Abstract
Introduction
The rapid and wide-scale environmental spread of multidrug-resistant bacteria in different ecosystems has become a serious issue in recent years.
Objectives
To investigate the epidemiology of antimicrobial resistance and extended spectrum beta-lactamase (ESBL) in Bangladeshi wild birds and aquatic environments, samples were taken from Open Bill Stork (Anastomus oscitans) (OBS) and the nearby water sources.
Methods
Water and fresh fecal samples were collected from several locations. All samples were processed and cultured for Escherichia coli and tested for antibiotic susceptibility against commonly used antibiotics. ESBL producers were characterized at genotypic level using polymerase chain reaction (PCR), sequencing, multilocus sequence typing, and rep-PCR.
Results and discussion
A total of 76 E. coli isolates from the 170 OBS and 8 E. coli isolates from three river sources were isolated. In total, 29% of E. coli isolated from OBS and all of the E. coli isolated from water sources were resistant to at least one of the tested antimicrobials. Resistant phenotypes were observed with all antimicrobials except tigecycline, gentamicin, imipenem, and chloramphenicol. Multidrug resistance was observed in 2.6% of OBS and 37.5% of the water isolates. Also, 1.2% of the ESBL-producing E. coli were isolated from OBS, whereas 50% of the E. coli isolated from water sources were ESBL producers possessing the CTX-M-15 gene. The most concerning aspect of our findings was the presence of human-associated E. coli sequence types in the water samples, for example, ST156-complex156, ST10-complex10 and ST46.
Conclusion
This study reports the presence of multidrug-resistant ESBL-producing E. coli in OBSs and nearby aquatic sources in Bangladesh.
Emergence and rapid dissemination of antibiotic resistance in bacteria is now a global concern. The antibiotic resistance acquired by organisms in one ecosystem can easily be transferred among organisms in different ecosystems (Citation1, Citation2). This, in turn, is responsible for wide-scale epidemic and endemic spreads of multidrug-resistant bacteria. Now, it is evident that resistant microbes are found in the different environmental compartments and reservoirs due to misuse and overuse of antibiotics and poor health-care infrastructures (Citation3, (Citation4). Surprisingly, antibiotic-resistant bacteria were found in the pristine environments where there was no direct human influence like habitation, farming, and hospitals (Citation5, Citation6).
Free-living water birds can act as reservoirs and vectors for antibiotic-resistant bacteria belonging to several species that have veterinary and medical importance (Citation3, Citation7). Due to their frequent mobility and the ease with which they pick up food from various environment, they can induce fecal contaminates in natural water reservoirs such as hoar, lakes, and rivers (Citation8, Citation9). Escherichia coli is a fecal flora in many species including birds, and the presence of E. coli in hosts differ in their ecological niches and life-history characteristics (Citation10. E. coli is an excellent indicator to study antibiotic resistance (Citation11). E. coli from wild birds were used to study environmental antibiotic resistance (Citation12, Citation13), and reasonably wild birds are considered as important environmental bioindicators and vectors of antibiotic resistance in bacteria (Citation2, Citation3, Citation12, Citation14).
Human activities, for example, generating waste water from human settlements, hospitals, farms, and pharmaceutical industries, can cause contamination of the natural water sources by resistant bacteria. Antibiotic-resistant bacteria were even found in the soil when raw cow dung and manure were used extensively as fertilizers (Citation15) and ultimately spread into natural and drinking water sources (Citation16).
Bangladesh is the seventh most populated country in the world having 58 transboundary rivers. Most of the rivers are connected with wetlands called beels and haor. These rivers, beels, and haor host thousands of local and migratory wild birds. Rivers and beel are also integrated parts of peoples’ daily lives in Bangladesh, and many human settlements are located beside or close to rivers and beel. Such settlements along with wild visitors contribute to pollution of natural water sources. The aim of this investigation was to study the extent antimicrobial resistance situation is reflected in birds and its associated ecological niches, for example, aquatic environments. Extended spectrum beta-lactamase (ESBL)-producing E. coli isolates were further characterized at genotypic level to understand the molecular epidemiology of ESBL in Bangladesh.
