Abstract
Background and Objective: Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most common organisms isolated from clinical samples, and has been associated with morbidity and mortality among hospitalized patients. The aim of this study was to evaluate the prevalence and antibiotic susceptibility patterns among MRSA and methicillin-sensitive S. aureus (MSSA) isolates collected from four hospitals in Iran. Material and Methods: A total of 183 isolates of S. aureus were collected from various clinical specimens of four hospitals in Iran. The isolates were identified by using the conventional biochemical tests. Three methods—oxacillin agar disk diffusion, oxacillin agar screening, and PCR— were applied to determine susceptibility to oxacillin. The conventional disk agar diffusion test was used to evaluate the antibiotic sensitivity of our isolates against 15 antibiotics, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Results: Of 183 isolates, 77 isolates (42.1%) were found to be MRSA, by the PCR method. The highest antibiotic resistance was found to be against penicillin, co-trimoxazole, erythromycin, and tetracycline respectively. All isolates were susceptible to vancomycin, according to the results of disk agar diffusion. Among other antibiotics, teicoplanin (84%) and fusidic acid (80.5%) were more active against MRSA isolates. For the different methods evaluated, the sensitivities and specificities were as follows: for disk agar diffusion (84.9% and 95.9%) and for agar screening test with oxacillin concentrations of 0.6 μg/ml (70.8% and 97.4%), 4 μg/ml (96.1%and 97.2%) and 6 μg/ml (96% and 96.3%), respectively. Conclusion: The results of our study showed that 47% of S. aureus isolates were MRSA. Overall, in this research study, resistance to all test antimicrobial agents in MRSA isolates were higher than that of MSSA isolates. Our results also revealed that 85% of mecA-positive isolates and 15% of mecA-negative isolates were resistant to methicillin; while 96% of mecA-negative isolates were sensitive to methicillin. Meanwhile 4% of mecA-positive isolates were also sensitive to methicillin.
Introduction
Staphylococcus aureus is an opportunistic pathogen often carried asymptomatically on the human body, and is a major cause of various community- and hospital-acquired infections that lead to a broad spectrum of diseases, ranging from diseases of the skin and soft tissues, surgical infections, to endocarditis and fatal pneumonia (CitationCasey et al. 2007, CitationIppolito et al. 2010, CitationAkpaka et al. 2011). Hospital-acquired methicillin-resistant S. aureus (HA-MRSA) strains are no longer limited to hospitals, but have now spread to other healthcare settings such as long-term care facilities (LTCFs) (CitationFuruno et al. 2008). Community-acquired MRSA (CA-MRSA) strains have also been associated with a variety of clinical manifestations, ranging from mild skin infections to lethal pneumonia and sepsis. Like methicillin-sensitive S. aureus (MSSA) clinical isolates in the community, the majority of CA-MRSA clinical isolates are recovered from skin or soft tissues (CitationMaltezou and Giamarellou 2006). This strain of bacteria has the considerable ability to adapt to different antibiotics, and now, with the emergence of multi-drug resistant (MDR) bacteria, S. aureus is a concern in the treatment of infectious diseases (CitationAkpaka et al. 2011, CitationDiep et al. 2008). MRSA includes those strains that have acquired the gene mecA for methicillin resistance and essentially for all other beta-lactam antibiotics (CitationFitzgerald et al. 2001, CitationLee 2003). MRSAs were initially isolated in healthcare facilities, for example, from patients in hospital intensive care units, nursing homes, and other chronic care facilities (CitationSaïd-Salim et al. 2003). MRSA organisms are commonly resistant to antibacterial agents such as cephalosporins, aminoglycosides, macrolides, tetracyclines, fluoroquinolones, chloramphenicol, clindamycin, trimethoprim/sulfamethoxazole, beta-lactamases, heavy metals, and antiseptics, because they have acquired the gene mecA, thereby making antibiotic resistance a global problem in public health, with considerable economic and social costs (CitationCapitano et al. 2003, CitationCosgrove et al. 2005, CitationMuto et al. 2003) . In the recent years, the widespread use of antibiotics has increased MRSA and led to the emergence of strains that have acquired multiple resistance genes(CitationStefani and Varaldo 2003). Many of these MRSA isolates are becoming multidrug-resistant and are susceptible only to glycopeptide antibiotics such as vancomycin and teicoplanin (CitationQureshi et al. 2004). Therefore, awareness of the prevalence of MRSA and their present antimicrobial profile becomes necessary in the selection of convenient empirical treatment methods for these infections. The purpose of this study was to evaluate the prevalence and antibiotic resistance patterns among MRSA and MSSA isolates collected from 4 hospitals in Iran.
