Abstract
Objectives:
To estimate the clinical and economic trade-offs involved in using a molecular assay (92-gene assay, CancerTYPE ID) to aid in identifying the primary site of difficult-to-diagnose metastatic cancers and to explore whether the 92-gene assay can be used to standardize the diagnostic process and costs for clinicians, patients, and payers.
Methods:
Four decision-analytic models were developed to project the lifetime clinical and economic impact of incorporating the 92-gene assay compared with standard care alone. For each model, total and incremental costs, life-years, quality-adjusted life-years (QALYs), incremental cost–effectiveness ratios (ICERs), and the proportion of patients treated correctly versus incorrectly were projected from the payer perspective. Model inputs were based on published literature, analyses of SEER (Surveillance Epidemiology and End Results) data, publicly available data, and interviews with clinical experts.
Results:
In all four models, the 92-gene assay increased the proportion of patients treated correctly, decreased the proportion of patients treated with empiric therapy, and increased quality-adjusted survival. In the primary model, the ICER was $50,273/QALY; thus, the 92-gene assay is therefore cost effective when considering a societal willingness-to-pay threshold of $100,000/QALY. These findings were robust across sensitivity analyses.
Conclusions:
Use of the 92-gene assay for diagnosing metastatic tumors of uncertain origin is associated with reduced misdiagnoses, increased survival, and improved quality of life. Incorporating the assay into current practice is a cost-effective approach to standardizing diagnostic methods while improving patient care. Limitations of this analysis are the lack of data availability and resulting modeling simplifications, although sensitivity analyses showed these to not be key drivers of results.
Introduction
Despite recent advances in technology and diagnostic approaches, a significant number of new cancer patients present with metastatic cancers in which the primary site of tumor origin remains uncertain after a standard clinical and pathological workupCitation1–3. These cases are often associated with poorly differentiated or undifferentiated tumors, atypical clinical presentation, and/or limited tissue samplesCitation1. Although some patients are identified as having cancer of unknown primary, many others do not fit the definition of unknown primary but have differential diagnoses of two or more possible tumor types. Such diagnostic uncertainty impacts clinicians’ abilities to determine the optimal treatment regimen. Standard treatment for patients without definitive diagnoses varies widely, often relying on nonspecific, broadly acting, empiric chemotherapy regimens, and resulting in poor prognosisCitation1,Citation2. These patients are also at risk for misdiagnosisCitation4, which may lead to incorrect primary treatment approaches. Clinical practice guidelines recommend comprehensive diagnostic testing and clinical evaluation to identify the tumor site of origin and thereby inform the optimal treatment pathwayCitation3. Multiple studies have demonstrated that, compared with empiric approaches, therapeutic strategies based on accurate identification of the primary tumor type and on targeted molecular pathways can improve patient outcomesCitation5–8.
Clinical evaluation for metastatic cancer patients with uncertain diagnoses depends on clinical presentation and may include a wide range of complex procedures. Many such diagnostic procedures are invasive, may be burdensome, and pose significant risk to patients; furthermore, physicians use a wide variety of diagnostic techniques when evaluating patients with metastatic cancerCitation2,Citation9. The financial implications of such a diagnostic odyssey can be significant: one study reported an average cost of nearly $18,000 per patient, not including newer imaging technologies such as positron emission tomography scansCitation10. The benefits of using these procedures are unclear; for example, in a recent study assessing the use of colonoscopy to identify the primary tumor site in metastatic cancer patients, colon cancer was identified in two of 160 patients tested, and one patient died from procedure-related complicationsCitation11.
