Abstract
Vibrio cholerae is an estuarine bacterium associated with a single peak of cholera (March–May) in coastal villages of Bangladesh. For an unknown reason, however, cholera occurs in a unique dual peak (March–May and September–November) pattern in the city of Dhaka that is bordered by a heavily polluted freshwater river system and flood embankment. In August 2007, extreme flooding was accompanied by an unusually severe diarrhea outbreak in Dhaka that resulted in a record high illness. This study was aimed to understand the unusual outbreak and if it was related to the circulation of a new V. cholerae clone. Nineteen V. cholerae isolated during the peak of the 2007 outbreak were subjected to extensive phenotypic and molecular analyses, including multi-locus genetic screening by polymerase chain reaction (PCR), sequence-typing of the ctxB gene, and pulsed-field gel electrophoresis (PFGE). Factors associated with the unusual incidence of cholera were determined and analysis of the disease severity was done. Overall, microbiological and molecular data confirmed that the hypervirulent V. cholerae was O1 biotype El Tor (ET) that possessed cholera toxin (CT) of the classical biotype. The PFGE (NotI) and dendrogram clustering confirmed that the strains were clonal and related to the pre-2007 variant ET from Dhaka and Matlab and resembled one of two distinct clones of the variant ET confirmed to be present in the estuarine ecosystem of Bangladesh. Results of the analyses of both diarrheal case data for three consecutive years (2006–2008) and regional hydroclimatology over three decades (1980–2009) clearly indicate that the pattern of cholera occurring in Dhaka, and not seen at other endemic sites, was associated with flood waters transmitting the infectious clone circulating via the fecal-oral route during and between the dual seasonal cholera peaks in Dhaka. Circular river systems and flood embankment likely facilitate transmission of infectious V. cholerae throughout the year that leads to both sudden and off-season outbreaks in the densely populated urban ecosystem of Dhaka. Clonal recycling of hybrid El Tor with increasing virulence in a changing climate and in a region with a growing urban population represents a serious public health concern for Bangladesh.
Introduction
Cholera is a rapidly dehydrating diarrhea that results from ingestion of toxigenic Vibrio cholerae serogroups O1 or O139. V. cholerae O1 comprises two biotypes, classical (CL) and El Tor (ET), both of which are responsible for seven cholera pandemics. Studies show that the fifth and sixth cholera pandemics were caused by V. cholerae CL biotype, while V. cholerae ET biotype is associated with the current seventh pandemic having completely replaced the CL biotype that had been prominent in the early 1980s Citation1. Despite the fact that the two biotypes of V. cholerae O1 share the same O polysaccharide genes, genetic studies show a high degree of gene conservation since the V. cholerae seventh pandemic islands (VSP)-I and -II are unique to pandemic V. cholerae ET strains and do not occur in V. cholerae CL biotype strains Citation2–Citation4. In addition to these few phenotypic and genetic differences, there are also differences in the pattern of infection caused by the two biotypes. Although many studies have demonstrated V. cholerae ET to be better adapted to the environment, they have been less virulent Citation5. Notably, more asymptomatic carriers than symptomatic cases of V. cholerae ET have been reported. The V. cholerae ET asymptomatic individuals outnumber clinically symptomatic patients by a ratio of almost 50:1 Citation6. Persistence in both the environment and the human host, together with a more efficient host-to-host transmission, renders V. cholerae ET strains more effective in causing disease in humans than V. cholerae CL strains Citation7.
Recent molecular analysis of V. cholerae ET isolated from patients in Bangladesh with acute watery diarrhea Citation8 shows that, currently, V. cholerae O1 ET strains are hybrids of both V. cholerae CL and ET biotypes. These hybrid strains have been designated ‘Matlab variants’, since they were first isolated in Matlab, a rural endemic cholera area 50 km south-east of Dhaka Citation8. Further studies have shown that all circulating strains of V. cholerae ET biotype isolated since 2001 are hybrids of both V. cholerae CL and ET biotypes, while those isolated before 2001 contain attributes of the seventh pandemic V. cholerae ET Citation9. Although the consequence of a continual genetic shift among cholera bacteria is not fully understood, the severe dehydration caused by hybrid V. cholerae ET strains is an increasingly significant clinical phenomenon in Bangladesh Citation10.
Historically, cholera is known as a seasonal disease, with a variable pattern of infection. Seasonal outbreaks of cholera arise from multiple endemic foci in Bangladesh, generally occurring in a single annual peak, as in other cholera affected countries of the world. Major epidemics of cholera characteristically have originated in coastal regions, including both the epidemic in Peru that began in the coastal regions of Peru in 1991–1992 and the V. cholerae O139 Bengal outbreaks in India and Bangladesh. However, in Dhaka and Matlab, Bangladesh, cholera occurs in a distinct pattern of two observable seasonal peaks, one before the annual monsoon (March–May) and the other after (September–November) Citation1 Citation6 Citation11.
It is important to note that V. cholerae is an estuarine bacterium autochthonous in brackish and estuarine ecosystems Citation12 and is commensal to plankton Citation13. Although climate factors such as sea surface temperature have been shown to be correlated with the incidence of cholera in coastal villages of Bangladesh Citation14, precisely how climate factors may contribute to the characteristic dual peaks of cholera that occur in Dhaka and Matlab (a major city and a rural village, respectively) that are located almost 50 km apart and distant from the coast of the Bay of Bengal is not yet understood. In August 2007, Bangladesh suffered severe flooding accompanied by a significantly large outbreak of diarrhea in Dhaka. During this outbreak, the International Center for Diarrheal Disease Research, Bangladesh (ICDDR,B) hospital in Dhaka treated a record number of cholera patients, peaking at 1,045 patients per day with 70% suffering from severe dehydration, more than double the number that had been seen in the previous 3 years when flooding had also occurred Citation15. Although V. cholerae O1 ET is generally known to be the primary cause of the severe epidemic of diarrhea in 2007, we carried out a detailed microbiological, molecular, and phylogenetic study and analyzed the hydroclimatological data for 30 years (1980–2009) to understand the ‘off-season’ outbreak peak and to determine if the extreme intensity of the outbreak was caused by a new, more virulent clone of V. cholerae O1.
