Abstract
Introduction
The genus Brachyspira contains well-known enteric pathogens of veterinary significance, suggested agents of colonic disease in humans, and one potentially zoonotic agent. There are recent studies showing that Brachyspira are more widespread in the wildlife community than previously thought. There are no records of this genus in wildlife from the southern Atlantic region and Antarctica. Our aim was therefore, to determine whether intestinal spirochaetes of genus Brachyspira colonise marine and coastal birds in this region.
Method
Faecal samples were collected from marine and coastal birds in the southern Atlantic region, including sub-Antarctic islands and Antarctica, in 2002, 2009, and 2012, with the aim to isolate and characterise zoonotic agents. In total, 205 samples from 11 bird species were selectively cultured for intestinal spirochaetes of genus Brachyspira. To identify isolates to species level, they were subjected to phenotyping, species-specific polymerase chain reactions, sequencing of partial 16S rRNA, NADH oxidase (nox), and tlyA genes, and phylogenetic analysis. Antimicrobial susceptibility tests were performed.
Results
Fourteen unique strains were obtained from 10 birds of three species: four snowy sheathbills (Chionis albus), three kelp geese (Chloephaga hybrida subsp. malvinarum), and three brown skua (Stercorarius antarcticus subsp. lonnbergi) sampled on the Falkland Islands, Tierra del Fuego in Argentina, South Georgia, South Shetland Islands, and the Antarctic Peninsula. Five Brachyspira strains were closely related to potentially enteropathogenic Brachyspira sp. of chickens: B. intermedia (n=2, from snowy sheathbills), and B. alvinipulli (n=3, from a kelp goose and two snowy sheathbills). Three strains from kelp geese were most similar to the presumed non-pathogenic species ‘B. pulli’ and B. murdochii, whereas the remaining six strains could not be attributed to currently known species. No isolates related to human strains were found. None of the tested strains showed decreased susceptibility to tiamulin, valnemulin, doxycycline, tylvalosin, lincomycin, or tylosin.
Conclusions
This is the first report of intestinal spirochaetes from this region. Despite limitations of current diagnostic methods, our results, together with earlier studies, show that Brachyspira spp., including potentially pathogenic strains, occur globally among free-living avian hosts, and that this genus encompasses a higher degree of biodiversity than previously recognised.
The southern Atlantic region and, in particular, Antarctica with its surrounding sub-Antarctic islands are remote and isolated habitats where most organisms face unique challenges in order to survive. Research on vertebrate-associated bacteria in these habitats is rare, but some recent studies have targeted human-associated and zoonotic agents in wildlife and the environment around research stations and at locations regularly visited by tourists (e.g. (Citation1, Citation2). Even less is known about the occurrence of veterinary pathogens. Thus, our aim was to determine whether intestinal spirochaetes of genus Brachyspira colonise marine and coastal birds in this region. Based on previous findings of intestinal spirochaetes in waterfowl in Australia, northern Europe, and Canada (Citation3–Citation6), we hypothesised that we would obtain isolates for characterisation.
The bacterial genus Brachyspira includes seven recognised and some proposed species. All are oxygen-tolerant, fastidious anaerobes that are highly vertebrate-associated, and they colonise the large intestine of their hosts through the faecal–oral transmission route. The genus contains well-known veterinary enteric pathogens. In pigs, B. hyodysenteriae and B. pilosicoli cause swine dysentery and spirochaetal diarrhoea, respectively. In chickens, B. alvinipulli, B. intermedia, and B. pilosicoli have been associated with decreased egg production, diarrhoea, and faecal staining of eggshells (Citation7, Citation8). Other members of this genus such as B. innocens, B. murdochii, and the proposed species ‘B. pulli’ are considered to be commensals or low-grade pathogens of livestock and poultry. Colonic spirochaetosis in humans has been reported in association with colonisation by B. aalborgi, B. pilosicoli, and ‘B. hominis’ (Citation9, (Citation10). A variety of abdominal complaints including chronic diarrhoea, irritable bowel syndrome, and invasive spirochaetaemia in terminally ill patients have been described, but the aetiological role of spirochaetes in human patients has not yet been finally established (Citation10, (Citation11). Moreover, B. pilosicoli is likely to possess zoonotic potential (Citation12.