Gene expression profiling assays have been developed to aid clinicians in identifying the primary tumor type in cases that are difficult to diagnosis with traditional methodologiesCitation12–16. One such option, the 92-gene assay (CancerTYPE ID; bioTheranostics Inc., San Diego, CA, USA), measures the expression of 92 genes to classify 50 tumor types. In a large, blinded, multiinstitutional validation study, the 92-gene assay demonstrated 87% accuracy (95% CI: 84%–89%)Citation14, and in a blinded comparative effectiveness study the test demonstrated higher accuracy than standard-of-care pathologic review with immunohistochemistry (IHC)Citation17. Finally, in a large prospective clinical trial, previously untreated cancer patients with metastatic disease and an unknown primary site of origin were tested with the 92-gene assay and received site-specific treatment based on assay results. Median overall survival was significantly improved in patients receiving assay-directed, site-specific treatment compared with those treated with empiric chemotherapy regimens (12.5 versus 9.1 months, respectively)Citation8.
Although the clinical value of incorporating the 92-gene assay into the diagnostic pathway to help inform treatment planning has been demonstratedCitation8,Citation13,Citation14,Citation17,Citation18, the cost effectiveness of this approach compared with standard care methods has not previously been assessed. With increasing medical expenditures in the US, it is critical to know which diagnostic and treatment options offer clinical benefit at a reasonable cost compared with viable alternatives. In this analysis, we used decision-analytic methodology to estimate the clinical and economic trade-offs involved in using the 92-gene assay to aid in identifying the primary site of tumors of uncertain origin. We also explored whether the 92-gene assay could be used to standardize the diagnostic process and costs for clinicians, patients, and payers.
Methods
Overview
We developed four separate deterministic decision-analytic models to project, from the payer perspective, the effectiveness and cost of using the 92-gene assay compared with standard care methods to identify the primary tumor site among patients with metastatic cancer of uncertain origin. The models each evaluated a cohort of hypothetical patients whose diagnosis was based on an initial oncology consultation and a hematoxylin and eosin (H&E) stain. Clinical outcomes and cost implications were evaluated given patients’ progression through pathways of diagnostic procedures and eventual treatment. The models, including one primary model and three additional models, differed in their assumptions about current standard diagnostic methods and assessed the benefits and costs of introducing the 92-gene assay under each such interpretation compared with standard care methods. Our study did not involve human participants, and therefore did not require institutional review board approval.
We projected lifetime costs, life-years (LYs), quality-adjusted life-years (QALYs), incremental cost–effectiveness ratios (ICERs), and the percentage of patients treated with correct site-specific therapy. Data regarding cancer incidence and survival, proportions of patients receiving diagnostic procedures and cancer treatments, health-related quality of life, and costs were estimated based on published sources and expert opinionCitation1,Citation3,Citation6,Citation7,Citation18–36. Outcomes were undiscounted because of the short metastatic cancer survival duration of approximately 1 year. The models were developed using TreeAge Pro 2012 (TreeAge Software Inc., Williamstown, MA, USA), and costs were inflated to 2012 US dollars (USD) using the annual medical care inflation indexCitation37.
Structures
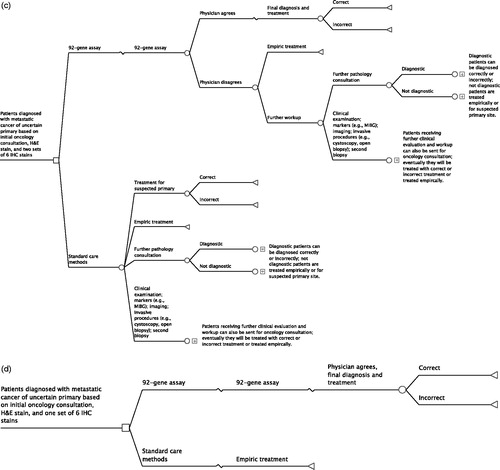
The primary model begins with patients with confirmed metastatic cancer with an uncertain site of tumor origin based on an initial oncology consultation, pathology review, hematoxylin and eosin (H&E) stained section, and an initial set of six IHC stains (). As patients progressed in the model through diagnosis, their experiences differed depending on the number of the following procedures that they received: pathology consultation, oncology consultation, or further clinical evaluation and workup; marker testing; imaging; invasive procedures; and 92-gene assay testing. All patients were eventually considered either ‘diagnosed’ (and were treated accordingly with site-specific therapies) or ‘undiagnosed’ (and were treated with site-specific therapy for a suspected primary site or with an empiric chemotherapy regimen). A portion of treatments was defined as ‘correct’, in which the site of origin was correctly identified and site-specific treatment was administered accordingly. Some patients receiving the 92-gene assay were treated according to the assay’s prediction, whereas others were not and instead continued to receive further diagnostic procedures.
