Abstract
Background and Aims. Molecular markers have contributed to species authentication by flagging mislabeling and the misidentification of commercial landings. Such tools are of great value since the market substitution of fish of lower value for highly commercialized species is expected to become more pronounced due to a shortage of natural stocks.
Materials and Methods. Here we report on the molecular identification 4results from processed fish products (i.e. fillets) and whole fishes sold in Brazilian markets under the common name surubim (Pseudoplatystoma spp.).
Results. DNA barcoding revealed the incorrect labeling of around 80% of all samples analyzed, with mislabeling being more pronounced within fillets rather than whole fish.
Conclusion. To our knowledge, this is the first report correlating the rate of fraud with processed fish products. The establishment of an official list of acceptable common names for freshwater fish and seafood is urgently needed in Brazil for further trade regulations to take place.
Introduction
The effects of species substitution include economic fraud, health hazards, and the illegal trade of protected species. Therefore, the detection of species substitution has become an important topic within the food industry, and there is a growing need for rapid, reliable, and reproducible tests to verify species in commercial fish and other animal products (Rasmussen and Morrissey Citation2008). In addition, species identification is useful in ensuring honest trading exchanges for correct consumer information. Toward this end, regulatory organizations such as the European Union have established labeling laws for fish and aquaculture products, in which labels should bring traceability information (i.e. identification, origin of fish, and production method; Moretti et al. Citation2003; CitationMartinez et al. 2005).
The substitution of a less valuable fish species and products (i.e. fillets and eggs) for a more valuable one represents a common commercial fraud (DeSalle and Birstein Citation1996; Civera Citation2003; Marko et al. Citation2004). It is known that fluctuations in the supply and demand of different fish species together with increases in international trade and fish consumption are important factors in intentional product mislabeling (Rasmussen and Morrissey Citation2008). However, the way fishes are processed for commercialization has not yet been correlated with the rate of fraud.
Moreover, it has become clear that most populations of fish are declining dramatically, and aquaculture efforts are not expected to compensate (Pauly et al. Citation2002). In this scenario, trade in animal species has contributed greatly to overall biodiversity crisis (Manel et al. Citation2002). For example, 77% of the fish sold in the USA as the highly threatened red snapper (Lutjanus campechanus) are in fact another species (Marko et al. Citation2004). Reliable and rapid molecular identification methods could help both to protect citizens from fraud and to protect endangered species from overexploitation and illegal trafficking (DeSalle and Birstein Citation1996; Teletchea et al. Citation2005).
To avoid mislabeling and commercial fraud, the US Food and Drug Administration has compiled an online Regulatory Fish Encyclopedia (http://www.fda.gov) listing acceptable market names, isoelectric focusing protein electrophoresis patterns, and DNA barcoding data (Yancy et al. Citation2008). These molecular data are also correlated with high-resolution images of whole fish and filleted fish species, plus geographic, taxonomic, and nomenclature information for imported and domestical species. All this information is on hand to assist government officials and purchasers in the correct identification and detection of species substitution and economic fraud. Another similar initiative is the European FishTrace Consortium (http://www.fishtrace.org), comprising 53 members from several institutions (Sevilla et al. Citation2007). Besides its rich biodiversity, no such initiative is yet currently ongoing in Brazil. Therefore, to implement laws against poaching and the trade of overexploited species and enforcing labeling regulations to prevent product substitution, there is a need for sensitive and reliable analytical methods that can be applied to determine the species of a fish, even when no detectable external features are present (Baker et al. Citation2000; Kyle and Wilson Citation2007). Not to mention the detrimental effects that fish adulteration can have on the commercial market, it can also put consumers at risk of purchasing potentially harmful and mislabeled products (Cohen et al. Citation2009; Lowenstein et al. Citation2010). The effectiveness of fish conservation and management programs can also be improved which aid in the protection of aquatic habitats and endangered species (Teletchea et al. Citation2005).
Molecular tools are advantageous for fish and fish products identification for three main reasons: large number of fish species from distinct live history stages (eggs, fry, and adults) can be examined; in addition, processed fish products lacking the morphological characteristics, such as frozen fillets and precooked fish, are also accessible (typically, these cannot be identified using the traditional identification procedure); and there are insufficient specialists in alpha taxonomy for fish identification, especially in the Neotropics.