Materials and methods
Bacterial strains
The V. cholerae serogroup O1 strains characterized and compared in the present study are shown in with their source, including place and year of isolation.
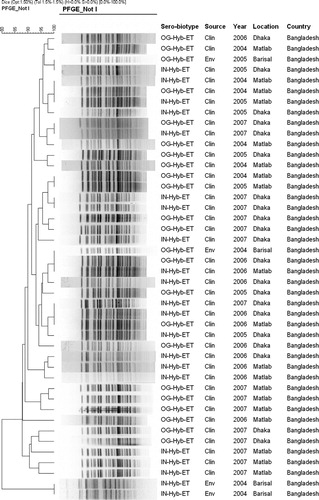
Fig. 1. Molecular fingerprinting analysis using pulsed-field gel electrophoresis (PFGE) of NotI-digested genomic DNA obtained from V. cholerae serogroup O1 biotype El Tor strains isolated during the peak of the Dhaka epidemic, 2007, and pre-existing V. cholerae hybrid El Tor strains isolated between 2004 and 2007 in Dhaka and Matlab, two inland (freshwater) cholera endemic sites located 50 km apart and Barisal, a cholera endemic coastal ecosystem of Bangladesh. The dendrogram was constructed using digital images of PFGE patterns and Dice similarity coefficient and UPGMA clustering methods.
Isolation of V. cholerae strains
The V. cholerae O1 isolates included in this study were obtained from rectal swabs of cholera patients admitted to the ICDDR,B hospital in Dhaka during the second week of August 2007 when the number of diarrhea cases exceeded the daily record reaching 1,000/day Citation15. Other V. cholerae O1 strains were included in the study for comparison that had been isolated in Dhaka, Matlab, and Barisal, Bangladesh between 2004 and 2007. Dhaka and Matlab are located 50 km apart and both are densely populated cholera endemic sites bordered by rivers and ponds. Dhaka and Matlab are situated 250 km upstream from the coastal villages of Barisal, where cholera is also endemic Citation16. Colonies were confirmed to be V. cholerae using standard culture methods and further identified using a combination of biochemical, serological, and molecular methods as described previously Citation16 Citation17.
Biotyping
Biotype determination involved the following tests: chicken erythrocyte agglutination, hemolysis of sheep erythrocytes, Voges–Proskauer reaction, sensitivity to polymyxin B, and susceptibility to Mukerjee classical (CL) phage IV and Mukerjee ET phage V Citation18. To complement the biotype characterization, polymerase chain reaction (PCR) assays targeted to detect tcpA (CL and ET variants) Citation19 and to determine the type of rstR gene encoding the phage transcriptional regulator were carried out using previously described methods Citation20.
Genomic DNA preparation
For extraction of genomic DNA, cells harvested from 3.0 ml of overnight LB broth were subjected to alkaline lysis followed by phenol-chloroform extraction, as described elsewhere Citation21. The DNA was stored at –20°C for subsequent PCR analysis.
Serogroup analysis by PCR
All strains were re-confirmed using V. cholerae species-specific ompW PCR, as described previously Citation22. Serogroup of the strains was confirmed using polyvalent O1 and monovalent Inaba and Ogawa antisera and multiplex-PCR targeted to genes encoding O1 (wbeO1) and O139 (wbfO139)-specific O biosynthetic genes, as well as the cholera toxin gene (ctxA) as described previously Citation23.
MAMA-PCR for determination of ctxB gene type
A mismatch amplification mutation assay (MAMA)-PCR was performed to test for presence of ctxB genes specific to V. cholerae CL and ET biotypes, employing previously described methods Citation24. The V. cholerae O1 isolates O395 CL and N16961 ET, both reference strains, served as controls.
DNA sequencing of ctxB gene
Nucleotide sequencing of the ctxB genes of six randomly selected strains of the hypervirulent V. cholerae O1 ET that had been isolated during the peak of the 2007 Dhaka cholera epidemic was carried out using an ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit (Perkin-Elmer Applied Biosystems, Foster City, CA, USA) on an ABI PRISM 310 automated sequencer as described previously Citation9. The nucleotide sequences of the reference strains were compared with the corresponding sequences of V. cholerae ET N16961 (GenBank accession no. NC-002505) and V. cholerae CL 569B (GenBank accession no. U25679) retrieved from GenBank by Basic Local Alignment Search Tool (BLAST) search.
DNA sequence and proteomic analysis
The chromatogram sequencing files were inspected using Chromas 2.23 (Technelysium). Nucleotide sequences of the 2007 isolates were compared with corresponding sequences of V. cholerae N16961 ET (NC_002505) and V. cholerae 569B CL (U25679) retrieved from GenBank using BLAST Citation25. Multiple sequence alignments were developed using CLUSTALX 1.81.13 and the DNA sequences were translated using GeneDoc version 2.6.002 alignment editor.
PFGE
Pulsed-field gel electrophoresis (PFGE) was carried out using a contour-clamped homogeneous electrical field (CHEF-DRII) apparatus (Bio-Rad), following previously described methods Citation26. Genomic DNA of the test strains was digested with the NotI restriction enzyme (Invitrogen, USA) and Salmonella braenderup was digested using XbaI, with the fragments serving as molecular size markers. The restriction fragments were separated in 1% pulsed-field-certified agarose in 0.5X TBE (Tris-borate-EDTA) buffer. In the post-electrophoresis gel treatment step, the gel was stained and de-stained. The DNA was visualized using a UV transilluminator and images were digitized using the 1D Gel documentation system (Bio-Rad). The images were processed using Bionumeric Software (Version 4.6, Applied Maths). The test fingerprint image was normalized according to the standard and the molecular weights of the DNA fragments were determined using Bionumeric Software (Version 4.6, Applied Maths). Digital images of PFGE fingerprint patterns were analyzed by Dice similarity coefficient and the UPGMA clustering method (Bio-Rad), as recommended by the manufacturer, and these were graphically represented as dendrograms.