Materials and methods
Isolation and identification of bacterial strains
A total of 183 S. aureus isolates were collected from various clinical specimens including the blood, urine, wound, body fluids, and sputum. The specimens were collected from 4 different hospitals, namely Tehran (Children’s Hospital), Tabriz (Imam Reza and Aalinasab Hospital), and Shabestar (Fatemiyyeh Hospital).
Identification of bacterial isolates was confirmed by using conventional tests such as gram staining, catalase production, growth on mannitol salt agar, DNase production, and coagulase test (CitationKateete et al. 2010). In this study, S. aureus ATCC 29213 (oxacillin-susceptible) and S. aureus ATCC 33591 (oxacillin-resistant) were used as negative and positive controls, respectively.
Antimicrobial Susceptibility Testing
Susceptibility of S. aureus isolates to oxacillin (1 μg/disk), penicillin (5 μg/disk), erythromycin (15 μg/disk), clindamycin (2 μg/disk), cefazolin (30 μg/disk), rifampicin (5 μg/disk), ceftriaxone(30 μg/disk), sulphamethoxazole/trimethoprim (1.25/23.75 μg/disk), tetracycline (30 μg/disk), fusidic acid (10 μg/disk), ciprofloxacin (30 μg/disk), vancomycin(30 μg/disk), teicoplanin (30 μg/disk), amikacin (30 μg/disk), and gentamicin (10 μg/disk) (MAST Group) was determined by the disc diffusion method, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (CitationWayne 2008).
Oxacillin agar screening
All isolates were placed on Mueller-Hinton agar (Conda) supplemented with 4% (w/v) NaCl comprising oxacillin (Sigma) at concentrations of 0.6, 4, and 6 μg/ml. The plates were inoculated with a swab dipped into a 0.5 McFarland standard suspension of each isolate, according to the procedures outlined in the CLSI guidelines for S. aureus (CitationShrestha et al. 2009). Oxacillin resistance was confirmed by bacterial growth after 24 and 48 h of incubation at 35°C. Moreover, for quality control in this experiment, the standard strains, S. aureus ATCC 33591 (MRSA) and S. aureus ATCC 29213 (MSSA), were used for the positive and negative controls, respectively.
DNA Extraction
Total DNA was extracted from individual colonies after growth in nutrient agar plates for 24 h at 37°C, following the protocol described by Sambrook (CitationSambrook et al. 2001). The loopful of bacteria was added to 1.5 ml of sterile distilled water, mixed gently, and was centrifuged at 1000 × g for 10 min. The supernatant was discarded, and 270 μl of T/E buffer, along with 30 μl of SDS 10% and 5 μl of proteinase K were added to the microtube and then incubated for 1 h at 65°C. Following this, 100 μl of 5 M NaCl solution was added to the microtube and mixed well. Next, 80 μl of prewarmed CTAB/NaCl solution (65°C) was added to the microtube and was incubated at 65°C for 10 min. Next, 700 μl of chloroform/isoamyl alcohol (24/1) solution was added to the microtube and vortexed for 20 s. The suspension was centrifuged at 12 000 × g for 5–10 min at 10°C, and the aqueous phase was transferred into a new microtube. Then, 200–300 μl of isopropanol was added to each microtube, mixed gently, and incubated at − 20°C for 30 min, before finally being centrifuged at 12 000 g for 10 min. The supernatant was discarded and the pellet was resuspended in 1 ml of 70% cold ethanol, and then centrifuged at 12 000 × g for 5 min at 10°C. The supernatant was discarded, the DNA pellet was air-dried, and was then dissolved in 50 μl of T/E (10/1) buffer and incubated at 37°C for 30 min, then stored at 4°C overnight. Then, the DNA extract was stored at −20°C.
