Abstract
Background
Livestock animals have been the assumed source of several human epidemics in recent years, for example, influenza H1N1, rotavirus G8/G9, and MERS-CoV. Surveillance of novel viruses in animals is essential to evaluate the risk to human and animal health and to determine any economic impact, for example, failure to thrive. There is a paucity of data regarding detection and characterisation of gastroenteritis viruses, particularly novel viruses, in porcines in Ireland. Recently, a number of small novel porcine DNA viruses have emerged globally, for example, torque teno sus virus, porcine bocavirus, and parvoviruses 2 & 4, and little is known about the biology and potential pathogenicity of these viruses. Bocaparvovirus is a genetically distinct group of viruses which has been recently detected in humans and animals.
Methods
In this study, the presence of gastroenteritis viruses (rotavirus A, porcine circovirus, adenovirus, and porcine bocavirus) was investigated in a selection of archived faecal samples from asymptomatic piglets from a commercial farm in Ireland. A total of 104 specimens were pooled and screened using conventional molecular techniques (PCR and RT-PCR), a subset of specimens (n=44) were then examined individually. Viral diversity was then investigated using statistical and phylogenetic techniques.
Results
Initial screening showed a high prevalence of PBoV in this farm, with the formation of three distinct groups in phylogenetic analysis. Other viruses were also investigated in this study with the first report of PCV, PAdV and lineage I G5 RVA in Ireland. Some specimens contained >1 virus, with statistical analysis indicating a strong correlation for mixed infections of PBoV and PAdV on this farm.
Conclusion
Investigating the diversity of circulating enteric viruses on Irish porcine farms is important to improve the prophylactic tools available and to facilitate the early detection of changes in circulating viruses.
Viral diseases of animals have been under increasing scrutiny in recent years, particularly from the perspective of food safety and public health (Citation1). Livestock animals and humans have shared an interdependent relationship (albeit more in human's favour) for centuries, and constant interaction with these animals, from farm to fork, creates many opportunities for zoonotic events to occur (Citation2). Early detection and characterisation of new or emerging viruses allows information to be gathered and diagnostic tools to be developed to allow prevalence of agents to be established, with potential future benefits to human and animal health, diagnostics, and vaccine production.
Parvoviridae is a group of small, non-enveloped single-stranded DNA viruses that can infect mammals and insects. Sub-family Parvovirinae infects mammals and consists of the genera Protoparvovirus, Erythroparvovirus, Dependoparvovirus, Amdoparvovirus, Tetraparvovirus (hokovirus), and Bocaparvovirus. Members of this group can cause diseases which are considered important to human and animal health, for example, fifth disease in humans (HPV B19) and stillbirth, mummification, embryonic death, and infertility (SMEDI) in pigs (PPV). Bocaviruses (BoV) are a recently described group of DNA viruses; members are characterised based on their genetic homology to bovine parvovirus and canine minute virus. BoVs were first described in humans (Citation3) and have also been detected in canine, feline, and porcine hosts (Citation4). They are a genetically diverse group, distinct from the other members of Parvovirinae as they contain three major open reading frame (ORFs), coding for four genes (NS1, NP1, and VP1/2) (Citation5, Citation6). The presence of ORF3 is a near exclusive trait of bocavirus species (shared with PPV4), coding for NP1 protein, which is essential in viral propagation (Citation7, Citation8).
PBoV was first detected by Blomström et al. (Citation9), in piglets suffering from post-weaning multi-systemic failure with porcine circovirus 2 (PCV2) aetiology. There have been many studies to characterise this group of viruses in pigs, and, to date, five putative species of PBoV species have been described, but classification of these isolates is poorly defined (Citation10). The ninth ICTV report (International Committee of Taxonomy of Viruses) defined a new bocavirus species as any isolate with less than 95% identity in non-structural gene sequences (Citation11). However, with the surge of sequence data for this virus family, classification of isolates has become confused. For consistency, PBoV isolates will be discussed as the groups as outlined by Yang et al. and Xiao et al. (Citation10, Citation12), throughout this report. In addition, recently proposed rationalisation and extension of the taxonomy of the family Parvoviridae ((www.ictvonline.org/virusTaxonomy.asp) is also used (Citation13).