Hydroclimatological analysis
Precipitation data (1980–2009) over the upper catchment areas of the Ganges in Central India and Nepal (Latitude 24–30°N; Longitude 76–88°E), and the Brahmaputra (Latitude 26–30°N; Longitude 88–96°E) basin areas in Northeast India, Bhutan, and parts of China were obtained from the NCEP/NCAR (National Center for Environmental Prediction/National Center for Atmospheric Research, National Oceanic and Atmospheric Administration, USA) atmospheric dataset. River discharge volumes for the same rivers, recorded at Paksey (Ganges) and Bahadurabad (Brahmaputra), were obtained from the Bangladesh Water Development Board (BWDB). Seasonal mean values of rainfall for the monsoon period (June–September), as well as monthly rainfall values for the peak rainfall months, July and August, were converted to corresponding anomalies for effective comparison against climatology (30-year mean: 1980–2009). Anomalies are calculated as deviations from the 30-year mean (1980–2009) and normalized with the respective standard deviations, an usual practice in hydrologic and climate science Citation27.
Results
Diarrheal outbreak during August 2007
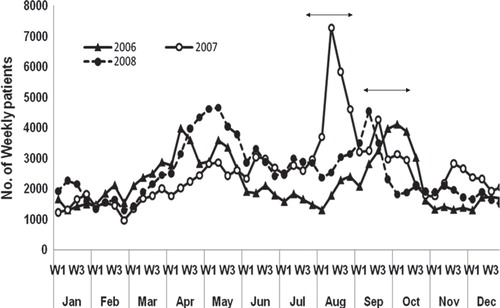
In August 2007 Bangladesh suffered severe flooding, caused by unusually heavy monsoon rains in the upstream catchments of the regional large rivers. At the same time, an unusually severe diarrhea outbreak took hold in Dhaka. The disease outbreak was remarkable in that it occurred between the usual two annual seasonal diarrheal peaks and set a new daily record of extremely ill patients, most of whom were severely dehydrated. In Dhaka, diarrhea occurs almost year-round with two distinctive peaks, one in spring and the other in the autumn Citation1 Citation11. As shown in , the number of patients rose sharply during the first week of August 2007. In the second week, the epidemic peaked with daily admissions reaching a record number of 1,045 patients Citation15. During the month of August 2007, the ICDDR,B Dhaka hospital alone treated 21,401 diarrhea patients, almost three times the number seen over the same period in 2006 and 2008.
Fig. 2. Comparative yearly data showing weekly admissions of diarrheal patients to the ICDDR,B hospital in Dhaka (2006–2008) during the unusual flood-related diarrheal epidemic that broke out in August 2007. The graph shows number of cases per week, based on actual numbers of diarrheal patients admitted to the ICDDR,B hospital. Number of cases for 2008 are included up to the third week of August.
During this outbreak, analysis of 2% of clinical samples systematically collected showed Vibrio cholerae to be the most commonly identified cause of diarrhea (35%), followed by Rotavirus (12%), and Escherichia coli (10%); however, no pathogen was identified for 45% of samples Citation15.
Biochemical and serological tests
All V. cholerae O1 strains included in the present study produced characteristic colonies when grown on selective agar media and all were confirmed by biochemical, serological, and molecular methods to be V. cholerae O1 Citation16 Citation17.
Phenotypic and genotypic characteristics
All 25 V. cholerae O1 strains including those concluded to be hypervirulent showed resistance to polymyxin B (50 U), an ability to agglutinate chicken erythrocytes, and a sensitivity to ET-specific phage V but not to CL phage IV Citation3, indicating V. cholerae ET ancestry. Furthermore, all strains amplified the primers for V. cholerae species-specific ompW gene, serogroup-specific wbe gene, cholera toxin (CT) encoding ctxA gene, and biotype-specific genes tcpA (ET) and rstR2 (ET). These results were confirmed using simplex- and multiplex-PCR assays () Citation6. Hypervirulent V. cholerae associated with the 2007 Dhaka epidemic of cholera was, indeed, toxigenic and belonged to V. cholerae serogroup O1 biotype ET.
Table 1. Phenotypic and molecular characteristics of Vibrio cholerae Ol strains isolated during the flood-related Dhaka epidemic in 2007 and V. cholerae O1 strains isolated between 2004 and 2007 in Dhaka and Matlab. V. cholerae O1 classical and El Tor strains were included as controls
MAMA-PCR–
All hypervirulent V. cholerae O1 strains including V. cholerae hybrid-ET amplified primers specific for ctxB gene of the CL biotype, as did the V. cholerae CL control. The V. cholerae ET control did not amplify these primers indicating presence of the ctxB gene of V. cholerae CL. The V. cholerae ET control strain amplified primers specific for the ctxB gene of the ET biotype but not that of V. cholerae CL and test strains. These results confirm specificity of the mismatch amplification mutation assay (MAMA)-PCR Citation24.
DNA sequencing and proteomic analysis of ctxB gene
Nucleotide sequence analysis of the ctxB genes and the deduced N′-terminal amino acid sequence revealed that all six strains had histidine at position 39, phenylalanine at position 46, and threonine at position 68, typical of CL 569B. This confirmed the MAMA-PCR assay results, in that the CT of the hypervirulent strains was of the CL biotype.
Genome analysis by PFGE and cluster analysis
The hypervirulent V. cholerae O1 ET strains isolated during the 2007 Dhaka outbreak were subjected to PFGE analysis and compared with V. cholerae hybrid-ET strains isolated in Dhaka during 2004 through 2006, Matlab (2004–2007), and Barisal (2004–2006) to determine clonal origin and genetic relatedness. The NotI restriction enzyme digested the genomic DNA into between 20 and 23 fragments (), the molecular sizes of which were between 20 to 350 kb. Except for the Barisal strains, which varied significantly, the overall PFGE patterns of the hypervirulent and the V. cholerae hybrid-ET from Dhaka and Matlab were more or less homogeneous () suggesting clonal relatedness.