Several protocols have been described for species identification of fish products, based on different technologies such as high-performance liquid chromatography, isoelectric focusing, and polyacrylamide gel electrophoresis (CitationMartinez et al. 2005; Rasmussen and Morrissey Citation2008). Among these, DNA-based methodologies are one of the most promising approaches since they can be applied to all the different life stages of fish species and fish products (Rasmussen and Morrissey Citation2008; Smith et al. Citation2008; Vinas and Tudela Citation2009).
The mitochondrial DNA (mtDNA) cytochrome c oxidase subunit I gene (COI) has provided numerous examples as a reliable and universal tool for the identification of species such as the flatfish (Terol et al. Citation2002; Espineira et al. Citation2008), tuna (Terol et al. Citation2002; Lowenstein et al. Citation2010), anchovy (Jerome et al. Citation2008), sharks (Barbuto et al. Citation2010), and also wildlife forensics investigations (Dawnay et al. Citation2007; Nelson et al. Citation2007). Its application in molecular taxonomy has been criticized due to introgressive hybridization, mitochondrial pseudogenes in the nucleus, and the retention of ancestral polymorphisms (e.g. Rubinoff Citation2006; Rubinoff et al. Citation2006). However, species assignment failure rates do not typically exceed 5–10% (Hebert and Gregory Citation2005; Ward et al. Citation2005; Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009). Therefore, a global effort to assemble a standardized reference DNA sequence library (using the COI region) for all fishes has been proposed. This initiative led to the establishment of international research collaboration, named the Fish Barcode of Life Campaign (FISH-BOL; Ward et al. Citation2009). The barcode data for thousands of freshwater fishes have been uploaded to the Barcode of Life Data Systems database (BOLD), including freshwater species from the Neotropics (data available on BOLD; Carvalho et al. Citation2011), allowing their application for the analysis of commercial fraud cases in Brazil.
This work was conducted to identify which species have been sold in Brazil labeled as surubim or pintado using DNA barcode data. These vernacular names are applied to Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum (former Pseudoplatystoma fasciatum; CitationFroese and Pauly 2011); however, Brazil does not have an official list of commercial and Latin names for these fishes. P. corruscans is considered the most valuable commercial and recreational freshwater fish in the São Francisco River (fourth biggest river in Brazil); however, surubim harvest has shown clear indications of collapse, since harvesting of P. corruscans has declined from 10.3 to 0.8 kg captured fish per day from 1987 to 1999 (Godinho et al. Citation2007). Moreover, the genus Pseudoplatystoma consists of at least eight species, five of which were recently described (Buitrago-Suarez and Burr Citation2007). P. reticulatum and P. corruscans are sympatric in the Paraná basin (southern Brazil), whereas only the latter has been described in the São Francisco basin. A recent molecular phylogeny of the genus Pseudoplatystoma did not find any correlation between morphological and molecular data (Control Region) in two species (P. reticulatum and Pseudoplatystoma punctifer; Torrico et al. Citation2009). For all other species, including P. corruscans (surubim), the molecular data supported the morphological classification.
From this work, we reported a fish market DNA barcode survey of processed (i.e. fillets) and whole fishes sold in Brazil under the common name surubim. We tested the hypothesis that substitutions would be more frequent among fillets than within whole fish because of the difficulty of visual identification. Our results showed a strong correlation between the methods in which fishes are commercialized (i.e. fillets or whole fish) and fraud, since the highest number of substitutions was detected within fillets.
