are found among the spines; the genital pore was located ventrally about mid-length of the left cecum; and eggs were operculated.
Figure 1. Morphological comparison of two nominal species of identified Langeronia and SEM photomicrographs. (I) General morphology: L. macrocirra (A–C) and L. cf. parva (D–F). (A) Holotype, Lithobates sp. (locality unknown, CNHE 13885), Mexico. (B) R. marina Lago de Catemaco, Veracruz (CNHE 4875). (C) R. marina, Lago de Catemaco, Veracruz (CNHE 4875). (D) Paratype, Lithobates pipiens, Alburg, Vermont, USA (USNPC 70558) and L. vaillanti, Laguna Escondida, Veracruz (CNHE 4874). (E) L. vaillanti, Laguna Escondida, Veracruz (CNHE 4874). (F) L. vaillanti, Laguna Escondida, Veracruz (CNHE 4874). Reference bar: C = 0.5 mm and F = 0.3 mm. (II) SEM photomicrographs: L. macrocirra from Lago de Catemaco, Veracruz, R. marina: (A, B, F–I, K, and L). L. cf. parva from Laguna Escondida, Veracruz, L. vaillanti (C–E, and J).Whole worm without tuck (A and C) and with tuck (B and E). Details of the tuck (E and F). Ciliated dome-shaped papillae (type I) on ventral surface anterior part of the oral sucker (G). Excretory pore (H). Small non-ciliated dome-shaped papillae (type II) close to a papillae type I on ventral surface around the acetabulum (I). Egg rough elliptical and operculate (J). Tegumental spines allocated near to the acetabulum (K). Spines allocated near to the posterior end (L). Small papillae ciliated among spines (M).
ITS2 sequences
The alignment was 235 bp in length (length range: 229–233 bp) and comprised 13 sequences. ITS2 sequences were highly similar for all samples regardless of previous identification, host, or geographical origin (data not shown). The Langeronia sequences exhibited 17.09% maximum nucleotide divergence with respect to the outgroup, P. chabaudi. Identical sequences were obtained (e.g. L. cf. parva from L. vaillanti from Laguna Escondida, and L. macrocirra from R. marina from Lago de Catemaco), while populations differed by a 0.87% of uncorrected distance (e.g. L. macrocirra from L. vaillanti from Guanacaste vs. L. macrocirra from Lago de Catemaco). This locus is highly conserved and thus does not provide sufficient information at this taxonomic level for species delimitation.
Cox1 sequences and phylogenetic relationships
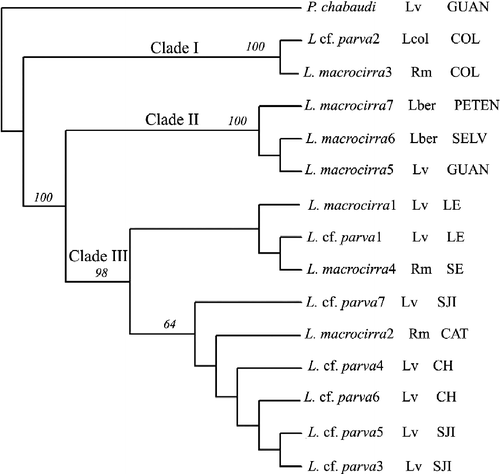
A total of 368 bp of cox1 were sequenced from 15 individuals and aligned unambiguously. The cox1 sequences differed by 21.19–22.01% with the outgroup, P. chabaudi (). Maximum parsimony cladistic analyses resulted in seven equally parsimonious trees (length = 121 steps, consistency index = 0.9339, retention index = 0.9292, rescaled consistency index = 0.8678). The strict consensus tree revealed three clades (), which are not consistent with the previous morphological identification or host infected: clade I with identical sequences from Colima, Mexico (on the Pacific slope, both morphotypes); clade II with sequences divergent by 0.27% from Costa Rica and Guatemala; and, finally, clade III with sequences divergent by 1.08% consisted of samples from Veracruz and Oaxaca (both morphotypes). Among clades, the divergence is 6.79–7.33% (II vs. III), 7.88–8.15% (I vs. II), and 8.15–8.69% (I vs. III). Within the same locality, sequences divergence ranged from 0 to 0.27% (e.g. from Colima or Oaxaca, respectively; see ).
Table II. Pairwise genetic distances (%) among cox1 sequences of analyzed specimens.
