Abstract
In this research, bone marrow mesenchymal stem cells (BMSCs) were isolated from mouse, and induced differentiation into myocardial cells in vitro after overexpression of miR-1a. The results showed that the BMSCs could induce differentiation into myocardial cells under the special condition medium, but when the miR-1a was over-expressed in BMSCs, the differentiation efficiency and induction time of myocardial cells from BMSCs could be promoted. This reason was demonstrated that Delta-like 1 (Dll-1) was a transcriptional repressor of myocardium gene expression during myocardium differentiation, miR-1a reduced Dll-1 levels, leading to the accumulation of myocardium gene mRNA and a dramatic increase in myocardium gene protein.
Introduction
The heart is the first organ to form in the embryo, and all subsequent events in the life of the organism depend on its uninterrupted contractility, and the heart is one of the least regenerative organs, loss of myocardium to infarction or other diseases often lead to heart failure (Laflamme et al. Citation2007, White et al. Citation1936). A novel emerging treatment for heart failure is cellular cardiomyoplasty whereby stem cells are delivered to the dysfunctional myocardium (Mishra et al. Citation2011, Yi et al. Citation2010). Stem cells offer the possibility of repairing damaged organs such as the heart from their component parts, and there is an intensive effort to develop stem cell-based strategies for cardiac repair. Mesenchymal stem cells (MSCs) are promising candidates for use in cellar therapies and tissue engineering repair. They are an adult stem cell population and derive their name from an intrinsic ability to differentiate into multiple cell lineages of mesodermal origin such as bone marrow. Bone marrow mesenchymal stem cells (BMSCs) are easy to culture and manipulate in vitro and have great plasticity, thus they have become an important tool in cell replacement therapy and are presently considered as candidates for different clinical applications.
Genes encoding microRNAs (miRNAs), which are found in most eukaryotes, produce short (18∼25 nt) RNAs that bind to mRNA transcripts and downregulate their expression either through mRNA destabilization or translational repression (Bartel Citation2009, Shukla et al. Citation2011). Therefore, miRNAs are post-transcriptional regulators of gene expression that have been shown to be central players in the establishment of cellular programs, often acting as switches that control the choice between proliferation and differentiation during development of organs (Gama-Carvalho et al. Citation2014). miR-1a is a special microRNA for development of muscle, including heart and skeletal muscle cells (Kwon et al. Citation2005, Takaya et al. Citation2012). In this research, miR-1a was transfected into BMSCs for differentiation of myocardial cells to research the functions in differentiation programs.
Materials and methods
Isolation and culture of mouse BMSCs
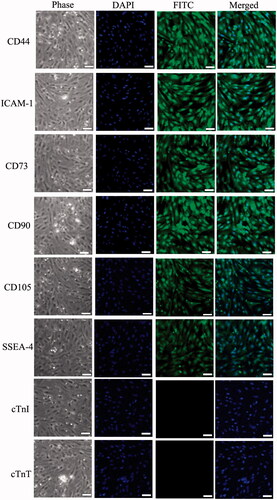
BMSCs were isolated from the bone marrow of BALB/c mouse, supplied by the Beijing HFK Bioscience CO. LTD, Beijing, PR China. The mice were sacrificed with a lethal dose of sodium pentobarbital. First, bone marrow samples were drained from the tibia of mice, and pelleted by centrifugation, then resuspended with DMEM medium and fractionated on density gradient (Ficoll, 1.077 g/ml) for 30 min at 400 g. The interface was collected and seeded at 1 × 104 cells/ml in 6-well culture plate with 1 ml medium containing l-DMEM (Gibco, USA) + 10% (v:v) fetal bovine serum (FBS) (Gibco, USA) + 10 U/ml penicillin + 10 μg/ml streptomycin. Cultures were maintained at 37.5°C in a humidified atmosphere containing 95% air and 5% CO2. The culture medium was changed every 3 days. The special markers of BMSCs were tested using Immunofluorescence, including CD44 (1:500, Santa Cruz, USA), ICAM-1 (1:500, Santa Cruz, USA), CD73 (1:500, Santa Cruz, USA), CD90 (1:500, Santa Cruz, USA), CD105 (1:500, Santa Cruz, USA), SSEA-4 (1:500, Santa Cruz, USA), CTnI (1:200, Abcam, USA) and CTnT (1:200, Abcam, USA).