Materials and methods
Samples labeled as surubim were purchased at nine supermarkets in the city of Belo Horizonte (Minas Gerais state, Brazil) in 2009–2010. Samples consisted of two types: whole fish (n = 30) and fillets (n = 33). Upon collection, tissues were stored in ethanol 95% and details of brand, price, and pictures of the product were taken for documentation purposes. DNA was isolated by homogenization and digestion with proteinase K at 37°C overnight, followed by standard phenol/chloroform purification (CitationSambrook et al. 1989). A fragment of 658 bp of COI was amplified using the primers FishF1 and FishR1 (Ward et al. Citation2005). The 25 μl PCR mixes included 19.5 μl ultrapure water, 2.5 μl of 10 × PCR buffer, 2.5 mM MgCl2, 0.35 μl each primer (10 mM), 2.5 μl dNTP (1 mM), 0.25μl PHT Taq polymerase (5 U/μl), and 1.0 μl DNA template (50–100 ng/μl). Thermal cycling conditions consisted of an initial denaturation step at 94°C for 2 min, 35 cycles at 94°C for 30 s, 54°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. PCR products (1–2 μl) were visualized on an agarose gel and selected for direct sequencing. Sequences were determined bi-directionally using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, California, USA), following the manufacturer's protocol on an ABI PRISM 310 Genetic Analyzer.
Data analysis
COI sequences recovered from samples labeled as surubim were identified by searching the GenBank database using the BLASTN algorithm (http://blast.ncbi.nlm.nih.gov) and by BOLD (http://www.boldsystems.org) identification engine to search DNA barcode records within BOLD (Ratnasingham and Hebert Citation2007). The best-scoring matches obtained from both databases for each sample were registered. Genetic divergence was calculated using Kimura's two-parameter (K2P) nucleotide substitution model (Kimura Citation1980). A neighbor-joining tree (Saitou and Nei Citation1987) of K2P distances was generated in MEGA 3 (Kumar et al. Citation2004) to provide a graphic representation of divergence among analyzed samples. Node support was assessed with 10,000 bootstrap replicates (Felsenstein Citation1985).
Brazil does not have a list of acceptable common names; therefore, the FishBase (http://www.fishbase.org) nomenclature was used to identify the scientific names that may correspond to the commercial name surubim (P. corruscans and P. reticulatum), especially since BOLD and GenBank rely on FishBase as a taxonomic authority for valid fish species names (CitationFroese and Pauly 2011). It is interesting to note that hybrids obtained by crossing a female P. reticulatum and a male P. corruscans (Carvalho et al. Citation2008; Bignotto et al. Citation2009) are of the most common freshwater catfishes commercialized in Brazil. A standard DNA barcode sequence for P. corruscans (Carvalho et al. Citation2011; BOLD record number BSB400-10 and GenBank accession number HM405206) was included as a reference in the alignment dataset.
Results
Genetic identification of samples commercialized as surubim using the GenBank and BOLD search engines
All amplified sequences that exceeded 600 nucleotides in length with no insertions, deletions, or stop codons were observed, thus reducing the possibility of mtDNA copies in the nucleus. The sequences obtained from the samples were deposited on GenBank (accession numbers HQ689323–HQ689385) and compared against the BOLD and GenBank databases. Successful matches varied from 89 to 100% pairwise sequence identity (). Only three samples could not be identified in the BOLD Species Reference database. Nonetheless, the BOLD Full database returned hits with a percentage of identity as high as GenBank, with the advantage of being a more reliable source of taxonomic identification. Our results showed that only one P. corruscans haplotype (sample BS1) was detected among the 63 commercial samples analyzed (). Most of the samples had 100% similarity with the catfish species Pseudoplatystoma tigrinum and P. reticulatum. However, high-identity matches were also obtained with marine species Genidens barbus (100% match) and Cynoscion virescens (89–100% match), clearly showing a case of mislabeling (). Lower identity scores were also returned, and samples were provisionally identified; for example, Netuma thalassina (sample 24, ) with a 90% match (BOLD database) and Cynoscion jamaicensis (sample 66) with an 89% match in BOLD. The latter species (sample 66) also had a GenBank top match with the spotfin croaker Roncador stearnsii (87%), a species from the eastern Pacific ocean ().
Table I. Identification of commercial surubim samples using the GenBank and BOLD search engines.