Figure 2. Strict consensus tree of seven equally parsimonious trees obtained from maximum parsimony analysis using partial cox1 gene sequences. Bootstrap percentages are shown above internal nodes. Geographical origin is designated as follows: Colima: Coquimatlán (COL); Veracruz: Lago de Catemaco (CAT), Laguna Escondida (LE), Salto de Eyipantla (SE); Oaxaca: San José Independencia (SJI), Chapultepec (CH); Costa Rica: La Selva Biological Station (SELV), Rio Pizote, Guanacaste (GUAN); Guatemala: El Petén (PETEN). Hosts: L. vaillanti (Lv), L. berlandieri (Lber), Lithobates sp. Colima (Lcol), R. marina (Rm).
Discussion, conclusions, and new prospects
The taxonomy of the genus Langeronia has been controversial due to morphological variation (Ubelaker Citation1965; Brenes et al. Citation1960). This variation has been observed in specimens of L. macrocirra from Mexico (e.g. CitationParedes-Calderón 2000). Herein, we corroborate the variability of some diagnostic characters, e.g. the presence and absence of pre-pharynx, the position of genital pore, and the shape and distribution of vitellaria. Variability was observed within the same morphotype and this could be partially owed to the process of fixation of the material (flattened or not) or a case of polymorphism. There are several studies exploring the tegumental surface in adult Pleurogenidae (Podyznaya Citation1986; Ferrer et al. Citation1996; Bogéa and Caira Citation2001), and observed tegumentary papillae are distributed around the oral sucker, acetabulum, and between the tegumentary spines. The same types are observed around the oral sucker, acetabulum, and among spines in specimens of Langeronia.
Defining species is not simple, except for organisms with clear morphological differentiation. This difficulty arises when certain groups exhibit a wide variability in morphology or phenotypic plasticity, such as in the case of Langeronia. Variability has been observed in specimens collected from different host species and can often be attributed to phenotypic plasticity and could be correlated with the degree of host specificity, or sometimes be the product of rate development (Brooks Citation2003).
Initially, the ITS region was used to distinguish species of digeneans when pairs of congeners showed divergence below 2%. Later on, cox1 was shown to be an accurate marker in studies of molecular taxonomy in parasites (Rollinson et al. Citation1986; Morgan and Blair Citation1998). The detection of cryptic species using mtDNA showed up to 21% divergence in congeneric species (Bowles et al. Citation1993; Morgan and Blair Citation1998; Bell et al. Citation2001; Razo-Mendivil et al. Citation2010), with the mean pairwise divergence between congeners of the barcode region at 19% (3.9–25%) (Moszczynska et al. Citation2009). Our ITS2 data showed that both morphotypes are less than 1% divergent. Compared with previous studies (e.g. Vilas et al. Citation2005), the observed genetic differences can be attributed to intraspecific variation of a single species of the genus, which in this case would correspond to L. macrocirra. However, the rate of change in a region of DNA may vary from one group to another, and it is likely that the ITS2 in this group does not provide information at the species level. Therefore, it was necessary to obtain information from a mitochondrial gene that harbors a higher evolutionary rate.
The analysis of cox1 in Langeronia showed three clades (), which are not consistent with the morphological identification. Nevertheless, there is a high level of divergence among the three clades, e.g. 8.15–8.69% in clade I vs. clade III. If the morphotypes of Langeronia are sympatric, parasitizing the same host species and individual at the same time, a plausible explanation for the morphological variability is related with the rate of development (heterochrony), which could explain that the changes in age generate changes in morphology, and is thought to be a source of evolutionary innovation (Brooks Citation2003).
The presence of three genetic and geographical clades of Langeronia that contradicts the morphological evidence in this preliminary analysis poses a new challenge in the study of the genus. Future work should explore the complete DNA barcode region (e.g. Folmer et al. Citation1994; Tautz et al. Citation2003; Casiraghi et al. Citation2004; Moszczynska et al. Citation2009), and perhaps other markers, as well as extend to the study of life cycles and detailed ultrastructural examination in order to find robust evidence for the disentanglement of the taxonomy of the group.