Expression of Pre-miR-1a by recombinant Lentivirus
The sequence of pre-miR-1a was synthesized by Sangon Biotech (Shanghai, China), cloned into lentiviral vector and then used to recombine lentivirus in HEK293F cells. The lentivirus designated as pCMV-SM30 also express enhanced green fluorescent protein (eGFP) as a marker for monitoring infection efficiency. Analogous lentivirus only expressing monomeric eGFP was used as a control. Real-time polymerase chain reaction (PCR) was used to evaluate the expression level of miR-1a after virus infection. miRNAs were isolated from cells using microRNA isolation kit (Applied Biosystems, Carlsbad, CA, US) according to the manufacturer’s instructions. cDNA synthesis was carried out with the High Capacity cDNA synthesis kit (Applied Biosystems, Carlsbad, CA, US) using 2 ng of RNA as template. The miRNA sequence-specific reverse transcription PCR primers for miR-1a and endogenous control U6 were purchased from Ambion (Ambion, Carlsbad, CA, US) () . Real-time PCR analysis was carried out using Applied Biosystems 7500 real-time PCR system. The gene expression threshold cycle (CT) values of miRNAs from each sample were calculated by normalizing with internal control U6 and relative quantitation values were plotted. And the targets of miR-1a were analyzed in BMSCs by Western Blotting.
Table I. Primer sequences used in real-time PCR assay.
Myocardial cells differentiation
BMSCs were plated and divided into four groups, the group A was incubated in serum-free cardiomyogenic medium containing 10 nM 5-Azacytidine (5-aza; Sigma) for 24 h, and then the medium was replaced with complete medium. The group B was also incubated in serum-free cardiomyogenic medium containing 10 nM 5-aza, but the miR-1a was overexpressed in cells before incubation of 5-aza. The group C was removed incubation of 5-aza from group B, group D as a control. The cells were harvested and the RNA from the four groups was extracted for further real-time PCR assays in every 2 days. The special markers of myocardial cells were analyzed in this research, including Cardiac Troponin I (cTnI), Cardiac Troponin T (cTnT) and Myosin, Heavy Chain 6 (MYH6).
Western blotting
The target genes of miR-1a were detected by western blotting. Cells were lysed using M-PER Protein Extraction Reagent (Pierce, USA) supplemented with protease inhibitor cocktail (DMSF). Protein concentrations of the extracts were measured with BCA assay (Pierce, Carlsbad, CA, US) and equalized with the extraction reagent. Equal amount of the extracts were loaded and subjected to SDS-PAGE, transferred onto nitrocellulose membranes, and then blotted as previously reported [15]. Specific antibodies and horseradish peroxidase-coupled secondary antibodies were purchased from Santa Cruz, USA. Membranes were probed using ultraenhanced chemiluminescence western blotting detection reagents. GAPDH was used as internal control.
Statistical analysis
Statistical analyses of the data were performed with a one-way ANOVA followed by the Tukey–Kramer honestly significant difference (HSD) test for the three sets of results. A p-value of <0.05 was considered significant. Statistical analyses were done with a JMPW Statistical Discovery Software (SAS Institute, Cary, NC).
Results
Characteristic of mouse BMSCs
The mouse BMSCs were cultured and proliferated in vitro from tibia, and the BMSCs were large, lucent and with strong refraction. Nuclei were oval and mixed with some blood cells. The nonadherent cells were removed on the third day and MSCs were fusiform and showed cell-like clone with growth being slower. Four days later, MSCs proliferated rapidly. These cells were maintained in culture for more than 100 days with no sign of senescence or differentiation. Immunofluorescence staining results showed that different passages of BMSCs expressed antigens CD44, ICAM-1, CD73, CD90, CD105 and SSEA-4, but did not express antigens cTnI and cTnT. cTnI is a cardiac troponin I expressed on the myofilaments, and cTnT is one of the cardiac troponins and expressed on the sarcoplasmic reticulum (). There was no significant difference in the positive rates of different passages (p > 0.05).
Function analyses of miR-1a in BMSCs
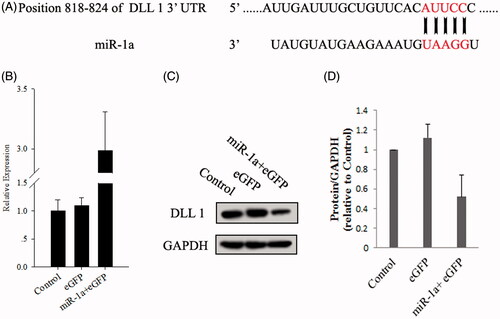
The expression level of miR-1a in BMSCs was quantified by real-time PCR after transfection 72 h. As shown in , miR-1a levels were significantly elevated after transfection. The target genes of miR-1a were predicted and analyzed using target Scan 6.2 (http://www.targetscan.org/) and previously report (Kwon et al. Citation2005), the miR-1a sequence and its target gene sequence, confirmed that DLL 1 was target genes for miR-1a. showed that miR-1a targeted DLL 1 at the 3′-UTR in mouse BMSCs to regulate expression (). Protein expression of DLL 1, putative target genes was performed on miR-1a transfected cells according to Image J tools comparative method.