Figure 1. The 92-gene assay model schematic. (a) Primary model. (b) Early model. (c) Late model. (d) Clinically simplified model. H&E, hematoxylin and eosin; IHC, immunohistochemistry; MIBG, iodine-131-meta-iodobenzylguanidine scintiscan.
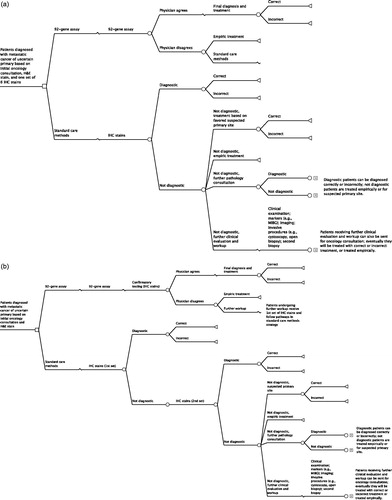
In addition to the primary model, three additional models were developed to assess alternative assumptions regarding the standard diagnostic methods. Two of these models evaluated incorporation of the 92-gene assay earlier and later in the diagnostic process. In the early model, the 92-gene assay was used before the first set of IHC stains in the patient population entering the model (). In the late model, the 92-gene assay was used after a second set of IHC stains in patients who were still undiagnosed at this time point (). Finally, a clinically simplified model was also developed. This model compared an approach using the 92-gene assay and treating immediately based on the assay result with an empiric chemotherapy treatment approach (). This also served as a validation exercise because these assumptions mimic the methods used in the prospective clinical trial of site-directed chemotherapy based on assay predictionCitation8.
Inputs
All models included patients with the following eight cancers, which comprised 80% of cancers in our patient population: breast; colon and rectum; kidney and renal pelvis; liver and intrahepatic bile duct; lung and bronchus; ovarian; pancreatic; and prostate cancerCitation6,Citation19. We analyzed Surveillance Epidemiology and End Results (SEER) data (SEER*Stat version 7.1.0) to calculate mean survival for each metastatic cancer and histologic subtypeCitation20. These estimates were adjusted for correct versus incorrect treatment based on published survival estimates associated with site-specific versus empiric therapyCitation1,Citation7 (Supplementary online appendix table 1). For patients receiving empiric treatment, survival (13.13 months) was estimated from published literature (Greco 2011Citation1). The proportion of cancer patients with one primary tumor site identified at different points along the diagnostic continuum was estimated based on published estimates and expert opinion ()Citation21. Additional workup increased the likelihood of correct treatment.
Table 1. The 92-gene assay model: event probabilities, costs, and utility weights.
Select model costs and utility estimates are shown in . The costs of IHC stains were estimated from the Physicians’ Fee and Coding Guide 2012Citation38. Use of pathology and oncology consultation was obtained from published estimatesCitation3,Citation18,Citation22,Citation23 and expert opinion, with related costs estimated from the Physicians’ Fee and Coding Guide 2012 and published studiesCitation24,Citation25,Citation38. The commercial list price was used for the 92-gene assay cost (bioTheranostics 2012). Costs of site-specific and empiric treatment were estimated from published literature ()Citation26,Citation27. These costs were based on Medicare data and consisted of all claims, including both inpatient and outpatient services. For site-specific therapy, we included separate costs for three time points: the first 12 months after diagnosis, the last 12 months of life, and all years in between.