Materials and methods
Sample collection
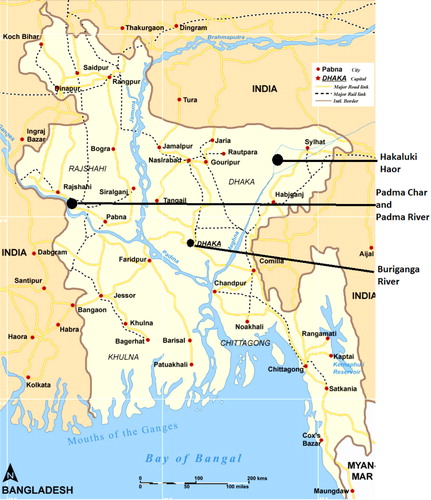
Fresh fecal droppings (n=170) were collected randomly from two different Asian Open Bill Stork (OBS) (Anastomus oscitans) colonies in Padma Char and Hakaluki haor areas () in Bangladesh from January to February 2010. Sterile cotton swabs were used to take the fresh fecal samples. All samples, immediately after collection in the field, were stored in sterile tubes containing bacterial freeze media Luria-Bertani broth (phosphate-buffered saline and 4.4% glycerol). The Padma River, previously named as the Ganges, is the biggest river in Bangladesh and on the banks of the Padma, major human settlement has developed. Due to unplanned settlement of the riverside cities, all wastewater from the city and livestock farms ended up into the rivers. The Buriganga River is another small river that passes through the capital city of Dhaka and is one of the most polluted rivers in Bangladesh (Citation17). Along with birds, water samples were also collected during that time from the Padma River, the Buriganga River, and Hakaluki haor. Forty milliliters of water samples was taken in a Falcon tube containing bacterial freeze media. From each location, 2–3 water samples were collected, having a distance of 1 km between the sampling sites. Water and fecal samples were stored in liquid nitrogen tanks and later stored at −80°C. All samples were shipped in an unbroken freeze chain for further analysis.
Bacterial isolation and identification
OBS fecal samples were taken by a sterile swab and spread over the solid surface of Cystine Lactose Electrolyte Deficient (CLED) plates (Lab M Ltd, Lancashire, UK) and incubated overnight aerobically at 37°C. At the same time, water samples were analyzed. Thirty-five milliliters of water samples was filtered by Millipore filters (0.45 µM; SAS, Molsheim, France) and immediately afterwards filter papers were placed on the CLED agar plate for overnight incubation at 37°C. Putative E. coli colonies were isolated from each CLED plate and confirmed at species level by API 20E biochemical strips (Biomerieux S.A., Marcy-l'Etoile, France).
Isolation and identification of ESBL producers
Fecal and water (15 ml) samples were enriched into the Brain–Heart Infusion broth (Becton Dickinson, Franklin Lakes, NJ, USA) containing vancomycin (16 µg/L, ICN Biomedicals, Inc., Aurora, OH, USA) and incubated overnight aerobically at 37°C. Enriched samples were spread on chromIDTM ESBL plates and incubated again overnight at 37°C. API 20E test was used to identify species of ESBL-producing E. coli. The isolates were confirmed for ESBL-production by cefpodoxime/cefpodoxime+clavulanic acid double-disk test according to EUCAST (European Committee on Antimicrobial Susceptibility Testing) and SRGA (Swedish Reference Group for Antibiotics) protocols.