There are several reasons to extend DNA barcoding to parasites, which can complement biodiversity studies (Moszczynska et al. Citation2009): facilitates the discovery and identification of new species (undescribed or unrecognized), the potential detection of parasites of biomedical and agricultural importance, biodiversity inventory initiatives associated with conservation, identification using developmental stages that would not be possible using only morphology (from several intermediate hosts or their free-living forms), the understanding of complex life histories, and the identification of cryptic and complex species will enhance and accelerate taxonomic efforts (Hebert et al. Citation2003; Casiraghi et al. Citation2004; Oceguera-Figueroa et al. Citation2005; Moszczynska et al. Citation2009; Junker et al. Citation2010). In general, we consider DNA taxonomy and DNA barcoding to be a good initiative complementing the traditional taxonomic practice of the genus in Mexico.
Acknowledgements
Patricia Escalante, Sergios-Orestis Kolokotronis, and Rob DeSalle invited us to participate in this special issue. The authors are grateful to Ma. Antonieta Arizmendi, Florencia Bertoni-Ruiz, Elisa Cabrera-Guzman, Sol Galicia, Jorge Falcon-Ordaz, Luis Jorge García, Agustin Jimenez-Ruiz, Georgina Lira, Rosario Mata, Berenit Mendoza-Garfias, Alejandro Oceguera-Figueroa, Laura Paredes-Calderón, Ulises Razo-Mendivil, Rogelio Rosas-Valdez, and Alejandro Zaldívar-Riverón for their help in the collection of specimens and their assistance in the field; Berenit Mendoza-Garfias, Instituto de Biología, UNAM, for assistance in processing samples for SEM; Laura Marquez-Valdelamar for provided technical assistance with the sequencer; and Rogelio Rosas-Valdez for assistance in the molecular analysis and figures. They thank Eric Hoberg and Patricia Pilitt, (USNPC, Beltsville, MD, USA), Luis Garcia-Prieto and Rafael Lamothe-Argumedo (CNHE, Instituto de Biología, UNAM), and Beatriz Rodriguez (Colección de Helmintos de, CHCR) for the loan of specimens. Daniel Brooks for provision samples of L. macrocirra and P. chabaudi from Guanacaste, Costa Rica for the present study; and Valerie McKenzie for providing samples of L. macrocirra from Guatemala and Costa Rica. Special thanks to Tyler Elliott for the detailed revision of English language. Sergios-Orestis Kolokotronis is thanked for his revision and suggestions that greatly improved the manuscript.
The present work was partially supported through funding from the National Science Foundation (grants DEB-0102383 and DEB-01613802) and CONACyT (grant no. 54475) to VLR. EAMS was funded by a scholarship from CONACyT and DGEP-UNAM, and a postdoctoral fellowship from CONACyT.
Declarations of interest: The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bell AS, Sommerville C. 2002. Molecular evidence for the synonymy of two species of Apatemon Szidat, 1928, A. gracilis (Rudolphi, 1819) and A. annuligerum (von Nordmann, 1832) (Digenea: Strigeidae) parasitic as metacercariae in British fishes. J Helminthol. 76:193–198.
- Bell AS, Sommerville C, Tellervo VE. 2001. A molecular phylogeny of the genus Ichthyocotylurus (Digenea, Strigeidae). Int J Parasitol. 31:833–842.
- Bogéa T, Caira JN. 2001. Ultrastructure and chaetotaxy of sensory receptors in the cercariae of Allassogonoporus sp. Olivier, 1938 (Digenea: Lecithodendriidae). Syst Parasitol. 50:1–11.
- Bowles J, Hope M, Tiu WU, Liu XS, MacManus DP. 1993. Nuclear and mitochondrial genetic marker highly conserved between Chinese and Philippine Schistosoma japonicum. Acta Trop. 55:217–229.
- Brenes RR, Arroyo SR, Delgado FE. 1960. Helmintos de la Republica de Costa Rica XI. Sobre la validez del genero Langeronia Caballero y Bravo, 1949 (Trematoda: Lecithodendriidae) y hallazgo de Ochetosoma miladelarocai Caballero y Vogelsang, 1947. Rev Biol Trop. 7:81–87.
- Brooks DR. 2003. Lessons from a quiet classic. J Parasitol. 89:878–885.
- Bursey CR, Goldberg SR. 2001. Falcaustra lowei n.sp. and other helminths from the Tarahumara frog, Rana tarahumarae (Anura: Ranidae), from Sonora, Mexico. J Parasitol. 87:340–344.