The pathogenicity of PBoV remains unclear, with viral isolates being detected in both diseased and healthy animals (Citation14–Citation16). In addition, viral DNA can be detected in a range of tissues, including lymph nodes (Citation17), serum (Citation18), lung (Citation19), saliva (Citation20), and faecal matter (Citation21). Furthermore, PBoV can be found in a high percentage of mixed infections leading to the question as to whether it is a true pathogen or opportunistic in nature, requiring activation through co-infection (Citation12, Citation22–Citation24). Most studies have used molecular techniques to detect and characterise PBoV (Citation17–Citation19, Citation21) (Citation25–Citation27), while some limited progress has been made using cell culture and immunological techniques for group 3 PBoV (Citation28, Citation29). Other gastroenteritis viruses such as group A rotavirus (RVA), porcine adenovirus (PAdV), and PCV are economically important viruses. PCV and PAdV have been associated with gastroenteritis, respiratory disease, or post-weaning multi-systemic wasting (PWMS) in piglets. RVA in pigs is attributed to severe gastrointestinal disease and accounts for 89% of diarrhoea in commercial pig farms (Citation30, Citation31); in addition RVA in pigs has been recognised to be both enzootic and epizootic, and many of the human RVA gene segments share a common origin with porcine RVA (Citation32, (Citation33).
In this study, the presence of bocavirus and other gastroenteritis viruses was investigated in archived faecal samples from a commercial pig farm in Ireland. Results highlight the diversity of gastroenteritis viruses present, also the diversity of BoV isolated from just one farm in Ireland and the detection of more than one BoV group in one host. In addition, other viruses such as adenovirus (PAdV), PCV1, and rotavirus A (RVA) were also detected and characterised. This is the first report of group 1 and 2 PBoV, PCV, and PAdV in Ireland.
Materials and methods
Sample preparation and extraction
Archived faecal specimens (n=104) which had been collected from a commercial farm in Ireland in 2007 and stored at 4°C were examined. Samples were taken from asymptomatic suckling and weaning piglets, aged from 2 to 6 weeks. Specimens were pooled prior to extraction and analysis (n=26). Nucleic acids were extracted from pooled specimens using Qiaamp DNA stool mini kit (Qiagen), following manufacturer's instructions. Viral nucleic acid was eluted in 100 µl of elution buffer and stored at −20°C prior to analysis.
For analysis of RVA in pooled specimens, nucleic acid was extracted using phenol-chloroform, with ethanol precipitation (Citation30). Viral RNA was re-suspended in 100 µl of DEPC H20 and stored at −20°C.
Detection and amplification of porcine bocavirus
PBoV was detected using three sets of primers targeting the VP1/2 region of PBoV; primers from Shan et al. (Citation34) were used to detect PBoV groups 1 and 2, while primers designed during this study were used to detect PBoV group 3 (Forward – 5′ – AAA TTG CGC CTG CGC TCA ACG – 3′ and Reverse – 5′ – TGC GTC CAA GGA AAG GCG TG – 3′). To characterise isolates, primers which amplify the NP1 gene were selected; previously published primers (Citation14) were used for group 1 PBoV; those reported by Choi et al. (Citation20) were used to characterise group 2 PBoV, while primers designed in this group were used for group 3 PBoV (Forward – 5′ – CAT ACT ACT ACC AGC GAC GG – 3′ and Reverse – 5′ – CTC GTC GGC TTT ATT GAC AGG – 3′). The PCR reactions were carried out in 50 µl volumes using the following reagents: 10 µl 5× Go Taq Flexi buffer®, 3 µl of 25 mM MgCl2, 8 µl 1.25 mM dNTP mix, 1 µl of each primer (50 pmol), 0.5 µl of 5 U/µl of Taq Polymerase (Promega, USA), and 5 µl of template.
Selected positive pooled samples were subsequently re-extracted using individual specimens, to further characterise and determine the genetic diversity of gastroenteritis viruses, in particular BoV, therein.
Detection of other DNA viruses and rotavirus
Specimens were also tested for the presence of other DNA viruses, namely adenovirus (Citation35) and PCV types 1 and 2 (Citation36), using the conditions described above. The presence of RVA was also investigated using primers specific to VP7 and VP4 genes (Citation30, Citation37). A one-step RT-PCR reaction was used for RVA using a similar reaction mix to that described previously: 10 µl 5× Go Taq Flexi buffer®, 3 µl of 25 mM MgCl2, 8 µl 1.25 mM dNTP mix, 1 µl of each primer (50 pmol), 0.5 µl of 5 U/µl Taq Polymerase (Promega, USA), 0.2 µl of 10 U/µl AMV-RT (Promega, USA), and 3 µl of template.