DNA barcode analysis of whole and filleted fishes commercialized as surubim
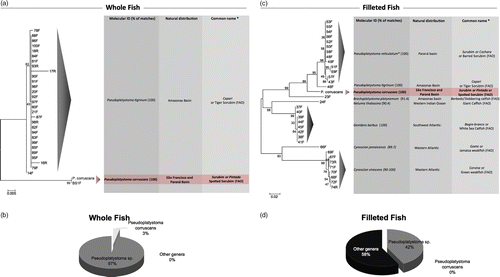
The careful inspection of the results presented in revealed a higher rate of substitution within species from genera other than Pseudoplatystoma among fishes sold as fillets rather than whole fish. To better evaluate this difference, a neighbor-joining tree using the K2P evolutionary model was built for each group (fillets and whole fishes) separately (). The overall K2P distance for the whole fish group () was 1.1%, and the divergence of commercial samples from the standard barcode sequence of P. corruscans varied from 0 to 8.4%. The overall K2P distance for the filleted fish group () was higher (15.5%), and the divergence of commercial samples from the standard barcode sequence of P. corruscans varied widely from 5.9 to 24.8%. Within whole fish only two clades were detected (99% bootstrap support value), one representing the Amazonian species Pseudoplatystoma tigrinus and the other representing P. corruscans clade, with only one sample grouping together with our standard barcode sequence for P. corruscans ( and ). When considering fillets, at least seven distinct clades with moderate to high-bootstrap value (99–68%) were recovered, characterizing at least seven different species (), with not even one haplotype belonging to P. corruscans ( and ). The pie charts () show more explicit graphical representations of the percentage of samples identified as P. corruscans within the whole fish and fillet categories. When considering the whole fish group (), one sample was identified as belonging to P. corruscans, whereas the remaining 97% of this group was identified as P. tigrinum (100% similarity with this Amazonian species). In contrast, only 42% of the filleted samples () belonged to Pseudoplatystoma sp., whereas 58% of them had higher similarity with other genera (i.e. Brachyplatystoma, Netuma, and Genidens).
Figure 1. Distance analysis of whole and filleted fishes commercialized as surubim. Neighbor-joining trees of K2P distances were generated among surubim samples commercialized as (a) whole and (c) filleted fishes. Common names in Portuguese and English are shown according to FishBase. Graphical representation of the percentage of surubim samples commercialized as (b) whole or (d) filleted that belong to P. corruscans species, to the genus Pseudoplatystoma (except for P. corruscans), or to other genera.
Discussion
The application of COI sequences in forensics has already been investigated for reproducibility, heteroplasmy, mixed DNA samples, chemical treatments, environmental conditions, and other factors showing consistent results in which a great range of reference data exist (Dawnay et al. Citation2007). Several examples have already highlighted the potential of such molecular tools to flag the mislabeling of fish and seafood products (e.g. DeSalle and Birstein Citation1996; Marko et al. Citation2004; Kyle and Wilson Citation2007; Jerome et al. Citation2008; Rasmussen and Morrissey Citation2008; Wong and Hanner Citation2008; Yancy et al. Citation2008; Barbuto et al. Citation2010; Filonzi et al. Citation2010; Lowenstein et al. Citation2010), identifying tuna sushi samples analyzed for mercury contamination (Lowenstein et al. Citation2010) plus the identification of smoked fish products (Smith et al. Citation2008). In Brazil, Ardura et al. (Citation2010) utilizing two mtDNA genes showed that at least seven distinct species have been sold under the common name “Acará”, making the real estimation of exploitation rates impossible.
In this study, we identified commercial samples labeled as surubim through the comparisons of COI mtDNA sequences using the BLAST engine to search GenBank. In addition, the BOLD identification engine was employed to search barcode records within BOLD, as well as using standard P. corruscans barcode sequences (Carvalho et al. Citation2011), Strikingly, analysis of surubim fillets revealed a 58% rate of substitution by fishes from the species that do not belong to the Pseudoplatystoma genus. These include marine species (G. barbus and C. virescens) that were identified with a great overall match (99–100%) as being the preferred substitutive species (). Other recovered haplotypes showed poorer matches and were only tentatively assigned to the species level (N. thalassina and Brachyplatystoma platynemum; ). Within whole fish, only P. tigrinum was detected. Nonetheless, if we consider the identification via FishBase, which lists only P. reticulatum and P. corruscans as valid names for surubim, selling P. tigrinum as surubim would also qualify as mislabeling.