Health-related quality of life was incorporated using utility weights associated with correct and incorrect treatment of each cancer; estimates were based on data from the published literature and constant across histologic subtypes (). Utility weights associated with stable, treated disease for each cancer were used for patients undergoing correct treatmentCitation28–35. For patients with incorrect treatment, utility weights associated with progressive disease were used when available by cancer type (for breast, colon and rectum, kidney and renal pelvis, and lung and bronchus); otherwise, proportionate decreases of utility weights associated with correct treatment were applied, as modeled by the National Institute for Health and Clinical Excellence (NICE)Citation36. The utility weight for patients treated with empiric treatment was calculated as the weighted average of values found in the NICE assessmentCitation36.
Analyses
In all models, ICERs were calculated as the ratios of incremental costs to incremental QALYs and incremental LYs. Proportions of patients diagnosed correctly and incorrectly and treated empirically were also predicted for each strategy.
For the primary model, one-way sensitivity analyses and probabilistic sensitivity analyses were conducted to evaluate the impact of parameter uncertainty on model outcomes. In one-way sensitivity analyses, each parameter was varied individually at ±25% of the base case value. In the probabilistic sensitivity analyses, all parameters were varied simultaneously for 1000 model iterations. Across iterations, clinical parameters (survival and diagnostic event rates) were normally distributed with a mean of the base case value, a standard deviation (SD) of 10% of the base case for estimates derived from the literature, and a SD of 25% of the base case for estimates based exclusively on expert opinion. Cost parameters followed a gamma distribution, with alpha and beta estimated for a mean of the base case value and a SD of 10% of the base case. Utility weights were uniformly distributed, ±25% of base case value, bounded by 0 and 1.
Results
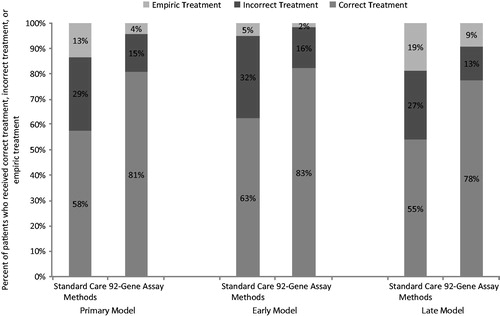
In the primary model, patients with metastatic cancer of uncertain origin who were diagnosed with the 92-gene assay compared with standard care methods alone were more likely to be treated with correct site-specific therapy (81% versus 58%) and less likely to be treated incorrectly (15% versus 29%) or treated with empiric therapy (4% versus 13%) (). Use of the 92-gene assay increased quality-adjusted survival by 1.15 months (10.34 vs. 9.20 months) and unadjusted survival by 0.92 months (15.56 vs. 14.64 months). Use of the assay added $4804 in diagnostic and treatment costs ($76,884 versus $72,080), resulting in ICERs of $50,273/QALY and $62,451/LY ().
Table 2. The 92-gene assay model: base case results.
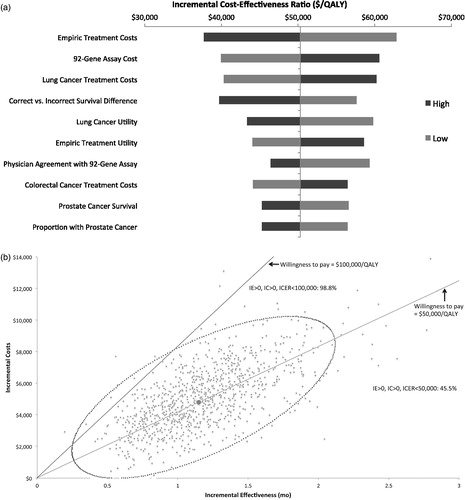
In one-way sensitivity analyses of the primary model, the ICER was most sensitive to the costs of empiric treatment and of the 92-gene assay. The ICER never exceeded $62,812 with any parameter variations (). In the probabilistic sensitivity analyses, at a societal willingness-to-pay (WTP) threshold of $100,000/QALY, the 92-gene assay would be considered cost effective 98.8% of the time ().