- Caballero CE, Bravo-Hollis M. 1949. Description d'un nouveau genre de Pleurogeninae (Trematoda: Lecithodendriidae) de grenouilles du Mexique: Langeronia macrocirra n.g., n. sp. Ann Parasitol Hum Comp. 24:193–199.
- Caballero CE, Caballero RCG. 1969. Un trématode nouveau parasite de Rana pipiens Schreber, 1872 de la République de Costa Rica (Amérique Centrale). Ann Parasitol Hum Comp. 44:539–546.
- Cabrera-Guzmán E, Garrido-Olvera L, León-Règagnon V. 2010. Helminth parasites of the leopard frog Lithobates sp. Colima (Amphibia: Ranidae) from Colima, Mexico. J Parasitol. 96:736–739.
- Casiraghi M, Bain O, Guerrero R, Martin C, Pocacqua V, Gardner SL, Franceschi A, Bandi C. 2004. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: Evidence for symbiont loss during evolution. Int J Parasitol. 34:191–203.
- Christian FA. 1970. Langeronia parva sp. n. (Trematoda: Lecithodendriidae) with revision of the genus Langeronia Caballero and Bravo-Hollis, 1949. J Parasitol. 56:321–324.
- Dailey MD, Goldberg SR. 2000. Langeronia burseyi sp. n. (Trematoda: Lecithodendriidae) from the California Treefrog, Hyla cadaverina (Anura: Hylidae), with revision of the genus Langeronia Caballero and Bravo-Hollis, 1949. Comp Parasitol. 67:203–240.
- Dallas JF, Irvine RJ, Halvorsen O. 2000. DNA evidence that Ostertagia gruehneri and O. arctica (Nematoda: Ostertagiinae) in rainndeer from Norway and Svalbard are conspecific. Int J Parasitol. 30:655–658.
- Espínola-Novelo JF, Guillén-Hernández S. 2008. Helminth parasites in Chaunus marinus and Cranopis valliceps (Anura: Bufonidae) from Lagunas Yalahau, Mexico. J Parasitol. 94:672–674.
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 39:783–791.
- Ferrer JR, Gracenea M, Trullols M, González O. 1996. Ultraestructural observations of the tegument of Postorchigenes gymnesicus (Digenea: Lecithodendriidae). J Helmintol. 70:13–19.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3:294–299.
- Gasser RB, Hoste H. 1995. Genetic markers for closely related parasitic nematodes. Mol Cell Probes. 9:315–320.
- Goodman JD. 1989. Langeronia brenesi, new species (Trematoda: Lecithodendriidae) in the mountain yellow-legged frog Rana muscosa from Southern California (USA). Trans Am Microsc Soc. 108:387–393.
- Guillén-Hernandez S, Salgado-Maldonado G, Lamothe-Argumedo R. 2000. Digeneans (Plathelminthes: Trematoda) of seven sympatric species of anurans from Los Tuxtlas, Veracruz, Mexico. Studies Neotrop Fauna Environ. 35:10–13.
- Hall TA. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98NT. Nucleic Acids Symp Ser. 41:95–98.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond Ser B Biol Sci. 270:313–321.
- Hillis DM, Mable BK, Larson A, Davis SK, Zimmer EA. 1996. Sequencing and cloning. In: Hillis DM, Moritz C, Mable BK. editors. Molecular Systematics. Sunderland, MA: Sinauer. p 321–378.
- Iruegas-Buentello FJ, Salinas-López N. 1989. Tremátodos de anfibios de Nuevo León, México I. Langeronia jimenezi nueva especie (Trematoda: Lecithodendriidae) en Rana pipiens. Southwest Nat. 34:369–373.
- Junker K, Lhermitte-Vallarino N, Barbuto M, Ineich I, Wanji S, Bain O. 2010. New species of Rhabdias (Nematoda: Rhabdiasidae) from Afrotropical anurans, including molecular evidence and notes on biology. Folia Parasitol. 57:47–61.
- León-Règagnon V, Brooks DR, Zelmer DA. 2001. Morphological and molecular description of Haematoloechus meridionalis n. sp. (Digenea: Plagiorchioidea: Haematoloechidae) from Rana vaillanti Brocchi of Guanacaste, Costa Rica. J Parasitol. 87:1423–1427.
- León-Règagnon V, Martínez-Salazar EA, Lazcano-Villareal D, Rosas-Valdez R. 2005. Helminth parasites of four species of anurans from Nuevo Leon, Mexico. Southwest Nat. 50:251–258.