Antibiotic susceptibility testing of E. coli
The disk diffusion method on Mueller Hilton plates was used to determine susceptibilities to antibiotics in accordance with EUCAST (www.eucast.org). E. coli isolates both with and without ESBL selection pressure were tested to multiple antimicrobials belonging to different chemical classes that are commonly used both in human and veterinary medicine such as gentamycin (30 µg), ciprofloxacin (5 µg), nalidixic acid (30 µg), tetracycline (30 µg), ampicillin (10 µg), chloramphenicol (30 µg), cefuroxime (30 µg) and nitrofurantoin (100 µg). Some of the antimicrobials used solely in human medicine were also included in the test; mecillinam (10 µg), tigecycline (15 µg), imipenem (10 µg) and aztreonam (30 µg). E. coli ATCC 25922 was used as a quality control strain, and breakpoints were determined according to the EUCAST (v 2.0, December 31, 2012).
DNA extraction
DNA was extracted from ESBL-producing isolates using the ‘Heating Block’ method. Two to three freshly cultured colonies were taken into a sterile Eppendorf tube containing 200 µl of double-distilled water and mixed properly. Then, this tube was heated for 10 min at 100°C and centrifuged at 13,000 rpm for 5 min. The supernatant was collected and stored at −20°C for future analysis.
Detection and genetic characterizations of ESBL producers
Polymerase chain reactions (PCRs) were performed to detect CTX-M groups I–IV using four different primers. Detection of ESBL-producing bla CTX-M genes was performed using PCR protocols described earlier (Citation18). Positive controls were K. pneumoniae U0503875 (CTX-M-I), C2 (CTX-M-II), C8 (CTX-M-III), and C9 (CTX-M-IV). PCR products that tested positive for CTX-M groups were purified using PCR product clean-up kits (Qiagen GmbH, Hilden, Germany). Purified PCR products were further sequenced by Eurofins MWG Operon (Ebersberg, Germany).
Detection of NDM-1 gene
To detect the blaNDM-1 gene, a PCR assay was used (Citation19). DNA thermal cycler GeneAmp® 9700 (Applied Biosystems Division, Foster City, CA, USA) was used with K. pneumonia CCUG60138 strain as the positive control.
Epidemiological typing of ESBL-producing E. coli
A previously described rep-PCR method was used to detect the clonal relationship among ESBL-producing isolates (Citation2, Citation20). The amplified PCR products were visualized in a 1% agarose gel stained with ethidium bromide. To determine the sizes of the PCR products, an express DNA ladder was used (Gene Ruler DNA Ladder; Ferment as, Hanover, Md.). The gel was photographed and the DNA bands were analyzed visually. Isolates differing by one strong band or more were assigned to different genotypes (Citation21), whereas isolates differing with one weak band from their genotype were assigned different subtypes.
Multilocus sequence typing
ESBL-producing E. coli strains were characterized by multilocus sequence typing (MLST; www.mlst.warwick.ac.uk/mlst/dbs/Ecoli), using specified primers for seven standard housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) according to the protocol described previously (Citation22). The PCR products were purified using a PCR purification kit (Fermentas, St. Leon-Rot, Germany). The PCR products were sequenced at Eurofins MWG Operon. Allele profiles and sequence types (STs) were determined and new allelic profile was assigned via the E. coli MLST database.
Results
We isolated 76 E. coli from 170 OBS fecal samples and 8 E. coli isolates found from 8 water samples. Of the76 E. coli isolates, 22 (28.94%) were resistant to one or more classes of the tested antimicrobials. Of them, 2.63% were multidrug resistant (resistant to three or more classes of antimicrobials) isolates. All of the E. coli isolates from water were resistant to at least one or more classes of antimicrobials and 50% were multidrug resistant. The most common resistant phenotypes for both water and OBS isolates were to ampicillin, tetracycline, nalidixic acid, and aztreonam. All of the isolates were sensitive to tigecycline, gentamycin, imipenem, and chloramphenicol. Detailed data regarding the prevalence of antibiotic-resistant phenotypes are presented in . Diversity of antibiotic resistance phenotypes among bird and water isolates is presented in . The common multiresistant pattern was CIP-NA-TE-AMP-SXT, which was found in both the rivers (Padma and Buriganga). There were two E. coli ESBL-producing bacteria found in OBS residing Padmar Char area. In total, four ESBL-producing E. coli were found in water of Padma and Buriganga Rivers. The common resistant antibiotic phenotypes were aztreonam, nalidixic acid, tetracycline, ampicillin, cefuroxime sodium, and sulfamethoxazole. There were no ESBL-producing bacteria found from the birds in Hakaluki haor and water. All the ESBL producers were multidrug resistant and CTX-M-15 positive (). E. coli STs observed between OBS and water isolates were completely different; however, the same ST (ST156 complex 156) was noticed in the Padma River. Surprisingly, several clinically relevant STs were found in rivers. The same clone profile (Clone D) was detected from OBS of Padmar Char and water from the Buriganga River.