Figure 3. Primary model. (a) One-way sensitivity analyses. Each parameter varied ±25%. (b) Probabilistic sensitivity analyses. Individual dots represent ICERs when all parameters are varied simultaneously for 1000 model simulations. The dotted circular line represents 95% confidence ellipse. IC, incremental cost, ICER, incremental cost-effectiveness ratio; IE, incremental effectiveness; QALY, quality-adjusted life-year.
In the early model – in which the 92-gene assay was used earlier in the diagnostic process (i.e., before a first set of six IHC stains) – more patients were diagnosed and treated correctly (). Life expectancy increased when using the 92-gene assay earlier in the diagnostic process, as did total cost, resulting in an ICER of $63,972/QALY (). When the 92-gene assay was used after the second set of nondiagnostic IHC stains (late model), the ICER increased a similar amount to $63,796/QALY; however, using the 92-gene assay later in the diagnostic process resulted in a slightly lower percentage of patients diagnosed and treated correctly compared with the primary model (). In the clinically simplified model, survival increased by 3.4 quality-adjusted months, and the ICER increased to $85,584/QALY ().
Appendix Table 1. Mean Survival for Eight Primary Cancer Sites by Histologic Subtype.
Discussion
Current methods for diagnosing patients with metastatic cancer of uncertain origin are highly heterogeneous, varying with clinical presentation and physician practiceCitation2,Citation9. Patient factors such as performance status, socioeconomic status, and treatment preferences must also be taken into account in diagnostic and treatment decision making. Initial evaluation will typically include a complete history and physical examination (including breast, genitourinary, pelvic, and rectal examinations as appropriate), routine laboratory studies (i.e., complete blood count, electrolytes, liver function tests, creatinine, and calcium), occult blood stool testing, and symptom-directed endoscopyCitation3. Further evaluation may include a wide range of imaging studies, IHC analysis, additional tumor marker tests, and invasive procedures, although use of such methods is not universal. Until now, the economic trade-offs associated with these procedures had not been formally assessed, and there was still controversy about the extent to which such high-cost and time-consuming diagnostic procedures optimized patient outcomes and resource allocationCitation1,Citation3,Citation10,Citation11.
Our analysis, the first of its kind to examine the clinical and economic trade-offs involved in using the 92-gene assay, indicates that the 92-gene assay may lead to efficient resource allocation by creating a standardized approach that improves clinical outcomes at a reasonable cost. In particular, the favorable $50,273/QALY found in the primary model highlights the value of the assay’s clinical benefit in situations that are closest to clinical reality. Recent evidence suggests that the societal willingness-to-pay threshold in oncology is at least $100,000/QALY, if not higherCitation39–46. Even when a range of assumptions regarding standard practice patterns are included, the 92-gene assay remained cost effective when considering these current standards. In all such cases, use of the 92-gene assay also increased the likelihood of correct diagnosis and treatment, leading to higher overall survival as well as quality-adjusted survival. These findings were stable when parameters were varied to explore uncertainty in model inputs.
Although these models were informed by the best available data and structured to represent clinical practice, the real world diagnostic process is complex and nonstandardized. In assessing the 92-gene assay, there was a need to define a standard care approach for comparison with the assay. As such, the primary model was developed to represent the experiences of a ‘typical’ patient, based on clinical practice guidelines and extensive input from practicing clinicians. To acknowledge and assess the impact of the significant heterogeneity in the diagnostic process, additional models were developed. All analyses showed that the cost effectiveness of the 92-gene assay was stable across differing assumptions of modality, extent, and intensity of diagnostic procedures. In addition, clinical results of the clinically simplified model – in which patients were all treated either according to the 92-gene assay or empirically – were similar to those of a prospective clinical trialCitation8. This further validates the model and findings of our analysis.