All reactions were carried out on a Biometra T3000 thermocycler. Amplified products were visualised on a 1.5% agarose gel stained with ethidium bromide using a UV transilluminator.
Sequencing and data analysis
Selected positive samples were cleaned using Roche High Pure PCR clean kit (Roche) and sequenced using a commercial service (GATC, Germany). For PBoV, seven specimens were sequenced; initially the VP1 gene was used in detection, while the NP1 gene was selected for sequencing as it is unique to BoV species. For group 1 PBoV, one specimen was sequenced (~1,040 bp) as this group typically displays little genetic diversity; likewise, two specimens were sequenced for group 2 PBoV (~820 bp) and four specimens were sequenced for group 3 PBoV (~1,000 bp), as these groups are more genetically diverse. For other gastroenteritis viruses (PCV, PAdV, and RVA), capsid genes were targeted, as these genes are more useful for detection purposes. Representative specimens were sequenced for PCV1 (~900 bp) and PAdV (~610 bp), as only one PCV1 positive was detected and PAdV3 has limited genetic diversity. For RVA, two specimens were sequenced to determine the G and P types, using VP7 (~1,070 bp) and VP4 (~876 bp) RT-PCR products, respectively, to examine and compare diversity of RVA to previous studies in Ireland.
Resulting sequence data were subsequently analysed and edited using BioEdit v7.0.9.0 (Citation38) and online BLAST tool (www.blast.ncbi.nlm.nih.gov/Blast.cgi). Alignment of BoV isolates was performed using MUSCLE, and the resulting alignment was used for phylogenetic analysis. The phylogeny for BoV was performed using maximum likelihood criterion as implemented in MetaPIGA v3.1 (Citation39). The dataset was evaluated for saturation and ambiguous sequences; furthermore, excessively gapped regions were trimmed using the TrimAl (Citation40) tool available in MetaPIGA. Bayesian Information Criterion was used to select the best model for the dataset; GTR model with rate heterogeneity and 1,000 bootstrap replicates was implemented and values less than 70% were collapsed.
Analysis of other viral isolates was performed using Clustal W alignment as implemented in BioEdit; the subsequent phylogenies were constructed using Neighbour-joining method as executed in Mega 6 (Citation41), using maximum composite likelihood model and 1,000 bootstrap replicates.
Taxa are displayed with accession number, virus and genotype, strain name, country of origin, and year of isolation, provided the data are available on GenBank. Isolates from this study are highlighted with a filled circle ().
Accession numbers
Sequence data obtained from this study are available on online nucleotide database GenBank, with the following accession numbers: porcine bocaparvovirus KJ923321–KJ923327, porcine adenovirus 3 KJ923328, porcine circovirus 1 KJ923329, RVA G9 KJ923330, RVA P[6] KJ923331, RVA G5 KJ923332, RVA P[13] KJ923333.
Results
Detection of PBoV and other porcine gastroenteritis viruses
In this study, 104 archived faecal specimens from a pig farm in Ireland were investigated for the presence of bocavirus and other selected gastroenteritis viruses. Initially, specimens were pooled (n=26, pools of 4) prior to analysis and results of initial screening were as follows: group 1 PBoV (2/26), group 2 PBoV (13/26), group 3 PBoV (18/26), PAdV (8/26), PCV1 (1/26), and RVA (10/26) (Table and 2 ). Subsequently, individual positive specimens were re-analysed and further characterised from 11 selected pools (n=44 individual samples). The 44 specimens were analysed individually using PCR/RT-PCR described previously; of these, 16% (n=7/44) were positive for RVA, 25% were positive for PAdV (n=11/44), 2% were positive for PCV1 and group 1 PBoV (n=1/44), 25% were positive for group 2 PBoV (n=11/44), and 57% were positive for group 3 PBoV (n=25/44). Many of the PBoV-positive specimens had more than one PBoV group present (48%, n=12/25), or were positive for other additional viruses; of these, 27% (n=12/44) had >2 viruses. RVA, PCV1, and PAdV were found in some of the PBoV-positive specimens (n=25), the most common additional virus was AdV (44%, n=11/25), while 18% of specimens appeared to have single PBoV infections. The frequency of mixed infections, including RVA, PAdV, group 2 or 3 PBoV, in individual specimens is represented using a mosaic plot (), correlation disparity is measured using Pearson residuals.