Recent work suggests that free-living wild birds, particularly ducks and geese act as natural reservoir species of a wide variety of Brachyspira spp. (Citation3–Citation5). It has been shown that mallards (Anas platyrhynchos) may host a wide range of genotypes that represent a majority of valid and hitherto proposed species (Citation3–Citation5). Many anseriform birds are migratory and hence have the ability to disperse bacteria over long distances, and may potentially come in close contact with livestock, poultry, and people. Interestingly, the finding of the two ‘novel’ porcine enteropathogens ‘B. suanatina’ and ‘B. hampsonii’ in migrating waterfowl in northern Europe and North America gives further support to birds as relevant carrier animals (Citation6, Citation13–Citation15).
The specific objectives of this study were to collect Brachyspira isolates from free-living wild birds in the southern Atlantic region and on and around the sub-Antarctic islands and the Antarctic Peninsula, and to use well-established phenotypic and molecular techniques for characterisation and comparison to strains from other parts of the world.
Methods
Sampling, cultures, and phenotyping
Fresh faecal samples for bacterial culture were obtained from the ground from apparently healthy birds. The samples were collected from 205 adult birds of 11 species during research expeditions to the south Atlantic region and the Antarctic Peninsula during the austral spring or summer of 2002, 2009, and 2012 (). Approval to retrieve samples was granted by the Swedish Polar Research Secretariat (www.polar.se/en/). Sampling was performed at breeding sites, except for one bird that temporarily landed on the research vessel (). Intervention with any birds and marine mammal species present in the areas of sampling was kept to a minimum to safeguard wildlife welfare. Hygienic measures were applied to reduce the risk of introducing new microbes into these relatively pristine areas. Appropriate protective gear was used by the staff retrieving the samples. The birds were identified to species level by experienced ornithologists. The birds were observed until they defecated, after which the faecal sample was immediately collected with care taken to avoid contamination. The samples were obtained with sterile cotton swabs, which were stored in Amies charcoaled medium (Copan Italia SPA), and refrigerated at 5–10°C for a duration of 1–5 weeks until arrival at the laboratories in Sweden where they were cultured within 24 h (National Veterinary Institute in 2002 and the Swedish University of Agricultural Sciences in 2009 and 2012).
Table 1 Summary of collected samples from birds in the southern Atlantic region, sub-Antarctic Islands, and Antarctica
All primary isolates were obtained by selective anaerobic culture for Brachyspira spp. as previously described (Citation16). Additionally, in 2009, the samples were cultured at a lower temperature (37°C) and at less stringent selective conditions as described for isolation of B. aalborgi (Citation17). The selective plates were investigated for spirochaetal growth every third day for up to 12 days, and 1–3 areas of swarming growth of different appearances were spread on non-selective, fastidious anaerobe agar (FAA) plates (LabM) supplemented with 10% equine blood. All isolates underwent 10-fold serial dilution to obtain pure genotypes as previously described (Citation5). The primary isolates and their subcultures (n=6–11) were tested phenotypically as described (Citation18). The isolates were stored in liquid nitrogen in nutrient broth with 30% glycerol. Naming of the subcultures, e.g. Ant10:1/1/09, was made as follows: Ant=Antarctic series; 10=unique bird number; :1=the first area of growth collected from the selective plate (only used when more than one sample was collected); /1=number of subculture obtained by 10-fold serial dilution; /09=year of isolation, in this case 2009.
Subcultures with different biochemical profiles and/or different growth patterns on FAA plates were selected for further characterisation. The ability to grow in broth was investigated in brain heart infusion broth (Difco Laboratories, MI, Detroit, USA) with 10% foetal calf serum (WVR) (BHIS) at 37°C on a shaking platform. For this test, a stock solution was prepared and 300 µl aliquots were inoculated in 30 ml broth in triplicate flasks at 106 CFU/ml and read spectrophotometrically at 620 nm every third day for 15 days of incubation. Spirochaetal growth was confirmed by spectrophotometry against a non-inoculated control flask and by phase contrast microscopy to confirm culture purity and cell motility. In vitro susceptibility to tiamulin, valnemulin, doxycycline, tylvalosin, lincomycin, and tylosin was tested by broth dilution in VetMIC™ Brachy panels (SVA) in BHIS as previously described (Citation19). The minimum inhibitory concentration (MIC) was read as the lowest concentration of the antimicrobial agent that prevented visible growth. Strain B78T (B. hyodysenteriae, ATCC 27164) was used as a control.