The high rate of substitution of this freshwater species could be due to the fact that the vernacular name surubim is well known within the Brazilian market. Therefore, by using this label traders might be able to sell their product for a better price. This becomes clear as we compare the market prices of the fishes sold as surubim identified in this study (). For instance, species labeled bagres, a Brazilian vernacular name for a less known catfish group, are sold at a 70% lower price than fishes under the surubim label. Therefore, we have strong evidence that intentional mislabeling of cheaper fish products is a more frequent phenomenon mainly within processed fish ().
Table II. Common names and price per kilogram of commercialized fish mislabeled as surubim.
One limitation of DNA barcode analysis is the fact that, by using a mitochondrial gene, only the matrilineal lineage is examined. This limits the interpretation of results when hybridization within species is common, as is the case for the genus Pseudoplatystoma. Hybrids between P. corruscans males and P. reticulatum females have been reported (Bignotto et al. Citation2009), figuring as the most common hybrid catfish produced on Brazilian aquaculture farms in Brazil (Carvalho et al. Citation2008). Therefore, we were expecting a higher representation of P. reticulatum haplotypes among the commercial samples analyzed in this study. Surprisingly, most haplotypes recovered belong to Amazonian species. Whether these species represent a new type of hybrids is a subject of future research. Further analysis using nuclear markers is recommended for the identification of different types of commercial hybrids. That said, recovered haplotypes from distinct genera (even from marine species, e.g. Genidens and Cynoscion) are unlikely to be due to hybridization, reinforcing our hypothesis of intentional mislabeling of lower value species as surubim.
Conclusions
In this study, we have clearly shown the occurrence of substitutions of the freshwater catfish surubim by other species of lower commercial value, including marine species. If we consider FishBase, only the species P. corruscans and P. reticulatum have the common name surubim valid. Therefore, close to 80% of the fish sold in the surveyed markets, Brazilian markets of Belo Horizonte city are mislabeled. This figure is higher than those reported for North American seafood (25%; Wong and Hanner Citation2008) and Italian fish products (32%; Filonzi et al. Citation2010). The high rates of substitution of P. corruscans by other species could also be an indication that its wild stocks have not been coupled with market growth. In fact, overexploitation of P. corruscans might explain, in part, why it has been substituted by morphologically similar species when commercialized as whole fish, and in some cases (e.g. fillets) substituted by quite dissimilar species (). The establishment of conservation strategies and the normalization of vernacular names for native commercially important Brazilian fishes, together with the molecular inspection of fish products, have the potential to form an important tool for the preservation of the Brazilian fish fauna and protect consumers from mislabeled products.
We strongly recommend the establishment of a valid list of commercial and Latin names for the fishes commercialized in Brazil. Such a reference list would make possible for State Fish and Game Departments to be able to regulate and detect fraud, substitution, and the commercialization of threatened species. In addition, customs services will have the ability to regulate and inspect imported/exported items, for the purpose of taxation and to protect the consumer from misguidance. Such a list is currently in use together with Barcoding analysis to detect market substitution in North American seafood (Wong and Hanner Citation2008).
Acknowledgments
The authors are grateful to José Vanderval Melo Junior and Arno Soares Seerig for helping with the collection of fish to all personnel of the Laboratory of Animal Genetics at UFMG Veterinary College for their assistance and to Shannon Loughnan and Sven Becker for their comments on the manuscript. This study was supported by CNPq/FAPEMIG (INCT 573899/2008-8) and Instituto Estadual de Florestas. D.C.C. is also grateful to CNPq for the PDJ fellowship (150420/2009-9) and CAPES postdoctoral fellowship (#4095-09-0).
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E. 2010. DNA barcoding for conservation and management of amazonian commercial fish. Biol Conserv. 143:1438–1443.
- Baker CS, Lento GM, Cipriano F, Palumbi SR. 2000. Predicted decline of protected whales based on molecular genetic monitoring of Japanese and korean markets. Proc R Soc Lond B Biol Sci. 267:1191–1199.