Fig. 1. Mosaic plot of viral diversity within individual specimens as displayed in ; graph constructed using R package.
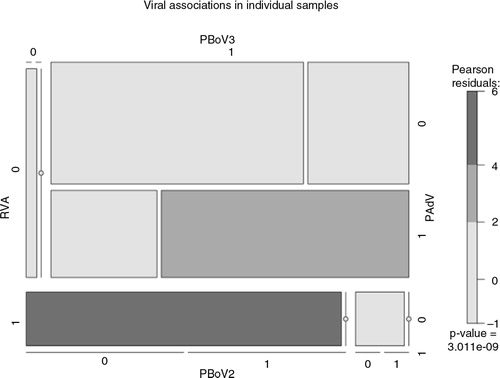
Table 1 Results of initial screening of pooled samples
Table 2 Selected pools containing positive results which were subsequently analysed individually
Phylogenetic analysis
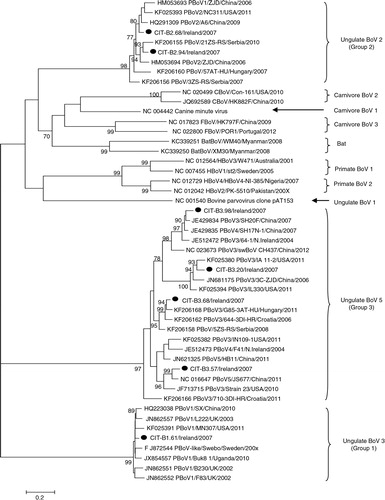
The phylogeny for PBoV isolates () reveals three major clades, similar to those described in Refs. (Citation10) and (Citation12). Group 1 and 3 PBoV form distinct monophyletic groups, in comparison to group 2 PBoV, which forms a paraphyletic group within a large monophyly, including other animal and human bocaviruses. Group 2 PBoV were more related to other animal BoV than to HBoV. Subgroups for group 3 PBoV have been proposed in previous publications (Citation10, Citation12), but, depending on the gene being investigated, these subgroups do not consistently cluster together.
Fig. 2. Maximum likelihood phylogeny of porcine bocavirus, including human and animals isolates. Isolates from this study are indicated with a filled circle ().
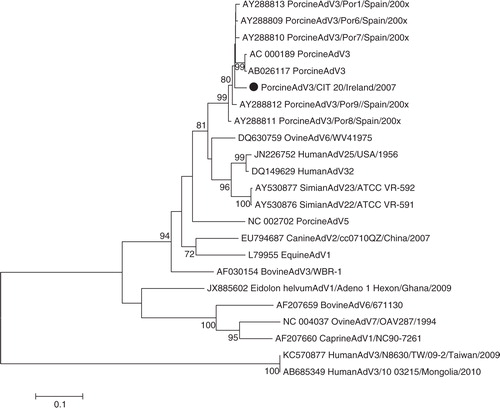
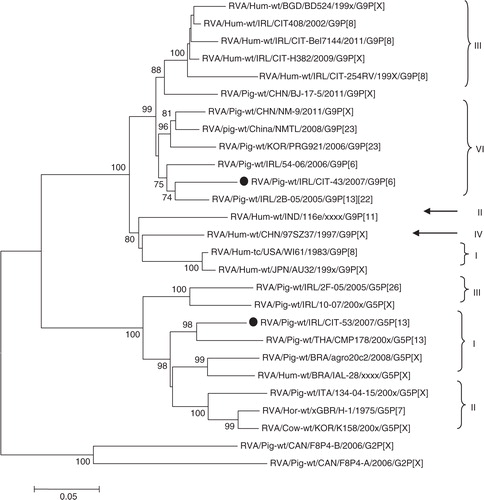
Phylogenies for other viruses were also determined; indicates that the PCV1 isolated in this study is closely related to the Hungarian wild boar isolate WB-H-8, although the corresponding bootstrap value does not support this relationship (<70%). reveals that PAdV from this study is related to PAdV 3, similar to the reference strain IAF and wild-type isolates from Spain. Two RVA isolates were sequenced to determine the G and P types, these isolates were genotyped as G5P [13] and G9P [6] ( and ). Phylogenetic analysis revealed greater genetic diversity of G5P[13] than in the G9P[6] strain. For G9P[6], the VP7 gene tree shows that G9 has a strong relationship with the Irish porcine RVA from a previous study; clustering within lineage VI, the VP4 gene tree indicates that P[6] is similar to other porcine and human strains within lineage I. For G5P[13], the tree shows more marked genetic diversity within the VP7 gene tree, G5 clusters within lineage I, a different lineage to previously described Irish isolates, and P[13] also displays greater diversity to previously isolated Irish strains.