Molecular characterisation
Boiled bacterial lysates were prepared from washed bacterial cells of the 14 studied isolates () and were used as templates at 1:1 and 1:10 dilution for polymerase chain reaction (PCR) detection of the tlyA gene of B. hyodysenteriae (Citation20), the 16S rRNA gene of B. pilosicoli (Citation20, Citation21), the nox gene of B. intermedia (Citation21, (Citation22), and the nox gene of B. innocens and/or murdochii (Citation23 with some modifications as described previously (Citation5). Appropriate negative and positive controls of porcine and avian origin and water were included. All PCR reactions were duplicated. To obtain the near-complete 16S rRNA (1,433–1,434 nts) and partial nox (893 nts) and tlyA (487 nts) sequences, PCR, DNA purification, and sequencing were performed as previously described (Citation5, Citation24–Citation26). The data were edited with the CLC Main Workbench version 6.8.3 (CLC bio, www.clcbio.com/index.php?id=28). The selection of strains used for comparative phylogeny and sequence analyses was based on NCBI GenBank nBLAST results (April 2015) and downloaded. For ‘B. hampsonii’, sequences were retrieved from the available contigs by blasting the B. hyodysenteriae WA1 16S rRNA and nox gene sequences against them. Accession numbers to sequence data downloaded from GenBank are given in . All sequences were aligned using MUSCLE (Citation27) and conserved blocks were selected using Gblocks (Citation28) with default parameters. Phylogenetic inference was performed using the maximum-likelihood method and the general time reversible model for the 16S rRNA gene, and the Jukes–Cantor model for the nox gene. Analyses were performed in MEGA6 (Citation29). Bootstrap analyses were performed using 1,000 replicates ( and ).
Fig. 1. Phylogenetic tree based on 34 nucleotide sequences of 1,267 positions each of the 16S rRNA gene showing relationships between Brachyspira isolates from the southern Atlantic region (this study, shown in bold) and previously described type, reference and field strains of this genus. The tree was rooted against the type strains of Treponema denticola (ex Flügge 1886) (aT), Leptospira interrogans serovar Icterohaemmorrhagiae (RGAT), and Borrelia burgdorferi (B31T). The evolutionary history was inferred by the maximum-likelihood method based on the general time reversible model. Evolutionary analysis was conducted in MEGA6 (Citation29). The tree is drawn to scale with branch lengths measured in the number of substitutions per site. Bootstrap values >0.5 of 1,000 replications are shown at branch points. The length of the scale bar represents 0.02 substitutions per nucleotide position.
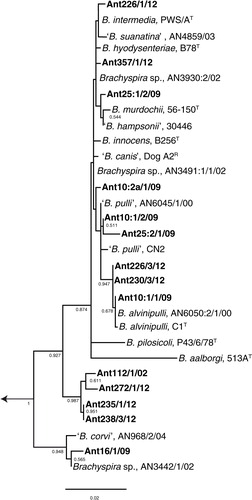
Fig. 2. Radial unrooted tree based on nox gene sequences of 27 Brachyspira strains including 12 from this study (shown in bold). There were a total of 655 positions in the final dataset. The tree was inferred by the maximum-likelihood method based on the Jukes–Cantor model. The tree is drawn to the scale with branch lengths measured in the number of substitutions per site and the analysis was conducted in MEGA6 (Citation29). Bootstrap values >0.7 of 1,000 replications are shown at branch points. The length of the scale bar represents 0.05 substitutions per nucleotide position.
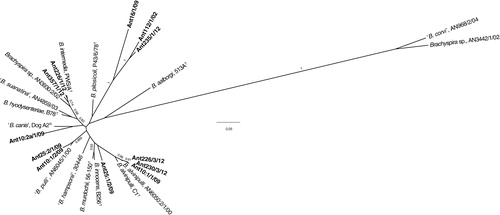
Table 2 List of investigated Brachyspira strains
Table 3 Sequence data downloaded from NCBI web page (www.ncbi.nlm.nih.gov/nucleotide) used for sequence comparisons
Results
Cultures and phenotypes
Results of culture and phenotyping are shown in Table and 2 . Intestinal spirochaetes were isolated from samples collected during all three expeditions, and from 10 out of 205 sampled birds (5%). Isolates were obtained from 3 out of 11 sampled avian species. Among culture-positive avian species, isolates were obtained from 4 out of 24 snowy sheathbills (17%), 3 out of 11 brown skuas (27%), and 3 out of 5 kelp geese (60%). At least one culture-positive bird was found at all geographical locations (Falkland Islands, Tierra del Fuego in Argentina, South Georgia Archipelago, South Shetland Island west of the Antarctic Peninsula, and on the Antarctic Peninsula itself), but not necessarily from all breeding sites.