The results of this analysis must be considered in light of its limitations. Eight cancer sites were included in the model, and the distribution of patients in the model between these cancers may not reflect reality. Because of the lack of published data, expert opinion was used to estimate model inputs such as resource utilization that are known to vary between providers. However, sensitivity analyses indicated that these inputs were not key drivers of model results. Survival and treatment cost data were based on retrospective SEER data and may underestimate the survival benefits and higher costs of newer therapies implemented beyond the data collection period as well as the costs faced by commercial payers. Finally, utility weights for each cancer were derived from different sources and may not accurately reflect relative differences in patient quality of life. Despite this uncertainty, these utility weights were found to have minimal influence on model results when varied in sensitivity analyses.
Conclusions
The lack of a standardized diagnostic process for patients presenting with metastatic cancer of uncertain primary poses significant clinical, economic, and quality-of-life burdens to patients, clinicians, and the healthcare system overall. The results of this study indicate that incorporating the 92-gene assay in the diagnostic paradigm will standardize the diagnostic process, improve treatment accuracy and clinical outcomes, and optimize resource allocation.
Transparency
Declaration of funding
This study was funded by bioTheranostics Inc. Partnership for Health Analytic Research LLC (PHAR LLC) was paid by bioTheranostics Inc. to conduct the research described in this manuscript.
Declaration of financial/other relationships
T.G.K.B., M.S.B., and J.D.O. have disclosed that they are employees of PHAR LLC and W.C.H. has disclosed that she was an employee when this research was conducted. B.E.S., C.A.S., and M.G.E. have disclosed that they are employees of bioTheranostics Inc.
JME peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Supplementary Appendix Table 1: Mean Survival for Eight Primary Cancer Sites by Histologic Subtype
Download PDF (110 KB)Acknowledgments
The authors thank Dr. Fadi Braiteh of Comprehensive Cancer Centers of Nevada, Dr. Lawrence M. Weiss of Clarient Pathology Services, Inc., and Dr. Federico A. Monzon of Texas Children's Hospital for contributions of clinical expertise during development of this model. These services were funded by bioTheranostics, Inc.
References
- Greco AF, Hainsworth JD. Cancer of unknown primary site. In: DeVita VT Jr, Lawrence TS, Rosenberg SA, eds. DeVita, Hellman, and Rosenberg’s Cancer: Principles & Practice of Oncology, 9th edn. Philadelphia, PA: Wolters Kluwer Health/Lippincott Williams & Wilkins, 2011:2033-51
- Pavlidis N, Briasoulis E, Hainsworth J, et al. Diagnostic and therapeutic management of cancer of an unknown primary. Eur J Cancer 2003;39:1990-2005
- National Comprehensive Cancer Network (NCCN). NCCN Guidelines: occult primary (cancer of unknown primary [CUP]). Version 1.2013. Available at: http://www.nccn.org/professionals/physician_gls/pdf/occult.pdf [Last accessed 13 June 2013]
- Anderson GG, Weiss LM. Determining tissue of origin for metastatic cancers: meta-analysis and literature review of immunohistochemistry performance. Appl Immunohistochem Mol Morphol 2010;18:3-8
- Varadhachary GR, Raber MN, Matamoros A, et al. Carcinoma of unknown primary with a colon-cancer profile-changing paradigm and emerging definitions. Lancet Oncol 2008;9:596-9
- Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer 2007;43:2026-36