Figure 2. The target site of miRNA-1a and up-regulation of miR-1a in pre-miR-1a-transfected BMMSCs. (A) The miR-1a complementary sites with 3′-UTR of DLL 1. (B) BMMSCs were transfected with adenoviruses as described in the Methods, and the expression of miR-1a was quantified by real-time PCR. (C) Effect of miR-1a on protein level of DLL 1. (D) Quantification of DLL 1 in BMMSCs transfected with miR-1a, eGFP or control for 72 h.
Myocardial cells differentiation
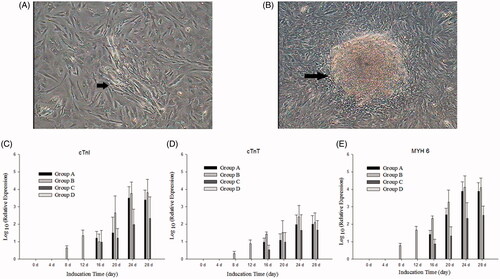
For differentiation of myocardial cells, BMSCs were divided into four groups. In group A, after incubation in cardiomyogenic medium, the cells polymerized to form myotubules, and the myotubules increased and fused to form fascicle. Autopulse of the myotubes was observed after about 28 days (date not show). The real-time PCR showed that the special genes of myocardial cells were expressed after 18 days. In group B, after expression of miR-1a, the cells were observed the changes in morphology after 10 days; the real-time PCR showed that the special genes of myocardial cells were expressed after 8 days. Autopulse of the myotubes was found after 18 days. In group C, the morphology of cells were not significant change, but the special genes of myocardial cells were expressed after 16 days, and autopulse of the myotubes were not found after about 50 days. And there were no obvious morphological changes in the control group (group D).
Discussion
Cardiomyocytes proliferate and form the heart in the embryonic period, but proliferation stops soon after birth. Therefore, it is difficult to cure heart failure, particularly in cases accompanied by myocardial injury, such as myocardial infarction and cardiomyopathy. Drug therapy for heart failure using β-blockers and angiotensin-converting enzyme inhibitors improves the short-term prognosis, but the 5-year survival rate of patients with severe heart failure is <50% (Takaya et al. Citation2012). To overcome this situation, regenerative therapy has been investigated worldwide wherein cardiovascular cell differentiation of stem cells has been induced and applied to supplement and regenerate the affected regions. The therapeutic application of various types of cells, including progenitor cells derived from adult bone marrow or adipose tissue has been shown to have cardioprotective effects in experimental models of heart failure and in some clinical trials (Dimmeler et al. Citation2014, Matsa et al. Citation2014). However, the establishment of successful cell therapies is challenging owing to poor homing and survival of the implanted cells, and a limited cardiac differentiation capacity of most types of adult progenitor cells. miRNAs have been shown to modulate such processes, and might be used to enhance the therapeutic benefits of cell therapy. The most important microRNAs for cardiogenesis are the miR-1/133 family specifically expressed at a high level in heart and skeletal muscle cells. However, miR-1 and miR-133 had opposing effects on further adoption of muscle lineages, with miR-1 promoting and miR-133 blocking differentiation into either cardiac or skeletal muscle fates. Delta-like 1 (Dll-1), a Notch ligand expressed in ES cells, was translationally repressed in miR-1-expressing ES cells, and depletion of Dll-1 from ES cells resulted in a bias toward the cardiac lineage while suppressing endoderm and neuroectoderm differentiation, similar to miR-1-expressing ES cells (Ivey et al. Citation2008). miR-1 is encoded by two genes (miR-1a and miR-1b), and each gene is simultaneously transcribed with one of two miR-133a genes (Chen et al. Citation2006). In cardiogenesis, miR-1 is considered to control the balance between the differentiation and proliferation of cardiomyocyte precursors through Hand2. miR-1 targets Hand2 to negatively control cardiogenesis, in addition to forming a feedback loop through targeting HDAC 4 to inhibit downstream MEF2 (Liu and Olson Citation2010). But miR-1 promoted myocardial differentiation in stem cells by targeting Notch ligand Delta-like 1 (Ivey et al. Citation2008).