- Barbuto M, Galimberti A, Ferri E, Labra M, Malandra R, Galli P, Casiraghi M. 2010. DNA barcoding reveals fraudulent substitutions in shark seafood products: The italian case of “Palombo” (Mustelus spp.). Food Res Int. 43:376–381.
- Bignotto TS, Prioli AJ, Prioli SMAP, Maniglia TC, Boni TA, Lucio LC, Gomes RA, Oliveira VN, Prioli AV, Júlio HFJr, Prioli LM. 2009. Genetic divergence between Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum (Siluriformes: Pimelodidae) in the parana river basin. Braz J Biol. 69:681–689.
- Buitrago-Suarez UA, Burr BM. 2007. Taxonomy of the catfish genus Pseudoplatystoma Bleeker (Siluriformes: Pimelodidae) with recognition of eight species. Zootaxa. 1512:1–38.
- Carvalho DC, Seerig AS, Melo DC, Sousa AB, Pimenta D, Oliveira DAA. 2008. Identificação molecular de peixes: O caso do surubim (Pseudoplatystoma spp.). Rev Bras Reprod Anim. 32:215–219.
- Carvalho DC, Oliveira DAA, Pompeu PS, Leal CG, Oliveira C, Hanner R. 2011. Deep barcode divergence in brazilian freshwater fishes—the case of the São Francisco river basin. Mitochondrial DNA. 22 Suppl 1 Epub.
- Civera T. 2003. Species identification and safety of fish products. Vet Res Commun. 27:481–489.
- Cohen NJ, Deeds JR, Wong ES, Hanner RH, Yancy HF, White KD, Thompson TM, Wahl M, Pham TD, Guichard FM, Huh I, Austin C, Dizikes G, Gerber SI. 2009. Public health response to puffer fish (tetrodotoxin) poisoning from mislabeled product. J Food Prot. 72:810–817.
- Dawnay N, Ogden R, McEwing R, Carvalho GR, Thorpe RS. 2007. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci Int. 173:1–6.
- DeSalle R, Birstein VJ. 1996. PCR identification of black caviar. Nature. 381:197–198.
- Espineira M, Gonzalez-Lavin N, Vieites JM, Santaclara FJ. 2008. Development of a method for the genetic identification of flatfish species on the basis of mitochondrial DNA sequences. J Agric Food Chem. 56:8954–8961.
- Felsenstein J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 39:783–791.
- Filonzi L, Chiesa S, Vaghi M, Nonnis Marzano F. 2010. Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Res Int. 43:1383–1388.
- Froese R, Pauly D, Editors. 2011. FishBase. World Wide Web electronic publication. www.fishbase.org, version (02/2011).
- Godinho AL, Kynard B, Godinho HP. 2007. Migration and spawning of female surubim (Pseudoplatystoma corruscans, Pimelodidae) in the São Francisco river, Brazil. Environ Biol Fish. 80:421–433.
- Hebert PDN, Gregory TR. 2005. The promise of DNA barcoding for taxonomy. Syst Biol. 54:852–859.
- Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, Watkinson D, Dumont P, Curry A, Bentzen P, Zhang J, April J, Bernatchez L. 2008. Identifying canadian freshwater fishes through DNA barcodes. PLoS ONE. 3:e2490.
- Jerome M, Martinsohn JT, Ortega D, Carreau P, Verrez-Bagnis V, Mouchel O. 2008. Toward fish and seafood traceability: Anchovy species determination in fish products by molecular markers and support through a public domain database. J Agric Food Chem. 56:3460–3469.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Kumar S, Tamura K, Nei M. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150–163.
- Kyle CJ, Wilson CC. 2007. Mitochondrial DNA identification of game and harvested freshwater fish species. Forensic Sci Int. 166:68–76.
- Lowenstein JH, Burger J, Jeitner CW, Amato G, Kolokotronis SO, Gochfeld M. 2010. DNA barcodes reveal species-specific mercury levels in tuna sushi that pose a health risk to consumers. Biol Lett. 6:692–695.