Fig. 3. Neighbour-joining phylogeny of porcine circovirus types I and II, based on partial REP and capsid genes. Isolates from this study are indicated with a filled circle ().
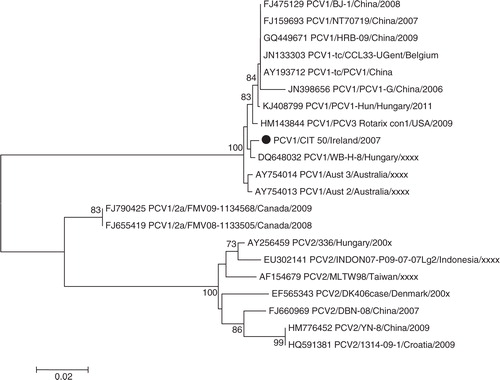
Fig. 4. Neighbour-joining phylogeny of porcine adenovirus, including other animal and human adenoviruses, based on partial hexon gene. Isolates from this study are indicated with a filled circle ().
Discussion
In this study, archived porcine faecal specimens were investigated for the presence of viruses proven or implicated in having a role in gastroenteritis, including the novel DNA virus, PBoV. Other porcine viruses (rotavirus A, porcine circovirus, adenovirus) were also investigated as there is still ongoing debate as to whether PBoV is a true pathogen or an opportunistic infection (Citation22, Citation42). In other animals, there is increasing evidence that BoV is a true pathogen found in respiratory and gastrointestinal disease cases, but the pathology is still unknown (Citation18, (Citation20) (Citation42, Citation43). In this study, 48% of PBoV-positive specimens contained more than one group of PBoV, while 48% were positive for other viruses. The most commonly detected additional virus was AdV (48%), followed by PCV and RVA, which were each detected in 4% of PBoV-positive specimens. A mosaic plot representing the frequency of mixed infections including RVA, PAdV, group 2 or 3 PBoV was constructed (), Pearson residuals suggest that there is a strong correlation between PBoV and PAdV, but further large-scale investigations are necessary to confirm this relationship. As only a subset of the total number of specimens were examined individually using PCR/RT-PCR, these figures are not completely representative but are indicative of the wide diversity of gastrointestinal viruses detected in the samples.
Since the discovery of PBoV in 2009, five putative species have been identified through the use of ICTV guidelines (<95% homology) (Citation11). Members of genus parvovirus have been suggested to have mutation rates similar to that of RNA viruses (Citation44, Citation45); in addition, BoV have been noted to undergo a high incidence of genetic recombination (Citation5, Citation21). Considering these factors, a <95% homology value was too low for accurate identification of new species. Recent release of ICTV guidelines stipulate new Parvoviridae species are identified by <85% homology of the NS1 gene. In addition, the nomenclature of taxonomic groups has also been changed and this has also been incorporated in this manuscript. Typically, three major monophyletic groups can be identified as outlined in previous reports (Citation10, Citation12); group 1 contains isolates from species 1, including original isolate Swebo; group 2 consists mainly of species 2 isolates, and group 3 contains species 3–5; in addition, group 3 contains viruses from the same species that do not cluster together.
In the BoV phylogeny displayed here (), three major monophyletic groups can be identified; two of these groups are exclusively PBoV groups 1 and 3, containing no other human or animal isolates. Group 2 PBoV, however, is part of a large monophyletic group which contains other animal and human BoV; in addition, PBoV in this group are more closely related to other animal BoV than to HBoV (). In general, Irish isolates cluster together with isolates previously described globally, displaying a high degree of diversity. For group 1 PBoV, only one isolate was sequenced; as seen from previous studies, there is very low genetic diversity in this group. CIT-B1.61 has 98.4–99.8% sequence identity to other isolates in group 1 PBoV. Group 2 PBoV isolates from this study (CIT-B2.68 and CIT-B2.94) have 93.9% identity to each other; overall, isolates within this group share 87.8–96% identity. There was greater diversity observed in group 3 PBoV in comparison. Irish strains share 73.3–83.3% sequence identity; overall, group 3 PBoV has 73–98.5% identity to global isolates within this group. Group 3 PBoV has been previously described in Northern Ireland (Citation28) and isolates CIT-B3.98 and CIT-B3.57 appear to be more closely related to these isolates; isolate CIT-B3.98 has 95.4% identity to the Irish isolate 64-1 (Accession no. JF512472), while CIT-B3.57 has 82.2% identity to isolate F41 (Accession no. JF512473).