5-Azacytidine (5-aza), an analogue of cytidine, causes hypomethylation of some cytosine in DNA which may be involved in activating phenotype-specific genes. Additionally, it was a conventional inducer for differentiation of myocardial cells from stem cells (Liu et al. Citation2003), but the molecular mechanism was not clear. Previous research reported that the optimal concentration for cardiomyogenic differentiation is 10 mM for 24 h, and at 4 weeks after induction, myotube-like structures were formed (). The changes in morphology with the characteristic appearance of “stick-like” cells may be associated with the expression of proteins maintaining cytoskeleton (Antonitsis et al. Citation2007). Therefore, BMSCs were treated with the demethylating agent 5-aza to induce cardiomyogenic differentiation in group A of our study. Although the group A were obtained autopulse of the myotubes, the group B were cut down induced time when miR-1a were transfected into BMSCs. miR-1a strongly enhanced cardiomyogenic differentiation combining with 5-aza, as indicated by an increase in expression of the cardiomyogenic markers cTnI, cTnT and MYH 6, and decrease in induced time. However, the miR-1a was transfected into BMSCs without incubation of 5-aza in group C, although the special genes of myocardial cells were positive, but the autopulse of the myotubes were also not observed after 50 days of transfection. Therefore, the overexpression of miR-1a combining with incubation of 5-aza is the best method for myocardial cells from BMSCs.
Figure 3. Cardiomyogenic differentiation of BMMSCs. (A) The cells polymerized to form myotubules after culture in group B for 12 days (arrow). (B) About 18 days later, the myotubules increased and fused to form fascicles (arrow). (C–E) Myocyte-specific genes – cTnI, cTnT and MYH 6 – were expressed after incubation in different induced groups for different induction days, whereas these genes were not detected in control cells (group D). Myocyte-specific genes showed a time-lapse increase in group A, group B and group C.
In conclusion, our research shows that the miR-1a promoted the differentiation of myocardial cells from BMSCs, and this mechanism was miR-1a reduced the expression of Dll-1 and increased the expression of myocardium gene.
Declaration of interest
The authors report no declarations of interest. The authors alone are responsible for the content and writing of the paper.
References
- Antonitsis P, Ioannidou-Papagiannaki E, Kaidoglou A, Papakonstantinou C. 2007. In vitro cardiomyogenic differentiation of adult human bone marrow mesenchymal stem cells. The role of 5-azacytidine. Interact Cardiovasc Thoracic Surg. 6:593–597.
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233.
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genet. 38:228–233.
- Dimmeler S, Ding S, Rando TA, Trounson A. 2014. Translational strategies and challenges in regenerative medicine. Nat Med. 20:814–821.
- Gama-Carvalho M, Andrade J, Bras-Rosario L. 2014. Regulation of cardiac cell fate by microRNAs: implications for heart regeneration. Cells. 3:996–1026.
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, et al. 2008. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2:219–229.
- Kwon C, Han Z, Olson EN, Srivastava D. 2005. MicroRNA1 influences cardiac differentiation in Drosophila and regulates notch signaling. Proc Natl Acad Sci USA 102:18986–18991.
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, et al. 2007. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 25:1015–1024.
- Liu N, Olson EN. 2010. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 18:510–525.
- Liu Y, Song J, Liu W, Wan Y, Chen X, Hu C. 2003. Growth and differentiation of rat bone marrow stromal cells: does 5-azacytidine trigger their cardiomyogenic differentiation? Cardiovasc Res. 58:460–468.
- Matsa E, Sallam K, Wu JC. 2014. Cardiac stem cell biology: glimpse of the past, present, and future. Circ Res. 114:21–27.
- Mishra R, Vijayan K, Colletti EJ, Harrington DA, Matthiesen TS, Simpson D, et al. 2011. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 123:364–373.
- Shukla GC, Singh J, Barik S. 2011. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 3:83–92.
- Takaya T, Nishi H, Horie T, Ono K, Hasegawa K. 2012. Roles of microRNAs and myocardial cell differentiation. Prog Mol Biol Transl Sci. 111:139–152.
- White PD, Mallory GK, Salcedo-Salgar J. 1936. The speed of healing of myocardial infarcts. Trans Am Clin Climatol Assoc. 52:97–104.
- Yi BA, Wernet O, Chien KR. 2010. Pregenerative medicine: developmental paradigms in the biology of cardiovascular regeneration. J Clin Invest. 120:20–28.