- Manel S, Berthier P, Luikart G. 2002. Detecting wildlife poaching: Identifying the origin of individuals with bayesian assignment tests and multilocus genotypes. Conserv Biol. 16:650–659.
- Marko PB, Lee SC, Rice AM, Gramling JM, Fitzhenry TM, McAlister JS, Harper GR, Moran AL. 2004. Fisheries: Mislabelling of a depleted reef fish. Nature. 430:309–310.
- Martinez I, James D, Loréal H. 2005. Application of modern analytical techniques to ensure seafood safety and authenticity. FAO Fisheries Technical Paper. Rome: Food and Agriculture Organization of United Nations.
- Moretti VM, Turchini GM, Bellagama F, Caprino F. 2003. Traceability issues in fishery and aquaculture products. Vet Res Commun. 27:497–505.
- Nelson LA, Wallman JF, Dowton M. 2007. Using COI barcodes to identify forensically and medically important blowflies. Med Vet Entomol. 21:44–52.
- Pauly D, Christensen V, Guenette S, Pitcher TJ, Sumaila UR, Walters CJ, Watson R, Zeller D. 2002. Towards sustainability in world fisheries. Nature. 418:689–695.
- Rasmussen RS, Morrissey MT. 2008. DNA-based methods for the identification of commercial fish and seafood species. Compr Rev Food Sci F. 7:280–295.
- Ratnasingham S, Hebert PDN. 2007. BOLD: The barcode of life data system (www.Barcodinglife.Org). Mol Ecol Notes. 7:355–364.
- Rubinoff D. 2006. Utility of mitochondrial DNA barcodes in species conservation. Conserv Biol. 20:1026–1033.
- Rubinoff D, Cameron S, Will K. 2006. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J Hered. 97:581–594.
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular Cloning: A Laboratory Manual. Cold Springs Harbor, NY, Cold Springs Harbor Laboratory Press. 1659p.
- Sevilla RG, Diez A, Noren M, Mouchel O, Jerome M, Verrez-Bagnis V, Van Pelt H, Favre-Krey L, Krey G, Consortium TF, Bautista JM. 2007. Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol Ecol Notes. 7:730–734.
- Smith PJ, Mcveagh SM, Steinke D. 2008. DNA barcoding for the identification of smoked fish products. J Fish Biol. 72:464–471.
- Teletchea F, Maudet C, Hanni C. 2005. Food and forensic molecular identification: Update and challenges. Trends Biotechnol. 23:359–366.
- Terol J, Mascarell R, Fernandez-Pedrosa V, Perez-Alonso M. 2002. Statistical validation of the identification of tuna species: Bootstrap analysis of mitochondrial DNA sequences. J Agric Food Chem. 50:963–969.
- Torrico JP, Hubert N, Desmarais E, Duponchelle F, Rodriguez JN, Montoya-Burgos J, Montoya-Burgos J, Garcia Davila C, Carvajal-Vallejos FM, Grajales AA, Bonhomme F, Renno JF. 2009. Molecular phylogeny of the genus pseudoplatystoma (Bleeker, 1862): Biogeographic and evolutionary implications. Mol Phylogenet Evol. 51:588–594.
- Valdez-Moreno M, Ivanova NV, Elias-Gutierrez M, Contreras-Balderas S, Hebert PDN. 2009. Probing diversity in freshwater fishes from mexico and guatemala with DNA barcodes. J Fish Biol. 74:377–402.
- Vinas J, Tudela S. 2009. A validated methodology for genetic identification of tuna species (genus Thunnus). PLoS ONE. 4:e7606.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc B. 360:1847–1857.
- Ward RD, Hanner R, Hebert PDN. 2009. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 74:329–356.
- Wong EHK, Hanner RH. 2008. DNA barcoding detects market substitution in north american seafood. Food Res Int. 41:828–837.
- Yancy HF, Zemlak TS, Mason JA, Washington JD, Tenge BJ, Nguyen NL, Barnett JD, Savary WE, Hill WE, Moore MM, Fry FS, Randolph SC, Rogers PL, Hebert PD. 2008. Potential use of DNA barcodes in regulatory science: Applications of the regulatory fish encyclopedia. J Food Prot. 71:210–217.