- Hainsworth JD, Schnabel CA, Erlander MG, et al. A retrospective study of treatment outcomes in patients with carcinoma of unknown primary site and a colorectal cancer molecular profile. Clin Colorectal Cancer 2012;11:112-18
- Hainsworth JD, Rubin MS, Spigel DR, et al. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon research institute. J Clin Oncol 2013;31:217-23
- Natoli C, Ramazzotti V, Nappi O, et al. Unknown primary tumors. Biochim Biophys Acta 2011;1816:13-24
- Schapira DV, Jarrett AR. The need to consider survival, outcome, and expense when evaluating and treating patients with unknown primary carcinoma. Arch Intern Med 1995;155:2050-4
- Saliminejad M, Bemanian S, Ho A, et al. The yield and cost of colonoscopy in patients with metastatic cancer of unknown primary. Aliment Pharmacol Ther 2013;38:628-33
- Ma XJ, Patel R, Wang X, et al. Molecular classification of human cancers using a 92-gene real-time quantitative polymerase chain reaction assay. Arch Pathol Lab Med 2006;130:465-73
- Erlander MG, Ma XJ, Kesty NC, et al. Performance and clinical evaluation of the 92-gene real-time PCR assay for tumor classification. J Mol Diagn 2011;13:493-503
- Kerr SE, Schnabel CA, Sullivan PS, et al. Multisite validation study to determine performance characteristics of a 92-gene molecular cancer classifier. Clin Cancer Res 2012;18:3952-60
- Pillai R, Deeter R, Rigl CT, et al. Validation and reproducibility of a microarray-based gene expression test for tumor identification in formalin-fixed, paraffin-embedded specimens. J Mol Diagn 2011;13:48-56
- Meiri E, Mueller WC, Rosenwald S, et al. A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist 2012;17:801-12
- Weiss LM, Chu P, Schroeder BE, et al. Blinded comparator study of immunohistochemical analysis versus a 92-gene cancer classifier in the diagnosis of the primary site in metastatic tumors. J Mol Diagn 2013;15:263-9
- Kim B, Schroeder BE, Schnabel CA, et al. Physician-reported clinical utility of the 92-gene molecular classifier in tumors with uncertain diagnosis following standard clinicopathologic evaluation. Personalized Med Onc 2013;2:68-76
- SEER Incidence Analysis; 2009 Incidence Rate Based on SEER 9 Regs Research Data, Nov 2011 Sub, Vintage 2009 Pops (1973–2009) <Katrina/Rita Population Adjustment> – Linked To County Attributes – Total U.S., 1969–2010 Counties. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch, April 2012
- SEER Survival Analysis; 5-Year (2005–2009) Observed Survival Based on SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2011 Sub (1973–2009 varying) – Linked To County Attributes – Total U.S., 1969–2010 Counties. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch, April 2012
- Losa Gaspà F, Germá JR, Albareda JM, et al. Metastatic cancer presentation: validation of a diagnostic algorithm with 221 consecutive patients [in Spanish]. Rev Clin Esp 2002;202:313-19
- Møller AK, Loft A, Berthelsen AK, et al. A prospective comparison of 18F-FDG PET/CT and CT as diagnostic tools to identify the primary tumor site in patients with extracervical carcinoma of unknown primary site. Oncologist 2012;17:1146-54
- Kopp RP, Stroup SP, Schroeck FR, et al. Are repeat prostate biopsies safe? A cohort analysis from the SEARCH database. J Urol 2012;187:2056-60
- Cao JQ, Rodrigues GB, Louie AV, et al. Systematic review of the cost-effectiveness of positron-emission tomography in staging on non–small-cell lung cancer and management of solitary pulmonary nodules. Clin Lung Cancer 2012;13:161-70