Table 1 Antibiotic-resistant pattern of E. coli in OBS and water
Table 2 Diversity of antibiotic-resistant phenotypes in E. coli from birds and water in different locations
Table 3 The genotype, MLST, and clone profile of ESBL producers
Discussion
The fecal carriage of E. coli in OBS was high, but the carriage of antibiotic-resistant bacteria was comparatively lower than that of other birds reported previously in Bangladesh (Citation2, Citation20, Citation23, Citation24). In total, 28.95% of OBS E. coli isolate showed resistance to at least one antibiotic. E. coli isolates, to a lesser extent, were resistant to antibiotics that were commonly used in human and veterinary medicine in Bangladesh. In our study, E. coli was resistant to antibiotics like ampicillin, tetracycline, and nalidixic acid, and these resistant phenotypes were common in livestock and poultry (Citation11, Citation23). Interestingly, E. coli from OBS were found to be resistant to broadspectrum antimicrobials such as aztreonam, ciprofloxacin, mecillinam, and cefuroxime sodium. It is a matter of concern how these birds acquired resistance to human-associated antibiotics. The human surroundings and environment in Bangladesh are heavily polluted by resistant bacteria (Citation2, Citation20), which might influence the birds’ fecal flora. Factors that can influence the fecal carriage of bacteria in birds are dependent on the feeding behaviors and lifestyles of birds (Citation25. The water samples were collected from the nearby river where the OBS were foraging. From all water samples, E. coli was isolated and these isolates showed resistance to at least one antimicrobial of the tested antibiotics with 50% multidrug-resistant phenotypes in general. Water isolates had resistance to several of the tested antimicrobials including ciprofloxacin, cefuroxime, nalidixic acid, and tetracycline.
The almost pandemic spreading of ESBL-producing bacteria is of great concern. The rate of ESBL-producing E. coli has been increasing in many countries, not only restricted to humans but also reported in environmental niches like wildlife, livestock, water, and in the grounds of different locations of the world (Citation26–Citation28). In this study, ESBL-producing E. coli was low and only found in an OBS colony from Padmar Char, but not from the OBS population from Hakaluki Haor.
All ESBL-producing E. coli from water and OBS were harboring CTX-M-15 genotypes. CTX-M-15 was previously detected in wild duck, gulls, pigeons, and poultry in Bangladesh (Citation2, Citation20, Citation24). Extensively distributed CTX-M-15 genes were found in supply water from the city of Dhaka in Bangladesh (Citation29. All CTX-M-15 ESBL-producing E. coli isolates had multidrug resistance phenotypes ranging from 4 to 7 classes of antibiotics. Therefore, it can be concluded that CTX-M-15 has disseminated widely in Bangladesh, which is reflected among isolates from wild birds and water.
NDM-1 evolved from the Indian subcontinent in humans (Citation30) and later spread into different ecological niches like in the water supply, lakes, ponds, and animals (Citation31–Citation33). In turn, NDM-1 has been reported in hospitals of Bangladesh (Citation34), but no NDM-1-producing E. coli has been found in OBS or the water, indicating the spread of NDM-1 producers is limited to the hospital and that there is no dissemination yet into the environment (Citation2, Citation23).