Detection of the mecA gene by the PCR method
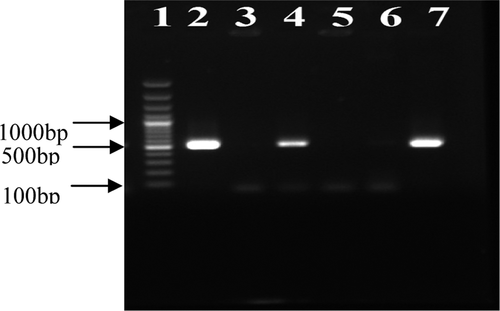
The mecA gene was detected by PCR with specific primers (mecF 5’-AAAATCGATGGTAAAGGTTGGC-3’ and mecR 5’-AGTTCTGCAGTACCGGATTTGC-3’) which were selected on the basis of a previous study that published the nucleotide sequence (CitationPetinaki et al. 2001). Briefly, the PCR reaction was prepared in 50 μl of a reaction mixture containing 1.5 mmol/L of MgCl2, 10 mmol/L of Tris-HCl, 25 pmol of each primer, 0.2 mmol/L of each deoxynucleotide, and 2.5 units of Taq polymerase (CinnaGen, Iran). The PCR cycles for the isolates were as follows: one cycle at 94°C for 3 min, followed by 30 cycles at 94°C for 30 sec, at 55°C for 30 sec, at 72°C for 30 sec, and finally at 72°C for 3 min. For analysis of the mecA gene, these primers gave rise to a PCR product of 533 bp, in all strains. The PCR product was detected on 2% agarose gel electrophoresis in a 1X (TAE) buffer and visualized by ethidium bromide staining. The 100 bp plus DNA ladder (Fermentas Company, Iran) was used as the DNA molecular weight standard.
Results
In our study of 183 S. aureus strains, 91 strains were isolated from females and 92 strains from males. The number of S. aureus isolates which were found to be MRSA by the oxacillin disk diffusion method was recorded as 86 (47%). The results of the oxacillin agar screening test with three concentrations of oxacillin (0.6 μg/ml, 4 μg/ml, and 6 μg/ml) are shown in the . The mecA gene was also detected in 77 isolates (42.1%) by using the PCR method ().
Table I. Results of the oxacillin susceptibility patterns and mecA gene in S. aureus isolates.
Lane 1: 100–3000 bp DNA ladder, Lane 2: positive control (S. aureus ATCC 33591), Lane 3: negative control (S. aureus ATCC 29213), Lanes 4 and 7: clinical isolates of S. aureus mecA-positive, Lanes 5 and 6: clinical isolates of S. aureus mecA-negative.
shows the bacterial isolates from different clinical specimens. As shown in this table, methicillin resistance in S. aureus isolated from sputum samples was higher than in the other samples, while methicillin resistance in isolates collected from wounds was less than that in the other samples. Moreover, in this study, the prevalence of MRSA among different hospitals was recorded as: 65%, 57%, 43%, and 40% for Alinasab, Fatemiyyeh, Imam Reza, and Pediatrics hospital, respectively. Considerable differences were also observed when the distributions of MRSA isolates in outpatients (5.2%) and inpatients (94.85%) were compared (p < 0.05).
Table II. S. aureus isolated from various clinical specimens.
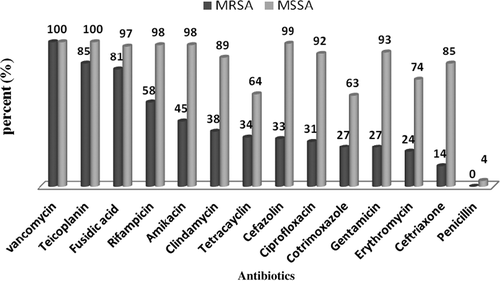
The antibacterial susceptibility patterns of both MRSA and MSSA isolates are shown in . Overall, in this research, resistance to all antimicrobial agents was higher in MRSA isolates than in the MSSA isolates.