Many of the PBoV-positive specimens described here contain either more than one group of PBoV or other potential pathogens. There is ongoing debate as to whether BoV is a true pathogen or a passenger in which co-infection is required for replication (Citation12, Citation22) (Citation23). The original report of PBoV (group 1) was in the presence of piglets that died from PWMS, with PCV2 aetiology (Citation9). Our study describes PCV1 with group 3 PBoV (CIT-20). Although PCV1 is not considered pathogenic, there is some limited data which suggest that PCV1 may have potential to cause disease (Citation46). RVA and PBoV have been reported previously in an investigation into co-infection and porcine diarrhoeal disease (Citation24), but the isolates were not genotyped. In our study, one specimen (CIT-43) was positive for both RVA (G9P[6]) and group 3 PBoV. RVA isolated from this study was for the most part similar to isolates previously isolated from Ireland ( and ) (Citation47), except for isolate CIT-53, which is the first report of lineage 1 G5 rotavirus in Ireland. To our knowledge, this is the first report of group 1 and group 2 PBoV in Ireland; additionally, PBoV in the presence PAdV. The majority of PBoV and PAdV 3 co-infections were with group 3 PBoV (44%). The issue of diarrhoeal co-infections in pigs has been investigated previously by Zhang et al. (Citation24), who reported that 75.1% of clinically diseased animals had co-infections with up to four pathogens. In our study, PBoV was isolated from asymptomatic animals, which has also been noted in previous studies (Citation14–Citation16).
The 2012 All-island Surveillance Report in Ireland on a wide variety of animals including bovines, porcines, equines, and ovines reported that enteritis was among the leading causes of mortality in pigs (Citation48). Currently, there is no vaccine licensed for the prevention of viral enteritis in piglets in Ireland and given the broad genetic heterogeneity of the strains detected in this study (and previous studies in Ireland), it remains unclear whether this diversity may pose a challenge for future prophylaxis programmes for the prevention of enteritis in piglets.
This study, along with other previous studies (Citation49), indicates that a wide variety of enteric viruses are circulating in piglets in Ireland; therefore, it will be important to establish surveillance for such viral pathogens, including porcine bocavirus, circovirus, and adenovirus, to gather information on the genetic diversity of these viruses and to plan more adequate measures of control for porcine enteritis. A clear understanding of viral diversity is still hampered by the fact that enteric viruses are not included routinely in the diagnostic tests of porcine enteritis, and the information is limited. It is likely that porcine enteritis has a multi-factorial nature, with viruses and other micro-organisms acting in synergy, yet under the influence of other factors, such as environmental and social stress due to overcrowding and/or incorrect management.
Pigs have been an assumed reservoir for previous human epidemics such as influenza H1N1 and rotavirus G9 infections. Bocavirus/Bocaparvovirus is a recently described virus genus which can be detected in humans and animals. Although animal BoVs are relatively distinct from HBoV, PBoV does appear to share greater genetic homology with HBoV, based on NS1 gene (Citation50). PBoV also appears to be much more genetically diverse in comparison to other animal and human BoV. To date, there have been no reported cases of BoV zoonosis. However, monitoring and surveillance of pathogens which can be isolated from humans and animals is important to understand the risks of zoonosis, approaches to vaccination, and implications for both human and animal health.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- Kahn LH. Confronting zoonoses, linking human and veterinary medicine. Emerg Infect Dis. 2006; 12: 556–61.
- Pulliam JRC. Viral host jumps: moving toward a predictive framework. EcoHealth. 2008; 5: 80–91.
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005; 102: 12891–6.
- Manteufel J, Truyen U. Animal bocaviruses: a brief review. Intervirology. 2008; 51: 328–34.
- Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, etal. Human Bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis. 2010; 11: 1633–43.