The hypervirulent and hybrid-ET V. cholerae strains from Dhaka and Matlab, including two of the four hybrid-ET V. cholerae from Barisal, formed a major cluster of closely related strains in the analyses indicating clonal lineage. The remaining two Barisal strains, which varied significantly in their PFGE patterns, formed a separate but distant cluster of clonally unrelated strains, confirming two different clones in Barisal, one of which showed high relatedness to hypervirulent and hybrid-ET V. cholerae strains from Dhaka and Matlab. Although minor variations were evident among the hypervirulent and hybrid-ET V. cholerae strains that formed the major cluster, the similarity coefficient as shown in the dendogram, suggests they are clonal and supports the hypothesis that continuous fecal-oral transmission allows recycling of a clone .
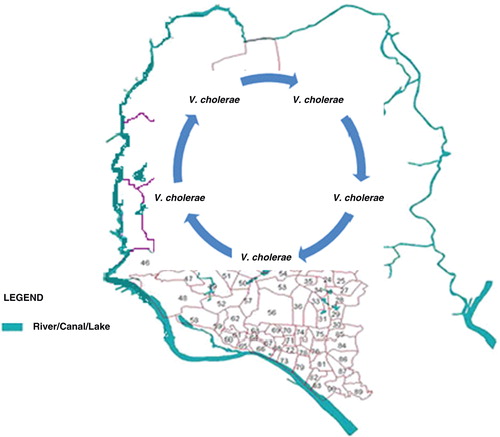
Fig. 3. Map of Dhaka city showing the circular river systems along its border that receive the sewage effluent of nearly 15 million inhabitants. The city has an extensive, but not fully effective, sewage network linked to ponds, lakes, and canals that flow into the surrounding river systems. The circular arrows at the center indicate clonal recycling of V. cholerae in Dhaka, a fresh-water ecosystem situated 250 km distant from the estuarine ecosystem of the Bay of Bengal, Bangladesh. Unlike the estuarine ecosystem, where V. cholerae comprises a portion of the autochthonous microflora (12) in association with plankton year round (13,14), the water bodies of Dhaka serve as a constant source of highly transmissible V. cholerae. During extreme climatic conditions such as flooding in the poor urban setting of Dhaka, fecal-oral transmission is enhanced leading to significant off-season outbreaks, as occurred during flooding in 2007 in Dhaka. Flooding churns the waters, bringing sediment and V. cholerae biofilm attached to particulates into the surface water, thereby enriching the V. cholerae population.
The flood of August 2007
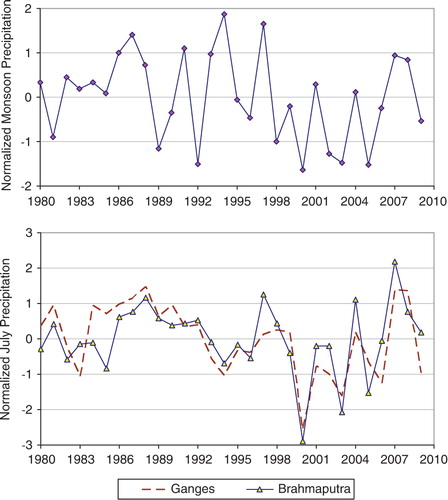
The aim of this analysis was to characterize the flood of 2007 and understand the evolution of this event with respect to rainfall and streamflow of the Ganges and Brahmaputra Rivers and their catchment regions between 1980 and 2009. A shows the seasonal anomaly of combined monsoon precipitation of these two river basins. The results suggest that 2007 did not rank as one of the record flood years over the history of this region. However, the monthly records for rainfall into individual rivers in 2007 show contrasting results. B shows that the July 2007 rainfall in the upstream Brahmaputra catchment area ranks as the highest amount over the previous 30 years. The Ganges rainfall also ranked very high (second highest in 30 years) for the analysis period (1980–2009). An important observation from these results is that the catchment regions of the two rivers received these high amounts of rainfall in the same month, July 2007, although typically the Ganges basin region experiences monsoon precipitation after about a month, compared to the Brahmaputra basin Citation28. These results are corroborated by similarly high combined monthly streamflow values for the two rivers (not shown). Thus, it is suggested that the strong flooding in Bangladesh during August 2007 was caused by the simultaneous and anomalously high rainfall in both the Ganges and Brahmaputra upstream regions in central and Northeastern India, respectively, even though the average strength of the monsoon season in 2007 did not qualify as a record year, with respect to 30-year climatology.
Fig. 4. (A) Climatological analysis of Bangladesh monsoon precipitation for 1980–2009 shows the relative magnitude of combined monsoon (JJAS: June–September) rainfall in 2007. The normalized anomaly values indicate 2007 was not a record year. (B) However, anomaly analysis of monthly rainfall shows that July 2007 was the highest monthly amount (for July) recorded for the Brahmaputra catchment areas in 30 years. In addition, the Ganges catchment areas also experienced very heavy rainfall in July 2007, as shown by the anomaly calculated against the 30 year base period (1980–2009).
Discussion
The floods in Bangladesh during 1988, 1998, and 2004 were associated with massive outbreaks of water-borne disease in Bangladesh Citation29. The flood-related diarrheal outbreak that struck Dhaka in August, 2007 was remarkable not only because of the record number of patients admitted to the ICDDR,B hospital in Dhaka, but also because more than 70% of the patients were affected with severe dehydration Citation15 Citation30. Although the severity of dehydration was more than double, compared with that seen during the previous 3 years when flooding occurred Citation29, deaths were prevented among the hospitalized patients by using aggressive rehydration therapy. Primary microbiological culture, coupled with biochemical and serological characterization, confirmed that a hypervirulent bacterium, Vibrio cholerae O1, possessing all conventional phenotypic traits typical of V. cholerae ET Citation2 Citation5, was responsible for the severe cholera disease. Genetic screening, using simplex- and multiplex-PCR assays Citation6 showed that all of the strains in this study that were tested amplified primers for the V. cholerae species-specific gene ompW Citation29, the ctxA gene encoding sub unit A of CT, and the surface antigen (serogroup O1-specific) encoding gene wbe Citation23 confirming our initial results, showing that the hypervirulent V. cholerae was both toxigenic and belonged to serogroup O1. Molecular data showed that the strains carried biotype-specific genes, e.g. tcpA-ET Citation19 and rstR2-ET Citation20, the genes encoding major pilin and a repressor, respectively. All of these findings led to the conclusion that both the hypervirulent V. cholerae ET that caused the 2007 cholera outbreak and the V. cholerae hybrid-ET isolated earlier in both Dhaka and Matlab shared phenotypic and genetic traits of the seventh pandemic V. cholerae ET prototype.