The results of our study showed that all S. aureus isolates were susceptible to vancomycin, and no vancomycin-resistant isolates were observed by the disk agar diffusion test, while almost all of the isolates were resistant to penicillin. According to the comparison of methicillin resistance patterns among mecA-positive and mecA-negative isolates, 85% of the mecA-positive isolates and 15% of the mecA-negative isolates were resistant to methicillin, while, 96% of the mecA-negative isolates were susceptible to methicillin and only 4% of the mecA-positive strains were susceptible to it.
In this study, we found several cases of multiple resistance, as 64 isolates were resistant to more than 10 antibiotics and 7 isolates were resistant to less than 5 antimicrobial agents simultaneously. We compared the frequency of MRSA strains isolated from different hospitals (). The MRSA strains were isolated with a high prevalence from Alinasab hospital and low prevalence from the Pediatric hospital.
Table III. Studies of antibiotic resistance in S. aureus isolates obtained from 4 hospitals.
Discussion
Over the last decade, MRSA strains have emerged as a serious threat to public health worldwide. Hospitalization costs related with MRSA infections are significantly greater than those related with MSSA infections, and MRSA has wider economic effects that involve indirect costs to the patient and to society (CitationIppolito et al. 2010). Moreover, there is some evidence showing that MRSA infections increase morbidity and the risk of mortality. In the recent study, we showed that the frequency of MRSA is 42.1%. Our findings are similar to other reports from India and Iran, as the prevalence of MRSA in these areas have been reported as 44.9% and 46.3%, respectively (CitationShrestha et al. 2009, CitationDibah et al. 2014).
However, some other studies have reported low prevalence for MRSA (29.7%). This variation in different geographical regions of Iran and other countries might be due to factors such as different patterns of drug usage, and different health care programs (CitationRahimi et al. 2013). Differences in the prevalence of MRSA in different areas emphasize the importance of local inspections in reporting local resistance, which can be used in empirical therapies.
We compared different phenotypic methods to evaluate the oxacillin susceptibilities of S. aureus. In this study, the agar screening test with an oxacillin concentration of 0.6 μg/ml showed the lowest sensitivity (70.8%), and the disk diffusion method showed a sensitivity of 84.9% (). We found four mecA-positive isolates to be susceptible and 13 mecA-negative isolates to be resistant to oxacillin by disk agar diffusion. This can be associated with the heteroresistance of our isolates to oxacillin, and the absence or presence of mecA gene expression in these isolates (CitationPlata et al. 2011).
In the current study, we found a high rate of resistance to ceftriaxone, erythromycin, co-trimoxazole, gentamicin, tetracycline, cefazolin, ciprofloxacin, and clindamycin in MRSA isolates. This may be explained by higher usage of these antibiotics. In the study conducted by Rahimi et al., the rates of resistance to ciprofloxacin, erythromycin, amikacin, tetracycline, clindamycin, fusidic acid, and vancomycin were 95%, 93%, 84%, 83%, 75%, 3% and 0% respectively (CitationRahimi et al. 2013).
This research indicated that resistance to fusidic acid and teicoplanin was low, and that they were the most effective antibiotics against MRSA isolates. This may be due to the low consumption of these antibiotics in Iran. Although fusidic acid and teicoplanin are the most effective antibiotics against MRSA isolates, the high price of these drugs limits their consumption.
Vancomycin is drug of choice for treatment of MRSA infections. The appearance and spread of vancomycin resistance is a significant threat to public health. Therefore, it is necessary to carefully monitor the prevalence of vancomycin-resistant S. aureus, especially in MRSA populations.
In conclusion, the results of this study have provided baseline information in assisting physicians, clinical microbiologists, and public health officials on critical issues regarding empirical and pathogen-specific therapy. Our study showed that 47% of S. aureus isolated in Iranian hospitals during a study period were MRSA. Accurate diagnosis of MRSA strains in patients and health care workers is necessary, and the distribution, in Iranian hospitals, of MRSA strains with a high rate of resistance to different antibiotics can be efficiently treated. Therefore, continuous monitoring and surveillance of antibiotic-resistant S. aureus is recommended.
Acknowledgements
We would like to thank the laboratory staff of the hospitals from which bacterial isolates were collected.
Declaration of interest
The authors report no declarations of interest. The authors alone are responsible for the content and writing of the paper.