Phenotypic results are shown in . Spirochaete growth was apparent within 3–6 days on selective plates as either poor growth of small pinpoint slow-growing colonies surrounded by a thin haze (Ant16/1/09, Ant235/1/12, and Ant238/3/12), or as moderate to profuse growth of swarming colonies (remaining isolates). At least three out of the 10 culture-positive birds were colonised by more than one spirochaetal genotype as suggested by different growth patterns, phenotypes, and molecular results of subcultures (bird number 10 and 25 in 2009, and bird 226 in 2012). All strains except Ant16/1/09, Ant25:2/1/09, Ant235/1/12, and Ant238/3/12 grew in BHIS at 37°C, and could therefore be tested for antimicrobial susceptibility by broth microdilution test. None of the 10 investigated strains showed decreased susceptibility to tiamulin, valnemulin, doxycycline, tylvalosin, lincomycin, or tylosin in vitro (). No signs of growth in BHIS were detected spectrophotometrically at any time point for up to 15 days of broth culture for the remaining four strains, and the result was confirmed by the finding of low numbers of non-motile and/or degenerated cells by phase contrast microscopy.
Table 4 Minimum inhibitory concentration (MIC) values obtained for 10 Brachyspira strains of avian origin from the southern Atlantic region, sub-Antarctic Islands, and Antartica
Molecular characterisation and species affiliation
Results of PCR analyses and GenBank accession numbers to sequence data of the 16S rRNA (n=14), nox (n=12), and tlyA (n=2) genes are shown in . A phylogenetic tree based on the 16S rRNA gene and an unrooted radial tree based on nox gene sequence data are shown in and . Two strains could not be amplified by primers targeting the nox gene despite repeated attempts (strains Ant238/3/12 and Ant272/1/12). All strains were identified as belonging to genus Brachyspira by BLASTn analysis.
The phylogenetic analysis () based on the 16S rRNA gene showed that nine of the 14 strains were evolutionary closely related to previously known Brachyspira spp. Two isolates (Ant226/1/12 and Ant357/1/12) with a phenotype consistent with the type strain PWS/AT of B. intermedia clustered together with representatives of B. hyodysenteriae, ‘B. suanatina’, B. intermedia, and a taxonomically unclassified strain from a mallard sampled in Sweden (AN3930:2/02). Because of the highly conserved nature of the 16S rRNA gene, isolates that belong to B. hyodysenteriae, ‘B. suanatina’, and B. intermedia form a common phylogenetic group and cannot be differentiated to species level based on sequence information. However, in the tree based on nox gene sequences, the two isolates (Ant226/1/12 and Ant357/1/12) formed a separate subcluster together with the B. intermedia strain and the mallard strain mentioned above. Based on a combination of phenotype, the fact that one (Ant226/1/12) was positive by the species-specific PCR for B. intermedia and the close evolutionary and nox gene sequence similarity, the two strains were considered to belong to the potential chicken enteropathogenic species B. intermedia (Ant226/1/12) or a closely related B. intermedia-like genotype (Ant357/1/12).