Another important point to make is that acute dehydrating diarrhea is a hallmark of cholera caused by CT encoded by ctxAB genes, components of the filamentous bacteriophage, CTX?, that is bio-specific and lysogenizes V. cholerae Citation31, mainly V. cholerae O1 and O139. Although CTX? is biotype specific, all V. cholerae ET isolated in Dhaka since 2001 carry the ctxB gene of the V. cholerae CL biotype, while V. cholerae ET isolated before 2001 have ctxB of the V. cholerae ET biotype Citation8 Citation9. The V. cholerae O1 ET harboring CL type CTX? was reported in Mozambique in 2004 Citation32. All V. cholerae tested in the present study had the CL type of CT as confirmed by MAMA-PCR assay Citation24, nucleotide sequencing, and analysis of the ctxB genes. This raised a key question; namely, was hypervirulence of the hybrid-ET V. cholerae due to V. cholerae CL biotype CT in a V. cholerae ET biotype? Increased severity of dehydration caused by hybrid-ET V. cholerae was demonstrated in a molecular epidemiological study carried out between 2004 and 2006 employing V. cholerae isolated from cholera cases in coastal villages of Bangladesh Citation10.
As stated earlier, V. cholerae CL and ET biotypes differ from each other in phenotypic and genotypic traits and also in the pattern of infection they cause Citation33–Citation35. Many studies have shown that V. cholerae ET is better able to adapt to the environment but is less virulent in clinical manifestation Citation6. It can be hypothesized that by adapting to change in the ecosystem and undergoing efficient host-to-host transmission, hybrid-ET V. cholerae are rendered significantly more robust Citation7. However, if more severe diarrhea and increased dehydration can be attributed solely to CL biotype CT in an ET biotype V. cholerae, the question then arises as to why the diarrhea and dehydration increased so suddenly, since all V. cholerae ET strains that had been isolated in Dhaka since 2001 have CL biotype CT Citation9. One can suggest that the increase in severity of dehydration derived from rapid passage within the very large population of Dhaka, increasing significantly the number of infectious hybrid-ET V. cholerae transmitted, notably during the August 2007 flooding Citation34 Citation35. More plausible, however, is extensive mixing of particulates from the sediment of the urban river and pond system of Dhaka that receives untreated sewage containing infectious V. cholerae, especially during intensively heavy flooding, as occurred in 2007, breaching the flood embankment and contaminating the drinking water sources. Cholera, known to be a dose dependent disease Citation36, with an increase in the number of cholera bacteria in the water caused by fresh release of rice-watery stool and sediment mixing during turbulent flood activity undoubtedly will lead to an extensive and intensive epidemic, such as that which occurred in 2007.
The overall results of PFGE, a tool useful in determining clonal origin Citation37, and the clustering of strains observed in the dendrograms, led to the conclusion that hypervirulent V. cholerae hybrid-ET strains isolated in Dhaka and Matlab in 2005 and 2006, represent the same clone of strains isolated in 2007 that, in turn, is the same as one of the two distinct clonal types of hybrid-ET V. cholerae isolated from Barisal located in the coastal region of Bangladesh. These results suggest a recycling of clonal V. cholerae that occurred in Dhaka and Matlab, where cholera is present throughout the year. Although the genetic fidelity of strains of V. cholerae causing cholera provides strong support for clonal recycling in Dhaka, the minor variations detected, as shown in the dendrograms, indicate that genetic re-assortment also occurs Citation9. In contrast, the two distant clonal types of V. cholerae hybrid-ET isolated from Barisal suggest that, unlike the urban ecosystem of Dhaka where year-round cholera can involve recycling of a clone, the genetically divergent clones that are present in the natural estuarine ecosystem of V. cholerae may also emerge but with less dramatic intensity Citation34 Citation35 .
Because V. cholerae is autochthonous to brackish and estuarine ecosystems, it persists throughout the year and during inter-epidemic periods of the year resides in a non-culturable, dormant state Citation12 Citation13 Citation16. Many studies have shown that V. cholerae is commensal to plankton and multiplies during plankton blooms, thereby contributing to the onset of seasonal cholera Citation14. The actively growing state of V. cholerae that initiates seasonal cholera has been shown to be climate driven with signals, such as temperature and conductivity, playing a role in a series of naturally occurring events Citation14 Citation16.
Dhaka is located in the fresh water zone of Bangladesh, but hundreds of tannery industries release huge amounts of Na+ into the local water bodies along with domestic waste from the households of approximately 15 million residents of the city. Also, it should be remembered that river ecosystems carry high concentrations of Ca+ + that can spare the requirement for Na+, with the consequence that V. cholerae can thrive in fresh water ecosystems Citation38. The pattern of two seasonal peaks can be related to the annual plankton blooms occurring in both the spring and fall with a lower incidence of the disease between the peaks Citation39.
Dhaka is situated in the downstream floodplains region of the Ganges–Brahmaputra–Meghna (GBM) basin, with rainfall catchments encompassing central and Northeastern India to the Himalayas. The hydroclimatology of this region is highly seasonal in nature with over 80% of the annual precipitation occurring during the four monsoon months of June through September Citation28. As a result, the region experiences an asymmetric nature of water availability, namely a severe scarcity of water during the winter months, followed by an abundance of water beginning in summer. Approximately 92% of the basin areas of these rivers lie outside the boundaries of Bangladesh; as a result, flooding typically occurs due to monsoon flow from upstream catchment regions compared to local rainfall events Citation28. On average, about 20% of the land area of Bangladesh is inundated every year and up to 60% in high flood years. A hydro-meteorological analysis of the record flood events in the GBM basin region shows that extreme events occur when peak flow volumes of the two major rivers, the Ganges and the Brahmaputra, coincide during the months of August or September Citation40.