References
- Akpaka PE, Monecke S, Swanston WH, Rao A, Schulz R, Levett PN. 2011. Methicillin sensitive Staphylococcus aureus producing Panton-Valentine leukocidin toxin in Trinidad & Tobago: a case report. J Med Case Rep. 5:157.
- Capitano B, Leshem OA, Nightingale CH, Nicolau DP. 2003. Cost effect of managing methicillin-resistant Staphylococcus aureus in a long- term care facility. J Am Geriatr Soc. 51:10–16.
- Casey A, Lambert PA, Elliott T. 2007. Staphylococci. Int J Antimicrob Agents. 29:S23–S32.
- Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. 2005. The Impact of Methicillin Resistance in Staphylococcus Aureus Bacteremia on Patient Outcomes Mortality, Length of Stay, and Hospital Charges. Infect Control Hosp Epidemiol. 26:166–174.
- Dibah S, Arzanlou M, Jannati E, Shapouri R. 2014. Prevalence and antimicrobial resistance pattern of methicillin resistant Staphylococcus aureus (MRSA) strains isolated from clinical specimens in Ardabil, Iran. Iran J Microbiol. 6:163–168.
- Diep BA, Chambers HF, Graber CJ, Szumowski JD, Miller LG, Han LL, et al. 2008. Emergence of multidrug-resistant, community- associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med. 148:249–257.
- Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci USA. 98:8821–8826.
- Furuno JP, Hebden JN, Standiford HC, Perencevich EN, Miller RR, Moore AC, et al. 2008. Prevalence of methicillin-resistant Staphylococcus aureus and Acinetobacter baumannii in a long-term acute care facility. Am J Infect Control. 36:468–471.
- Ippolito G, Leone S, Lauria FN, Nicastri E, Wenzel RP. 2010. Methicillin-resistant Staphylococcus aureus: the superbug. Int J Infect Dis. 14:S7–S11.
- Kateete DP, Kimani CN, Katabazi FA, Okeng A, Okee MS, Nanteza A, et al. 2010. Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann Clin Microbiol Antimicrob. 9:23.
- Lee JH. 2003. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 69:6489–6494.
- Maltezou HC, Giamarellou H. 2006. Community-acquired methicillin-resistant Staphylococcus aureus infections. Int J Antimicrob Agents. 27:87–96.
- Muto CA, Jernigan JA, Ostrowsky BE, Richet HM, Jarvis WR, Boyce JM, et al. 2003. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect Control Hosp Epidemiol. 24:362–386.
- Petinaki E, Rvaniti A, Dimitracopoulos G, Spiliopoulou I. 2001. Detection of mecA, mecR1 and mecI genes among clinical isolates of methicillin-resistant staphylococci by combined polymerase chain reactions. J Antimicrob Chemother. 47:297–304.
- Plata KB, Rosato RR, Rosato AE. 2011. Fate of mutation rate depends on agr locus expression during oxacillin-mediated heterogeneous-homogeneous selection in methicillin-resistant Staphylococcus aureus clinical strains. Antimicrob Agents Chemother. 55: 3176–3186.
- Qureshi A, Rafi S, Qureshi S, Ali A. 2004. The current susceptibility patterns of methicillin resistant Staphylococcus aureus to conventional anti Staphylococcus antimicrobials at Rawalpindi. Pak J Med Sci. 20:361–364.
- Rahimi F, Bouzari M, Katouli M, Pourshafie MR. 2013. Antibiotic resistance pattern of methicillin resistant and methicillin sensitive Staphylococcus aureus isolates in Tehran, Iran. Jundishapur J Microbiol. 6:144–149.
- Saïd-Salim B, Mathema B, Kreiswirth BN. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect Control Hosp Epidemiol. 24:451–455.
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 2001. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- Shrestha B, Pokhrel BM, Mohapatra TM. 2009. Phenotypic characterization of nosocomial isolates of Staphylococcus aureus with reference to MRSA. J Infect Dev Ctries. 3:554–560.
- Stefani S, Varaldo P. 2003. Epidemiology of methicillin-resistant staphylococci in Europe. Clin Microbiol Infect. 9:1179–1186.
- Wayne P. 2008. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement NCCLS document M100-S9 National Committee for Clinical Laboratory Standards.