- Lotz JM, Font WF. 2008. Family Pleurogenidae Looss, 1899. In: Bray RA, Gibson DI, Jones A. editors. Keys to the Trematoda. Vol. 3. London: CAB International and Natural History Museum. p 563–575.
- McManus DP, Bowles J. 1996. Molecular genetic approaches to parasite identification: Their value in diagnostic parasitology and systematics. Int J Parasitol. 26:687–704.
- Morgan JAT, Blair D. 1998. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda). Parasitology. 116:289–297.
- Moszczynska A, Locke SA, Mclaughlin JD, Marcogliese DJ, Crease TJ. 2009. Development of primers for the mitochondrial cytochrome c oxidase I gene in digenetic trematodes (Platyhelminthes) illustrates the challenge of barcoding parasitic helminths. Mol Ecol Resourc. 9:75–82.
- Oceguera-Figueroa A, León-Règagnon V, Siddall ME. 2005. Phylogeny and revision of Erpobdelliformes (Annelida, Arhynchobdellida) from Mexico based on nuclear and mithochondrial gene sequences. Rev Mex Biodivers. 76:191–198.
- Palumbi SR. 1996. Nucleic acids II: The polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK. editors. Molecular Systematics. Sunderland, MA: Sinauer . p 205–247.
- Paredes-Calderón L. 2000. Fauna helmintológica de Rana vaillanti en la región de Los Tuxtlas, Veracruz, México. Bachelor thesis, Mexico D.F.
- Paredes-Calderón L, León-Règagnon V, García-Prieto L. 2004. Helminth infracommunities of Rana vaillanti Brocchi (Anura: Ranidae) in Los Tuxtlas, Veracruz, Mexico. J Parasitol. 90:692–696.
- Podyznaya IM. 1986. Surface ultrastructure of two species of Allassogonopoine and Lecithodendriidae (Trematoda, Plagiorchiida). Trudy Zool Inst. 155:94–104 (Morfologiya, sistematika I faunistika paraziticheskikk Zhivotnykh).
- Razo-Mendivil U, Rosas-Valdez R, Pérez-Ponce de León G. 2008. A new cryptogonimid (Digenea) from the Mayan cichlid, Cichlasoma urophthalmus (Osteichthyes: Cichlidae), in several localities of the Yucatán Peninsula, Mexico. J Parasitol. 94:1371–1378.
- Razo-Mendivil R, Vázquez-Domínguez E, Rosas-Valdez R, Pérez-Ponce de León G, Nadler SA. 2010. Phylogenetic analysis of nuclear and mitochondrial DNA reveals a complex of cryptic species in Crassicutis cichlasomae (Digenea: Apocreadiidae), a parasite of Middle-American cichlids. Int J Parasitol. 40:471–486.
- Rollinson D, Walker TK, Simpson AJG. 1986. The application of recombinant DNA technology to problems of helminth identification. Parasitology. 92:s53–s71.
- Sullivan JJ. 1971. Pseudosonsinotrema echinophallus sp.n. (Digenea: Pleurogenidae) a new trematode from Rana pipiens in Costa Rica. Proc Helminthol Soc Wash. 38:34–37.
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Version 4. Sunderland, MA: Sinauer.
- Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AP. 2003. A plea for DNA taxonomy. Trends Ecol Evol. 18:70–74.
- Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequences weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
- Ubelaker JE. 1965. The taxonomic status of Langeronia Caballero and Bravo-Hollis, 1949 with the synonymy of Loxogenes provitellaria, Sacks, 1952 with Loxogenes macrocirra Caballero and Bravo-Hollis, 1949. Trans Kansas Acad Sci. 68:187–190.
- Vilas R, Criscione CD, Blouin MS. 2005. A comparison between mitochondrial DNA and the ribosomal internal transcribed regions in prospecting for cryptic species of platyhelminth parasites. Parasitology. 131:1–8.
- Yáñez-Arenas CA, Guillén-Hernández S. 2010. Helminth fauna of Lithobates brownorum (Anura: Ranidae) at three localities in the state of Yucatán, Mexico. Rev Mex Biodivers. 81:191–195.
- Zaldívar-Riverón A, León–Règagnon V, Nieto-Montes de Oca A. 2004. Phylogeny of the Mexican coastal leopard frogs of the Rana berlandieri group based on mtDNA sequences. Mol Phylogenet Evol. 30:38–49.