Three other strains, Ant10:1/1/09, Ant226/3/12, and Ant230/3/12 possessed the same phenotype as the type strain of B. alvinipulli (C1T) and clustered in both trees together with the type strain and an avian field strain suggested to belong to this species. This is another Brachyspira species with enteropathogenic potential in chickens. Another four strains (Ant25:1/2/09, Ant10:2a/1/09, Ant10:1/2/09, and Ant25:2/1/09) were each evolutionary closely related to one or more of the presumed commensal species B. innocens, B. murdochii, ‘B. canis’, or ‘B. pulli’. Strain Ant16/1/09 that was isolated from a kelp goose at the Falkland Islands () was phylogenetically closely related to two strains from earlier studies from Scandinavia: ‘B. corvi’ from corvid birds (Citation30) and Brachyspira sp. strain AN3442/1/02 from a mallard (Citation5) (). Common characteristics of these strains were poor growth with pinpoint-sized and only slightly swarming colonies on FAA, inability to grow in BHIS under conditions described in this study, and a positive result using both primer pairs targeting the species B. pilosicoli. As described in earlier studies (Citation5, Citation30), this positive reaction was caused by the presence of a B. pilosicoli signature hexa-T sequence in the 16S rRNA gene. When comparing nox sequence information (), strain Ant16/1/09 clustered together with two other strains from this study: Ant112/1/02 and Ant238/3/12. Finally, four isolates from this study formed a separate cluster in the phylogenetic tree with two subclusters (). One of these subclusters contained strains Ant112/1/02 and Ant272/1/12 from a snowy sheathbill and a brown skua, respectively, which had both been sampled on the South Shetland Islands but at different breeding sites. These two isolates showed moderate growth on agar and in BHIS, but differed in terms of phenotype and PCR results (). The other subcluster was formed by two isolates from brown skuas sampled at South Georgia Archipelago: Ant235/1/12 and Ant238/3/12. Both grew poorly on FAA, showed no growth in BHIS, but were similar in terms of phenotype and PCR results. None of the four isolates could be attributed to any known or proposed Brachyspira species.
Discussion
Here, we describe phenotypic and molecular characteristics of a unique collection of 14 spirochaetal isolates from free-living wild marine and coastal birds from the Falkland Islands, Tierra del Fuego in Argentina, sub-Antarctic islands, and the Antarctic continent. To the best of our knowledge, these isolates are the first to be described from this region. The isolates encompass seemingly well-known genotypes with enteropathogenic potential (B. intermedia and B. alvinipulli) as well as several new genotypes without taxonomic standing.
Together with earlier reports from Australia, Europe, and the Canadian Arctic region (Citation3, Citation4) (Citation6, Citation31), our results confirm that intestinal spirochaetes may be found in free-living birds in a wide variety of climate zones and habitats, and on several continents. Available data suggest that anseriform (ducks, geese, and swans) birds, especially mallards, are commonly colonised worldwide, but very little is currently known about variables that may influence the occurrence in different species. There may be host range limitations due to dietary or other physiological factors not yet revealed. In this study, only three out of the 11 bird species sampled were shown to be colonised. The sampled kelp geese likely represent a population of the subspecies Chloephaga hybrida subsp. malvinarum, which is endemic to the Falkland Islands where it mainly feeds on seaweed and algae (Citation32). Colonisation was expected because kelp geese belong to the anseriform order; however, kelp geese have a specialised diet that differs from that of mallards. The other two species, the snowy sheathbill and the brown skua, both belong to the charadriiform order (waders) of two different families: Chionidae and Stercorariidae, respectively. As far as we know, there are no previously published findings of intestinal spirochaetes in free-living wild waders, but in our laboratory we have also isolated intestinal spirochaetes from faecal samples of five ruffs (Philomachus pugnax) sampled in south-eastern Sweden (unpublished results, D.S. Jansson). Interestingly, the snowy sheathbill and the brown skua share some lifestyle traits. Both are found in sub-Antarctic and Antarctic zones and southern populations migrate to the north during the austral winter (Citation33, Citation34). Sheathbills are commonly found in seal rookeries and penguin colonies during the breeding season (Citation33). They are scavenging terrestrial birds that feed on krill and fish stolen from penguins, as well as eggs, chicks, faeces, seal placentas, carrion, and even on anthropogenic waste (Citation33). Brown skuas are mainly predatory birds that feed on eggs, nestlings, afterbirths, and carrion (Citation34). Through these opportunistic, scavenging, and kleptoparasitic feeding habits both species are likely to encounter a variety of host-associated bacteria, perhaps including Brachyspira sp.
Interestingly, in our study, only 5% of the 205 birds were culture-positive, and among the three culture-positive species between 27 and 60% of the birds were found to be colonised. However, these results should be interpreted with caution because of the limited number of samples and isolates from each location and avian species. Many Antarctic birds come to shore only to breed and therefore the only way to obtain samples is to capture them or collect their faeces at the breeding sites. Thus, our study represents a couple of snapshot situations in three different years. Another contributing factor for the seemingly low colonisation rate could have been the prolonged sample storage (1–5 weeks) before culture, which may have had a negative effect on bacterial survival. In a previous study (Citation35), prolonged sample storage of B. hyodysenteriae in Amies medium (at room temperature for 1–3 days and thereafter at 4°C) decreased the sensitivity of culture at 42 days post-sampling. Moreover, storage conditions in the field and during transport to the laboratory are likely to be more variable than during an assay performed under controlled laboratory conditions.