In Dhaka, a freshwater ecosystem situated at the confluence of several distributaries of the regional major rivers, cholera outbreaks are typically seen in the spring (Mar-Apr-May) and fall (Sep-Oct-Nov) months of the year. Akanda et al. Citation41 provided a hydroclimatological explanation of the biannual cholera peaks, showing that low flow in the Brahmaputra and the Ganges during the spring is associated with the first outbreaks of cholera in Bangladesh with elevated spring outbreaks observed in strong drought years. On the other hand, peak streamflow of these rivers during the monsoon months of July through September can create a different cholera transmission environment with peak flood volumes and the extent of flood-affected areas during monsoon being associated with the autumn cholera outbreaks Citation42.
The cholera outbreak of August 2007 occurred between the two known outbreak seasons. Despite 2007 not being one of the major monsoon years overall (A), rainfall amounts for the month of July were exceptionally high in upstream areas of both the Ganges (second highest in the last 30 years) and Brahmaputra (also highest in the last 30 years; B). More importantly, these two very high rainfall months coincided in this particular year, even though the Ganges basin typically experiences rainfall about a month later compared to the Brahmaputra Citation40. Considering the rainfall-runoff response time of about 3 to 4 weeks from the upstream catchment areas Citation28 Citation43, the anomalously high rainfall in July contributed to a sudden rush of streamflow in these two major rivers with scouring of the river bottom and increased turbidity caused by resuspension of sediments during August 2007. As a result, the combined flow volumes experienced a sudden rise and subsequently had a disastrous impact on points downstream of their confluence, such as Dhaka city, which usually is protected during non-flood years by a flood embankment. The floods dissipated quickly, with the lack of consistently high rainfall in upstream or local catchments. Thus, the magnitude of the cholera outbreaks in Dhaka city were controlled and did not increase as the flood water receded after August Citation15.
When compared to the cholera outbreak records over the last 30 years, 2007 was not an anomalously strong year in terms of average cholera incidence in Dhaka. That is, in the GBM basin region, record cholera outbreaks typically occur when strong drought events take place and spring cholera outbreaks are followed by a strong monsoon season Citation44. Based on climatological analysis of hydroclimatic variables, low flow and chlorophyll conditions were not conducive for a strong spring (MAM) outbreak. With the lack of a strong and persistent monsoon that year, there was no strong fall (SON) outbreak either. These results suggest that the sudden outbreak of cholera in the summer of 2007 did not have a coastal origin as observed in other record years. This evidence points to the contamination of the water resources of Dhaka city by infectious strains present in the local ecosystem, related to the sudden upsurge of flood water in the peripheral rivers and infringement of part of the flood embankment.
The August 2007 cholera outbreak in Dhaka was unusual because it began before the normal fall peak, occurring between the two annual seasonal peaks. The role of climate and the association of V. cholerae with plankton has previously been described in detail Citation13 Citation14 Citation45, but in 2007 the severe rain and floods that occurred in Bangladesh can be concluded also to have played a role in both the early onset and severity of illness. Sediment scouring by flood waters and mixing of large numbers of infectious cholera bacteria with drinking water are suggested to have contributed to intensity of the outbreak.
Construction of embankments and protection barriers against annual monsoon flooding was recently shown to be correlated with increased cholera prevalence in Matlab, Bangladesh Citation46. By analogy, flood embankment and the circular rivers bordering the urban slums can explain year-round cholera in Dhaka where continuous circulation of large numbers of cholera bacteria within a closed type of urban ecosystem can be maintained in the water throughout the year.
Based on molecular data, when cholera occurs simultaneously in different locations within an endemic area, it is initiated by one or more clones that prevail locally Citation47. The outbreak caused by the V. cholerae hybrid-ET in Dhaka and its suburbs during August 2007, did not spread beyond the city limits despite a major part of Bangladesh being under a constant flow of floodwater Citation15. Because cholera outbreaks characteristically begin by causing disease in individuals ingesting water containing an infective dose of cholera bacteria Citation48, an infective dose in flowing water can be achieved by massive scouring of fecal material contaminating the drinking water systems, as occurs in Dhaka city during extreme flooding. It is interesting that the Dhaka outbreak of 2007 had some of the characteristics of the Cryptosporidium outbreak in Milwaukee, Wisconsin in the United States, when severe rains caused an overflow of the sewage treatment system Citation49.
In summary, data and evidence provided here clearly suggest that, unlike the defined seasonal cholera, which is initiated by a cascade of climatic events Citation14 that include the activation of naturally occurring cholera bacteria from its non-culturable state Citation12 Citation13, the early epidemic in August 2007 was associated with flood waters transmitting the infectious V. cholerae clone circulating via the fecal-oral route during and between the dual seasonal cholera peaks in Dhaka. The hypervirulent V. cholerae hybrid-ET that was associated with this epidemic may prove to be harbinger of additional genetic drift and shift in V. cholerae, as observed by Chun et al. Citation50 with enhanced virulence in strains of V. cholerae to be reported. The role of urban sediment as a reservoir of V. cholerae is currently under study in light of predicted increases in monsoon rains for Bangladesh during a time of climate change.
