Abstract
Uropsilus sp. 4 is a new cryptic species, collected in Changyang county, Hubei province, China. In this study, the whole mitochondrial genome of Uropsilus sp. 4 was first determined and characterized. The genome is 16,542 bp in length, containing 13 protein coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a putative control region. Base on NJ, ML, and BI methods, we obtained the same topologies. U. sp. 4 clustered with U. gracilis and the divergence time was 1.78 Ma (95% CI 1.24–2.32 Ma), in concordance with the third period of last orogenic push of the Qinghai-Tibetan Plateau, might contribute to the speciation of U. sp. 4.
Keywords:
There are 10 recognized species within the genus Uropsilus: U. aequodonenia, U. andersoni, U. atronates, U. gracilis, U. investigator, U. nivatus, U. sorcipes, U. sp. 1, U. sp. 2, and U. sp. 3 (Liu et al. Citation2013; Wan et al. Citation2013). We collected a specimen, SAF_HB10N042, female, from Changyang county, Hubei province, China. By sequencing the cyt b gene and constructing a neighbor-joining (NJ) phylogenetic tree following the study of Wan et al. (Citation2013), we identified this individual as a new cryptic species, Uropsilus sp. 4. To date, mitogenomes of six species of Uropsilus have been reported: U. aequodonenia, U. andersoni, U. soricipes, U. nivatus, U. sp.1, and U. gracilis (Tu et al. Citation2015; Hou et al. Citation2016). More mitogenomes are required to better know phylogenetic relationships within Uropsilus.
In this study, we thus determined the complete mitochondrial genome of U. sp. 4 and also built the phylogenetic trees with other eleven related species based on 12 protein-coding genes (PCGs) using the NJ, maximum likelihood (ML), and Bayesian inference (BI) methods. NJ analysis was performed by MEGA 6.06 (MEGA Inc., Englewood, NJ) (Tamura et al. Citation2013) based Kimura-2-parameter model. ML was carried out online software PhyML 3.0 (Bioo Scientific, Austin, TX) (Guindon et al. Citation2010) with default parameters. BI analysis was done by MrBayes 3.1.2 (MrBayes Inc., La Jolla, CA) (Ronquist and Huelsenbeck Citation2003) and the process parameters were based on the study of Tu et al. (Citation2012). Neotetracus sinensis was set as outgroup. To estimate divergence time (with 95% confidential interval (CI)) between U. sp. 4 and its sister species, we performed the analysis using MEGA 6.06 (Tamura et al. Citation2013) based on the NJ tree of PCGs. Two calibration points were defined (Wan et al. Citation2013): (i) the oldest known Talpini was from the early Oligocene at approximately 33.9–32.6 million years ago (Ma); (ii) the oldest known U. soricipes was at 2.4–2.0 Ma.
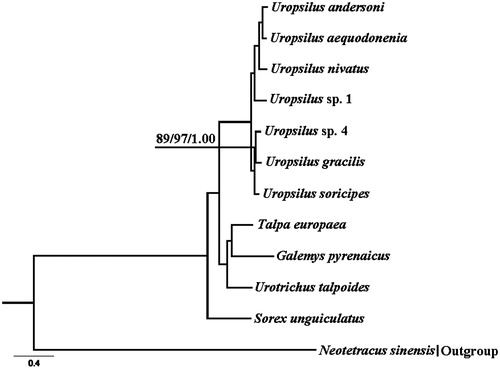
The complete mitochondrial genome of U. sp. 4 (GenBank Accession Number KM503088, 16,542 bp), consisting of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a displacement loop region (Table S1). Its genome structure is similar to other mammal mitogenomes (Chen et al. Citation2015; Tu et al. Citation2015; Wang et al. Citation2016). We generated the similar topologies (). Within the genus Uropsilus, U. sp. 4 clustered with U. graclis. Divergence time between U. sp. 4 and U. gracilis was 1.78 Ma (95% CI 1.24–2.32 Ma). U. gracilis, collected in the Jinfo Mountains, Chongqing (Hou et al. Citation2016), which is located in the eastern Qinghai-Tibetan Plateau (QTP). The divergence time between U. sp. 4 and U. gracilis is in concordance with the third phase of last orogenic push of the QTP occurring about 1.7 Ma (Li et al. Citation2001). Therefore, the uplifted event might contribute to the speciation of U. sp.4. However, further study needs to confirm this.
Figure 1. Bayesian 50% majority-rule consensus phylogenetic tree based on Bayesian analysis of 12 protein-coding genes. Neotetracus sinensis was used as an outgroup. The numbers on the internode branches from left to right were the NJ, ML and BI support values, respectively. Sequence data used in the study are the following: Uropsilus sp. 4 (KM503088), Uropsilus andersoni (JX945573), Uropsilus aequodonenia (KC516778), Uropsilus nivatus (JX945574), Uropsilus sp. 1 (JX034737), Uropsilus gracilis (KM379136), Uropsilus soricipes (JQ658979), Talpa europaea (NC_002391), Galemys pyrenaicus (NC_008156), Urotrichus talpoides (NC_005034), Sorex unguiculatus (NC_005435), and Neotetracus sinensis (JX519466).
TMDN_A_1060440_Supplementary_Information.zip
Download Zip (8.6 KB)Acknowledgements
The authors would acknowledge Prof. Shaoying Liu at Sichuan Academy of Forestry for providing sample and also thank Dr Chaochao Yan at Sichuan University for building phylogenetic evolutionary trees.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chen SD, Tu FY, Zhang XY, Li W, Chen GY, Zong H, Wang Q. 2015. The complete mitogenome of Stripe-Backed Shrew, Sorex cylindricauda (Soricidae). Mitochondrial DNA. 26:477–478.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 59:307–21.
- Hou QF, Tu FY, Liu Y, Liu SY. 2016. Characterization of the mitogenome of Uropsilus gracilis and species delimitation. Mitochondrial DNA Part A. 27:1836–1837.
- Li JJ, Fang XM, Pan BT. 2001. Late Cenozoic intensive uplift of Qinghai-Xizang Plateau and its impacts on environments in surrounding area. Quat Sci. 21:381–91.
- Liu Y, Liu SY, Sun ZY, Guo P, Fan ZX, Murphy RW. 2013. A new species of Uropsilus (Talpidae: Uropsilinae) from Sichuan, China. Acta Theriol Sin. 33:113–22 (in Chinese).
- Ronquist FR, Huelsenbeck JP. 2003. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 19:1572–74.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–9.
- Tu FY, Fan ZX, Chen SD, Yin YH, Li P, Zhang XY, Liu SY, Yue BS. 2012. The complete mitochondrial genome sequence of the Gracile shrew mole, Uropsilus gracilis (Soricomorpha: Talpidae). Mitochondrial DNA. 23:382–4.
- Tu FY, Fan ZX, Murphy RW, Chen SD, Zhang XY, Yan CC, Liu Y, et al. 2015. Molecular phylogenetic relationships among Asiatic shrew moles inferred from the complete mitogenomes. J Zool Syst Evol Res. 53:155–160.
- Wan T, He K, Jiang XL. 2013. Multilocus phylogeny and cryptic diversity in Asian shrew-like moles (Uropsilus, Talpidae): implications for taxonomy and conservation. BMC Evol Biol. 13:232.
- Wang Q, Fu CK, Chen SD, Yong B, Chen GY, Zong H. 2016. The complete mitogenome of Asiatic Short-tailed Shrew Blarinella quadraticauda (Soricidae). Mitochondrial DNA Part A. 27:282–283.