- Kapoor A, Slikas E, Simmonds P, Chieochansin T, Naeem A, Shaukat S, etal. A newly identified Bocavirus species in human stool. J Infect Dis. 2009; 15: 196–200.
- Sun B, Cai Y, Li Y, Li J, Liu K, Li Y, etal. The nonstructural protein NP1 of human bocavirus 1 induces cell cycle arrest and apoptosis in Hela cells. Virology. 2013; 440: 75–83.
- Zhang Z, Zheng Z, Luo H, Meng J, Li H, Li Q, etal. Human bocavirus NP1 inhibits IFN-β production by blocking association of IFN regulatory factor 3 with IFNB promoter. J Immunol Baltim Md 1950. 2012; 189: 1144–53.
- Blomström A-L, Belák S, Fossum C, McKillen J, Allan G, Wallgren P, etal. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 2009; 146: 125–9.
- Yang W, Yu J, Li J, Cheng W, Huang C, Duan Z. Genome characterization of a novel porcine bocavirus. Arch Virol. 2012; 157: 2125–32.
- King AMQ, Adams MJ, Lefkowitz EJ, Carstens EB. Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. 2012; 1463. Elsevier, San Diego.
- Xiao C-T, Halbur PG, Opriessnig T. Molecular evolutionary genetic analysis of emerging parvoviruses identified in pigs. Infect Genet Evol. 2013; 16: 369–76.
- Cotmore SF, McKenna MA, Chiorini JA, Gatherer D, Mukha DV, Pintel DJ, etal. Rationalization and extension of the taxonomy of the family Parvoviridae. International Committee on Taxonomy of Viruses; 2013. Available from: http://ictvonline.org/proposals/2013.001a-aaaV.A.v4.Parvoviridae.pdf [cited 2 February 2014]..
- McMenamy MJ, McKillen J, McNair I, Duffy C, Blomström A-L, Charreyre C, etal. Detection of a porcine boca-like virus in combination with porcine circovirus type 2 genotypes and torque teno sus virus in pigs from postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected farms in archival samples from Great Britain. Vet Microbiol. 2013; 164: 293–8.
- Meng XJ. Emerging and re-emerging swine viruses. Transbound Emerg Dis. 2012 59 85–102.
- Ndze VN, Cadar D, Cságola A, Kisfali P, Kovács E, Farkas S, etal. Detection of novel porcine bocaviruses in fecal samples of asymptomatic pigs in Cameroon. Infect Genet Evol. 2013; 17: 277–82.
- Blomström A-L, Ståhl K, Okurut AR, Masembe C, Berg M. Genetic characterisation of a porcine bocavirus detected in domestic pigs in Uganda. Virus Genes. 2013; 47: 370–3.
- Zhai S, Yue C, Wei Z, Long J, Ran D, Lin T, etal. High prevalence of a novel porcine bocavirus in weanling piglets with respiratory tract symptoms in China. Arch Virol. 2010; 155: 1313–17.
- Cságola A, Lo˝rincz M, Cadar D, Tombácz K, Biksi I, Tuboly T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch Virol. 2012; 157: 1003–10.
- Choi M-G, Park S-J, Nguyen V-G, Chung H-C, Kim A-R, Park B-K. Molecular detection and genetic analysis of porcine bocavirus in Korean domestic swine herds. Arch Virol. 2014; 159: 1487–92.
- Lau SKP, Woo PCY, Yip CCY, Li KSM, Fu CTY, Huang Y, etal. Co-existence of multiple strains of two novel porcine bocaviruses in the same pig, a previously undescribed phenomenon in members of the family Parvoviridae, and evidence for inter- and intra-host genetic diversity and recombination. J Gen Virol. 2011; 92: 2047–59.
- Schildgen O, Muller A, Allander T, Mackay IM, Volz S, Kupfer B, etal. Human bocavirus: passenger or pathogen in acute respiratory tract infections?. Clin Microbiol Rev. 2008; 21: 291–304.
- Schildgen O. Human bocavirus: lessons learned to date. Pathogens. 2013; 2: 1–12.
- Zhang Q, Hu R, Tang X, Wu C, He Q, Zhao Z, etal. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch Virol. 2013; 158: 1631–6.
- Cheng W, Li J, Huang C, Yao D, Liu N, Cui S, etal. Identification and nearly full-length genome characterization of Novel Porcine Bocaviruses. PLoS One. 2010; 5: e13583.