Conclusions
The aquatic environment plays a major role in cholera epidemics by serving as the reservoir of V. cholerae between epidemics and by initiating and transmitting the disease. The coastal link of cholera as an estuarine reservoir for V. cholerae has been established previously. Namely V. cholerae is a commensal of zooplankton with which it is associated mostly as dormant cells that can become actively growing in response to changes in the regional climate to initiate seasonal epidemics of cholera. However, concentrations of V. cholerae in natural water samples tend to be less than the infective dose for volunteer challenges and although the bacteria are present in and on plankton in significantly higher numbers, the direct role of environmental microorganisms in initiating and/or maintaining an epidemic has not been fully defined. Seasonal cholera, as in many other countries, occurs in a single annual peak in the coastal villages of Bangladesh. But, for reasons heretofore not addressed, cholera occurs in a unique dual peak pattern in Dhaka city and its suburb, a densely populated city bordered by fresh water river systems and flood embankment. Although significant advances have been made in the understanding of the molecular basis of V. cholerae pathogenicity and in determining its environmental reservoirs, flooding in August 2007 was accompanied by an unusually severe diarrhea outbreak only in Dhaka city, resulting in a record high severity of illness. Our molecular data on both the flood-related and the pre-flood V. cholerae strains, as well as analyses of diarrheal case data for the three consecutive years that include pre-flood and during the 2007 flood and regional hydroclimatology over three decades, clearly show that the cholera outbreak occurring in Dhaka and not seen at other endemic sites in 2007 was associated with flood waters. Sediment suspension during the massive flooding can be concluded to enhance the transmission of the infectious clone, circulating it via the fecal-oral route during and between the dual seasonal cholera peaks in Dhaka. The river systems and flood embankment of Dhaka facilitate transmission of infectious V. cholerae throughout the year with off-season outbreaks in the densely populated urban ecosystem of Dhaka. Thus clonal recycling of a newly emerged hybrid ET with increasing virulence in a time of changing climate and in a region with a growing urban population represents a serious public health concern for Bangladesh.
Conflict of interest and funding
The authors declare that they have no conflict of interests.
Acknowledgements
This research was supported in part by NIID, Tokyo and National Institutes of Health (NIH) Grant No. 1RO1A13912901 under collaborative agreements between the Johns Hopkins Bloomberg School of Public Health, the University of Maryland, College Park, and the International Center for Diarrheal Disease Research, Bangladesh (ICDDR,B), and NOAA Grant No. SO660009. This work was also supported by NIH research challenge grant (1RC1TW008587-01) under the American Recovery and Reinvestment Act (2009). The ICDDR,B acknowledges the following donors that provide unrestricted support to the Centre's research efforts: Australian Agency for International Development (AusAID), Government of the People's Republic of Bangladesh, Canadian International Development Agency (CIDA), Embassy of the Kingdom of the Netherlands (EKN), Swedish International Development Cooperation Agency (SIDA), and the Department for International Development, UK (DFID). This work was partially supported by a research challenge grant (1RC1TW008587–01) from the National Institutes of Health (NIH) under the American Recovery and Reinvestment Act (2009).
References
- Samadi AR, Huq MI, Shahid N, Khan MU, Eusof A, Rahman AS, et al.. Classical Vibrio cholerae biotype displaces El Tor in Bangladesh. Lancet. 1983; 1: 805–7. 10.3402/iee.v1i0.7273.
- Manning PA, Clark CA, Focareta T. Gene capture in Vibrio cholerae. Trends Microbiol. 1999; 7: 93–95. 10.3402/iee.v1i0.7273.
- Manning PA, Stroeher UH, Morona R. Molecular basis of O-antigen biosynthesis of Vibrio cholerae O1: ogawa-Inaba switching. Vibrio cholerae and cholera: molecular to global perspective. Wachsmuth K, Blake PA, Olsvik OASM Press. Washington DC, 1994; 77–94.
- Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002; 99: 1556–61. 10.3402/iee.v1i0.7273.
- Chanda PK, Chatterjee SN. Adsorption characteristic of a group I cholera phage. Bull Calcutta Sch Trop Med. 1973; 21: 7–8.
- Safa A, Bhuyian NA, Nusrin S, Ansaruzzaman M, Alam M, Hamabata T, et al.. Genetic characteristics of Matlab variants of Vibrio cholerae O1 that are hybrids between classical and El Tor biotypes. J Med Microbiol. 2006; 55: 1563–9. 10.3402/iee.v1i0.7273.
- Finkelstein RA. Cholera, Vibrio cholerae O1 and O139, and other pathogenic vibrios. 1996, In Baron S. Medical Microbiology. 4th ed.Texus: University of Texus.
- Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. 2002; 40: 3296–9. 10.3402/iee.v1i0.7273.
- Nair GB, Qadri F, Holmgren J, Svennerholm AM, Safa A, Bhuiyan NA, et al.. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006; 44: 4211–13. 10.3402/iee.v1i0.7273.
- Siddique AK, Nair GB, Alam M, Sack DA, Huq A, Nizam A, et al.. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol Infect. 2010; 138: 347–52. 10.3402/iee.v1i0.7273.
- Glass RI, Becker S, Huq MI, Stoll BJ, Khan MU, Merson MH, et al.. Endemic cholera in rural Bangladesh, 1966–1980. Am J Epidemiol. 1982; 116: 959–70.
- Colwell RR, Spira WM. The ecology of Vibrio cholerae. Cholera. Barua D, Greenough WB IIIPlenum Medical Book Co. New York, 1992; 107–27.
- Huq A, Colwell RR, Rahman R, Ali A, Chowdhury MA, Parveen S, et al.. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl Environ Microbiol. 1990; 56: 2370–3.
- Colwell RR. Global climate and infectious disease: the cholera paradigm. Science. 1996; 274: 2025–31. 10.3402/iee.v1i0.7273.
- Anonymous. Responding to the. 2007floods: record numbers of patients seek care at ICDDR,B's Dhaka Hospital. Health Sci Bull (English). 2007; 5: 1–5.
- Alam M, Hasan NA, Sadique A, Bhuiyan NA, Ahmed KU, Nusrin S, et al.. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl Environ Microbiol. 2006; 72: 4096–4104. 10.3402/iee.v1i0.7273.
- Alam M, Sultana M, Nair GB, Sack RB, Sack DA, Siddique AK, et al.. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl Environ Microbiol. 2006; 72: 2849–55. 10.3402/iee.v1i0.7273.
- Basu S, Mukerjee S. Bacteriophage typing of Vibrio eltor. Experientia. 1968; 24: 299–300. 10.3402/iee.v1i0.7273.
- Keasler SP, Hall RH. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993; 341: 1661. 10.3402/iee.v1i0.7273.