- Russo A, Elixhauser A, Steiner C, Wier L. Hospitalbased ambulatory surgery, 2007. HCUP Statistical Brief #86. February 2010. Rockville, MD: Agency for Healthcare Research and Quality. Available at: http://www.hcup-us.ahrq.gov/reports/statbriefs/sb86.pdf [Last accessed 14 March 2013]
- Yabroff KR, Lamont EB, Mariotto A, et al. Cost of care for elderly cancer patients in the United States. J Natl Cancer Inst 2008;100:630-41
- Tong KB, Murtagh KN, Hubert H, et al. Patient survival and health care utilization in Medicare beneficiaries diagnosed with cancer of unknown primary. J Clin Oncol 2006;24(18S; June 20 Suppl):6064
- Chong CAKY, Gulamhussein A, Heathcote EJ, et al. Health-state utilities and quality of life in hepatitis C patients. Am J Gastroenterol 2003;98:630-8
- Stein K, Sugar C, Velikova G, et al. Putting the ‘Q’ in quality adjusted life years (QALYs) for advanced ovarian cancer – an approach using data clustering methods and the internet. Eur J Cancer 2007;43:104-13
- Müller-Nordhorn J, Roll S, Böhmig M, et al. Health-related quality of life in patients with pancreatic cancer. Digestion 2006;74:118-25
- Bremner KE, Chong CAKY, Tomlinson G, et al. A review and meta-analysis of prostate cancer utilities. Med Decis Making 2007;27:288-98
- Lloyd A, Nafees B, Narewska J, et al. Health state utilities for metastatic breast cancer. Br J Cancer 2006;95:683-90
- Wang J, Zhao Z, Barber B, et al. A Q-TWiST analysis comparing panitumumab plus best supportive care (BSC) with BSC alone in patients with wild-type KRAS metastatic colorectal cancer. Br J Cancer 2011;104:1848-53
- Swinburn P, Lloyd A, Nathan P, et al. Elicitation of health state utilities in metastatic renal cell carcinoma. Curr Med Res Opin 2010;26:1091-6
- Nafees B, Stafford M, Gavriel S, et al. Health state utilities for non small cell lung cancer. Health Qual Life Outcomes 2008;6:84-99
- NICE Clinical Guidelines, No. 104. Diagnosis and Management of Metastatic Malignant Disease of Unknown Primary Origin. Cardiff, UK: National Collaborating Centre for Cancer (UK), July 2010
- US Department of Labor. Archived Consumer Price Index Detailed Report Information. Available at: http://www.bls.gov/cpi/cpi_dr.htm [Last accessed 30 March 2013]
- Physicians’ Fee & Coding Guide 2012. Atlanta, GA: MAG Mutual Healthcare Solutions, 2011
- Behl AS, Goddard KAB, Flottemesch TJ, et al. Cost-effectiveness analysis of screening for KRAS and BRAF mutations in metastatic colorectal cancer. J Natl Cancer Inst 2012;104:1785-95
- Braithwaite RS, Meltzer DO, King JT Jr, et al. What does the value of modern medicine say about the $50,000 per quality-adjusted life-year decision rule? Med Care 2008;46:349-56
- Gold HT, Hayes MK. Cost effectiveness of new breast cancer radiotherapy technologies in diverse populations. Breast Cancer Res Treat 2012;136:221-9
- Sharaf RN, Ladabaum U. Comparative effectiveness and cost-effectiveness of screening colonoscopy vs. sigmoidoscopy and alternative strategies. Am J Gastroenterol 2013;108:120-32
- Wang G, Kuppermann M, Kim B, et al. Influence of patient preferences on the cost-effectiveness of screening for Lynch syndrome. Am J Manag Care 2012;18:e179-85
- Tsoi DT, Inoue M, Kelly CM, et al. Cost-effectiveness analysis of recurrence score-guided treatment using a 21-gene assay in early breast cancer. Oncologist 2010;15:457-65
- Carlson JJ, Garrison LP, Ramsey SD, et al. The potential clinical and economic outcomes of pharmacogenomic approaches to EGFR-tyrosine kinase inhibitor therapy in non-small-cell lung cancer. Value Health 2009;12:20-7
- Le QA, Hay JW. Cost-effectiveness analysis of lapatinib in HER-2-positive advanced breast cancer. Cancer 2009;115:489-98