The rep-PCR typing method explains the dissemination of clones among different reservoirs (Citation2). In this study, dissemination of specific clones was found, which was reflected in different reservoirs in different locations. For example, certain ESBL-producing E. coli clones were common in the water and in birds in different locations. Clonal types C and D were found in OBS from Padmar Char and these clones were also found in water samples from the Buriganga River even though the distance between these two places was more than 300 km. However, OBS is often used to visiting the riverside of the Buriganga, and the presence of these clones in birds and water in two different locations indicate wide-scale spread of certain ESBL-producing E. coli clones in different ecological niches.
MLST analysis of ESBL-producing E. coli revealed five different STs. There was only one found as a new ST; however, remaining four STs were reported elsewhere before. E. coli ST10 complex 10, ST156 complex 156, and ST46 were found only in water from this study. These are human-associated STs reported in different countries, such as E. coli ST10 complex 10 found from a hospital in Spain (Citation35), ST156 complex 156 found from UK hospital (Citation36), and ST46 identified in Tanzanian clinical isolates (Citation37). Human-associated STs in water indicate hospital-related contamination of water sources. Interestingly, the OBS was also carrying birds associated ST2689, which was previously found in brown headed gulls (Chroicocephalus brunnicephalus) in Bangladesh (Citation20).
Conclusion
In conclusion, it can be assumed that the multidrug-resistant bacteria and their corresponding resistance genes along with human-associated STs have already spread into different ecological niches in Bangladesh. The environment in Bangladesh is highly contaminated with resistant bacteria. This is a reflection of wide-scale scenery of resistance, which is in turn reflected through natural water sources, wild birds, and OBS species fed on mollusks from the wetlands. Nationwide surveillance studies are needed in a plethora of ecological niches to better understand the full impact of human activities on antibiotic resistance ecology in Bangladesh.
Conflict of interest and funding
This work was supported by Marcus Borgström, Ester Lindahls, PO Lundells, Bergmark, Emil och Ragna Börjessons Stiftelse, Olle Engkvist Byggmästare, and Karin Korsner's foundations.
References
- Levy S. Antibiotic resistance: an ecological imbalance. Antibiotic resistance: origins, evolution, selection and spread. Ciba Foundation Symposium 207 . 1997; Chichester: Wiley.
- Hasan B, Drobni P, Drobni M, Alam M, Olsen B. Dissemination of NDM-1. Lancet Infect Dis. 2012; 12: 99.
- Bonnedahl J, Drobni P, Johansson A, Hernandez J, Melhus A, Stedt J, etal. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J Antimicrob Chemother. 2010; 65: 1939.
- Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008; 321: 365.
- Hernandez J, Stedt J, Bonnedahl J, Molin Y, Drobni M, Calisto-Ulloa N, etal. Human-associated extended-spectrum beta-lactamase in the Antarctic. Appl Environ Microbiol. 2012; 78: 2056.
- Sjolund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, Pinhassi J, etal. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg Infect Dis. 2008; 14: 70.
- Radhouani H, Poeta P, Goncalves A, Pacheco R, Sargo R, Igrejas G. Wild birds as biological indicators of environmental pollution: antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo). J Med Microbiol. 2012; 61: 837.
- Reed KD, Meece JK, Henkel JS, Shukla SK. Birds, migration and emerging zoonoses: west nile virus, lyme disease, influenza A and enteropathogens. Clin Med Res. 2003; 1: 5.
- Hubalek Z. An annotated checklist of pathogenic microorganisms associated with migratory birds. J Wildl Dis. 2004; 40: 639.
- Gordon DM, Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003; 149: 3575.
- Sorum H, Sunde M. Resistance to antibiotics in the normal flora of animals. Vet Res. 2001; 32: 227.
- Literak I, Vanko R, Dolejska M, Cizek A, Karpiskova R. Antibiotic resistant Escherichia coli and Salmonella in Russian rooks (Corvus frugilegus) wintering in the Czech Republic. Lett Appl Microbiol. 2007; 45: 616.
- Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, etal. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One. 2009; 4: e5958.
- Dolejska M, Cizek A, Literak I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from Black-headed Gulls in the Czech Republic. J Appl Microbiol. 2007; 103: 11.
- Sahoo KC, Tamhankar AJ, Sahoo S, Sahu PS, Klintz SR, Lundborg CS. Geographical variation in antibiotic-resistant Escherichia coli isolates from stool, cow-dung and drinking water. Int J Environ Res Public Health. 2012; 9: 746.
- da Costa PM, Loureiro L, Matos AJF. Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Res Public Health. 2013; 10: 278.
- Chakraborty C, Huq MM, Ahmed S, Tabassum T, Miah MR. Analysis of the causes and impacts of water pollution of Buriganga river: a critical study. Int J Eng Res Technol. 2013; 2: 245.
- Pitout JD, Hossain A, Hanson ND. Phenotypic and molecular detection of CTX-M-beta-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol. 2004; 42: 5715.
- Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011; 17: 1791.
- Hasan B, Melhus A, Sandegren L, Alam M, Olsen B. The gull (Chroicocephalus brunnicephalus) as an environmental bioindicator and reservoir for antibiotic resistance on the coastlines of the Bay of Bengal. Microb Drug Resist. 2014; 20: 466.
- van Belkum A, Kluytmans J, van Leeuwen W, Bax R, Quint W, Peters E, etal. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J Clin Microbiol. 1995; 33: 1537.
- Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, etal. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006; 60: 1136.
- Hasan B, Faruque R, Drobni M, Waldenstrom J, Sadique A, Ahmed KU, etal. High prevalence of antibiotic resistance in pathogenic Escherichia coli from large- and small-scale poultry farms in Bangladesh. Avian Dis. 2011; 55: 689.
- Hasan B, Islam K, Ahsan M, Hossain Z, Rashid M, Talukder B, etal. Fecal carriage of multi-drug resistant and extended spectrum beta-lactamases producing E. coli in household pigeons, Bangladesh. Vet Microbiol. 2014; 168: 221.
- Feare CJ, Sanders MF, Blasco R, Bishop JD. Canada goose (Branta canadensis) droppings as a potential source of pathogenic bacteria. J R Soc Promot Health. 1999; 119: 146.
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001; 14: 933.
- Ensor VM, Shahid M, Evans JT, Hawkey PM. Occurrence, prevalence and genetic environment of CTX-M beta-lactamases in Enterobacteriaceae from Indian hospitals. J Antimicrob Chemother. 2006; 58: 1260.
- Kamruzzaman M, Shoma S, Bari SM, Ginn AN, Wiklendt AM, Partridge SR, etal. Genetic diversity and antibiotic resistance in Escherichia coli from environmental surface water in Dhaka City, Bangladesh. Diagn Microbiol Infect Dis. 2013; 76: 222.
- Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, etal. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One. 2013; 8: e61090.
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, etal. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010; 10: 597.
- Isozumi R, Yoshimatsu K, Yamashiro T, Hasebe F, Nguyen BM, Ngo TC, etal. bla(NDM-1)-positive Klebsiella pneumoniae from environment, Vietnam. Emerg Infect Dis. 2012; 18: 1383.
- Wang Y, Wu C, Zhang Q, Qi J, Liu H, Wang Y, etal. Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One. 2012; 7: e37152.
- Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011; 11: 355.
- Islam MA, Huq M, Nabi A, Talukdar PK, Ahmed D, Talukder KA, etal. Occurrence and characterization of multidrug-resistant New Delhi metallo-beta-lactamase-1-producing bacteria isolated between 2003 and 2010 in Bangladesh. J Med Microbiol. 2013; 62: 62.
- Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, etal. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother. 2011; 66: 2002.
- Oteo J, Diestra K, Juan C, Bautista V, Novais A, Perez-Vazquez M, etal. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents. 2009; 34: 173.
- Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect. 2011; 17: 1279.