- Kimsey HH, Nair GB, Ghosh A, Waldor MK. Diverse CTXphis and evolution of new pathogenic Vibrio cholerae. Lancet. 1998; 352: 457–8. 10.3402/iee.v1i0.7273.
- Chowdhury NR, Chakraborty S, Ramamurthy T, Nishibuchi M, Yamasaki S, Takeda Y, et al.. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg Infect Dis. 2000; 6: 631–6. 10.3402/iee.v1i0.7273.
- Nandi S, Maiti D, Saha A, Bhadra RK. Genesis of variants of Vibrio cholerae O1 biotype El Tor: role of the CTXphi array and its position in the genome. Microbiology. 2003; 149: 89–97. 10.3402/iee.v1i0.7273.
- Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, et al.. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998; 20: 201–7. 10.3402/iee.v1i0.7273.
- Morita M, Ohnishi M, Arakawa E, Bhuiyan NA, Nusrin S, Alam M, et al.. Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol. 2008; 52: 314–17. 10.3402/iee.v1i0.7273.
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25: 3389–3402. 10.3402/iee.v1i0.7273.
- Cooper JE, Feil EJ. Multilocus sequence typing – what is resolved?. Trends Microbiol. 2004; 12: 373–7. 10.3402/iee.v1i0.7273.
- Jutla AS, Small DL, Islam S. A precipitation dipole in eastern North America. Geophys Res Lett. 2006; 33: L21703. 10.3402/iee.v1i0.7273.
- Chowdhury MR, Ward MN. Hydro-meteorological variability in the Greater Ganges-Brahmaputra-Meghna Basins. Intl J Climatol. 2004; 24: 1495–1508. 10.3402/iee.v1i0.7273.
- Schwartz BS, Harris JB, Khan AI, Larocque RC, Sack DA, Malek MA, et al.. Diarrheal epidemics in Dhaka, Bangladesh, during three consecutive floods: 1988, 1998, and 2004. Am J Trop Med Hyg. 2006; 74: 1067–73.
- Alam NH, Ashraf H. Treatment of infectious diarrhea in children. Paediatr Drugs. 2003; 5: 151–65.
- Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996; 272: 1910–14. 10.3402/iee.v1i0.7273.
- Ansaruzzaman M, Bhuiyan NA, Nair GB, Sack DA, Lucas M, Deen JL, et al.. The Mozambique Cholera Vaccine Demonstration Project Coordination Group. Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis. 2004; 10: 2057–9.
- Beltran P, Delgado G, Navarro A, Trujillo F, Selander RK, Cravioto A. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999; 37: 581–90.
- Karaolis DK, Lan R, Kaper JB, Reeves PR. Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect Immun. 2001; 69: 1947–52. 10.3402/iee.v1i0.7273.
- Karaolis DK, Lan R, Reeves PR. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995; 177: 3191–8.
- Cash RA, Music SI, Libonati JP, Snyder MJ, Wenzel RP, Hornick RB. Response of man to infection with Vibrio cholerae. I. Clinical, serologic, and bacteriologic responses to a known inoculum. J Infect Dis. 1974; 129: 45–52. 10.3402/iee.v1i0.7273.
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al.. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995; 33: 2233–9.
- Singleton FL, Attwell RW, Jangi MS, Colwell RR. Influence of salinity and organic nutrient concentration on survival and growth of Vibrio cholerae in aquatic microcosms. Appl Environ Microbiol. 1982; 43: 1080–5.
- Sack RB, Siddique AK, Longini IM-Jr, Nizam A, Yunus M, Islam MS, et al.. A 4-year study of the epidemiology of Vibrio cholerae in four rural areas of Bangladesh. J Infect Dis. 2003; 187: 96–101. 10.3402/iee.v1i0.7273.
- Mirza MMQ. Three recent extreme floods in Bangladesh: a hydro-meteorological analysis. Natural Hazards. 2003; 28: 35–64. 10.3402/iee.v1i0.7273.
- Akanda AS, Jutla AS, Islam S. Dual peak cholera transmission in Bengal Delta: a hydroclimatological explanation. Geophys Res Lett. 2009; 36: L19401. 10.3402/iee.v1i0.7273.
- Jutla AS, Akanda AS, Islam S. Tracking cholera in coastal regions using satellite observations. J Am Water Resour Assoc. 2010; 46: 651–62. 10.3402/iee.v1i0.7273.
- Jian J, Webster PJ, Hoyos CD. Large-scale controls on Ganges and Brahmaputra river discharge on intraseasonal and seasonal time-scales. Q J R Meteorol Soc. 2009; 135: 353–70. 10.3402/iee.v1i0.7273.
- Akanda AS, Jutla AS, Siddique AK, Alam M, Constantin de Magny G, Sack RB, et al. Hydroclimatic influences on seasonal and spatial cholera transmission cycles: implications for public health intervention in the Bengal Delta Water Resource Research. , 47, W00H07., 10.3402/iee.v1i0.7273.
- Jutla AS, Akanda AS, Griffiths J, Islam S, Colwell RR. Warming oceans, phytoplankton, and river discharge: implications for cholera outbreaks. Am J Trop Med Hyg 2011. 10.3402/iee.v1i0.7273.
- Carrel M, Voss P, Streatfield PK, Yunus M, Emch M. Protection from annual flooding is correlated with increased cholera prevalence in Bangladesh: a zero-inflated regression analysis. Environ Health. 2010; 9: 13. 10.3402/iee.v1i0.7273.
- Stine OC, Alam M, Tang L, Nair GB, Siddique AK, Faruque SM, et al.. Seasonal cholera from multiple small outbreaks, rural Bangladesh. Emerg Infect Dis. 2008; 14: 831–3. 10.3402/iee.v1i0.7273.
- Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet. 2004; 363: 223–33. 10.3402/iee.v1i0.7273.
- Rose JB. Environmental ecology of Cryptosporidium and public health implications. Annu Rev Public Health. 1997; 18: 135–61. 10.3402/iee.v1i0.7273.
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, et al.. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009; 106: 15442–7. 10.3402/iee.v1i0.7273.