- Li B, Ma J, Xiao S, Fang L, Zeng S, Wen L, etal. Complete genome sequence of a novel species of porcine bocavirus, PBoV5. J Virol. 2012; 86: 1286–7.
- Wang E, Liu W, Yang B, Liu J, Ma X, Lan X. Complete sequence and phylogenetic analysis of a porcine bocavirus strain swBoV CH437. Virus Genes. 2014; 48: 387–90.
- McKillen J, McNeilly F, Duffy C, McMenamy M, McNair I, Hjertner B, etal. Isolation in cell cultures and initial characterisation of two novel bocavirus species from swine in Northern Ireland. Vet Microbiol. 2011; 152: 39–45.
- McNair I, McNeilly F, Duffy C, McKillen J, McMenamy M, Welsh M, etal. Production, characterisation and applications of monoclonal antibodies to two novel porcine bocaviruses from swine in Northern Ireland. Arch Virol. 2011; 156: 2157–62.
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, etal. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990; 28: 276–82. [PubMed Abstract] [PubMed CentralFull Text].
- Will LA, Paul PS, Proescholdt TA, Aktar SN, Flaming KP, Janke BH, etal. Evaluation of rotavirus infection and diarrhea in Iowa commercial pigs based on an epidemiologic study of a population represented by diagnostic laboratory cases. J Vet Diagn Invest. 1994; 6: 416–22.
- Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol. 2010; 140: 246–55.
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, etal. Full genome-based classification of rotaviruses reveals a common origin between human wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol. 2008; 82: 3204–19.
- Shan T, Lan D, Li L, Wang C, Cui L, Zhang W, etal. Genomic characterization and high prevalence of bocaviruses in swine. PloS One. 2011; 6: e17292.
- Maluquer de Motes C, Clemente-Casares P, Hundesa A, Martin M, Girones R. Detection of bovine and porcine adenoviruses for tracing the source of fecal contamination. Appl Environ Microbiol. 2004; 70: 1448–54.
- Carman S, Cai HY, DeLay J, Youssef SA, McEwen BJ, Gagnon CA, etal. The emergence of a new strain of porcine circovirus-2 in Ontario and Quebec swine and its association with severe porcine circovirus associated disease – 2004–2006. Can J Vet Res Rev Can Rech Vét. 2008; 72: 259–68.
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, etal. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992; 30: 1365–73. [PubMed Abstract] [PubMed CentralFull Text].
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999; 41: 95–8.
- Helaers R, Milinkovitch MC. MetaPIGA v2.0: maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics. 2010; 11: 379.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009; 25: 1972–3.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–9.
- Schildgen O. Human bocavirus: increasing evidence for virulence. Pediatr Pulmonol. 2010; 45: 118–19.
- Malecki M, Schildgen V, Schildgen O. Human bocavirus: still more questions than answers. Future Virol. 2011; 6: 1107–14.
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008; 9: 267–76.
- Shackelton LA, Parrish CR, Truyen U, Holmes EC. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci USA. 2005; 102: 379–84.
- Saha D, Lefebvre DJ, Ducatelle R, Doorsselaere JV, Nauwynck HJ. Outcome of experimental porcine circovirus type 1 infections in mid-gestational porcine foetuses. BMC Vet Res. 2011; 7: 64.
- Collins PJ, Martella V, Sleator RD, Fanning S, O'Shea H. Detection and characterisation of group A rotavirus in asymptomatic piglets in southern Ireland. Arch Virol. 2010; 155: 1247–59.
- AFBI, DAFM. All-island Animal Disease Surveillance Report 2012. 2012. Available from: http://www.afbini.gov.uk/all-island_animal__disease_surveillance_report_2012.pdf [cited 8 July 2013]..
- Collins PJ, Cullinane A, Martella V, O'Shea H. Molecular characterization of equine rotavirus in Ireland. J Clin Microbiol. 2008; 46: 3346–54. 54.
- Zeng S, Wang D, Fang L, Ma J, Song T, Zhang R, etal. Complete coding sequences and phylogenetic analysis of porcine bocavirus. J Gen Virol. 2011; 92: 784–8.

![Fig. 6. Neighbour-joining phylogeny of rotavirus A VP4 gene for genotypes P[6] and P[13]. Isolates from this study are indicated with a filled circle ().](/cms/asset/d1c94f33-9c0b-4027-a183-63da5be082ef/ziee_a_11815316_f0006_ob.jpg)