In 2009, an alternative selective culture protocol was applied in addition to the routine Brachyspira selective culture protocol to allow isolation of strains that require less stringent culture conditions, like B. aalborgi. For this purpose, the samples were cultured at 37°C instead of 42°C without supplementation of the agar medium by vancomycin and colistin, but with spectinomycin and polymyxin, as described in the original publication of B. aalborgi (Citation36). Both selective culture conditions produced the same results in this study, which was expected as B. aalborgi has never been isolated from birds.
In several previous studies on Brachyspira strains of chicken and mallard origin, so-called species-specific PCRs have not performed well (e.g. 5, 22). In the present study, four strains were identified by PCR as B. hyodysenteriae, one as B. intermedia, one as B. pilosicoli, and one as B. innocens or B. murdochii (). Based on the intensity of haemolysis, enzyme profiles, and sequence analyses, only one strain (Ant226/1/12, B. intermedia) appeared to be correctly identified by PCR. Thus, in accordance with other studies, this work suggests that PCR results should not be relied upon as the sole source of information regarding species affiliation of Brachyspira isolates of avian origin.
Several genotypes of presumably unknown species were isolated in our study. Based on GenBank BLAST results they were most closely related to genus Brachyspira within the phylum Spirochaetes. Among these isolates was Ant16/1/09 from a kelp goose. This isolate shared characteristics with previously described isolates from free-living wild corvid birds (Citation30) and a mallard in Scandinavia (Citation5). They clustered together in the phylogenetic tree () and displayed the same phenotype (negative spot-indole and hippurate test and lacked α-galactosidase, but possessed β-glucosidase activity). They had similar growth characteristics on FAA (poor growth of small colonies) and showed inability to grow in BHIS under conditions described in this study. Furthermore, they all displayed the signature hexa-T region of B. pilosicoli in the 16S rRNA gene. Having been repeatedly described in avian isolates from Scandinavia and now in an isolate from the Falkland Islands in yet another avian species, we suggest that this sequence feature should no longer be considered a characteristic of B. pilosicoli. As expected, isolate Ant16/1/09 was positive by both PCRs targeting B. pilosicoli because they are both based on this locus for the forward primer, although at slightly different positions.
A sister cluster was formed in the phylogenetic tree () and the nox-based dendrogram () of four isolates from this study: Ant112/1/02, Ant235/1/12, Ant238/3/12, and 272/3/12, that were isolated from a snowy sheathbill and brown skuas from the South Georgia Islands and South Shetland Islands. BLAST analysis of the 16S rRNA gene sequences showed approximately 97% similarity to other Brachyspira isolates in GenBank, and in the two isolates that were amplified by the nox primers F1 and R1, the sequence similarity was even lower (83–84% or lower). These isolates may again represent one or several unrecognised species. Three out of these four isolates were positive in the PCR protocol that detects the tlyA gene of B. hyodysenteriae, but they differed in many ways from this species.
We did not find any strains of B. pilosicoli, B. aalborgi, or the tentatively proposed species ‘B. hominis’ in this study. From this, we can infer that human influence on the spirochaetal microbiota of free-living wild birds in this habitat is apparently small. Accordingly, our results do not support a human source, which is an issue often raised in connection with increased tourist and research activities in Antarctica. In case of B. pilosicoli, there is, however, a potential risk of transfer between humans and birds. Although the source of spirochaetes is unknown, one could speculate that there is an ongoing transmission cycle between bird species and possibly between mammals and birds. Mammalian samples were however not included in this study. Migratory birds like the snowy sheathbill or the brown skua could also become colonised from faecal sources at other sites during the austral winter season.
In this study, the results of combined phenotypic and molecular approaches of strain classification have generated a deeper insight into the biodiversity of intestinal spirochaetes, and has enabled preliminary identification of strains from aquatic birds in the southern Atlantic region and on and around the Antarctic Peninsula. Nevertheless, further efforts, such as whole-genome sequencing and investigations into the ability to colonise the chicken intestine, are needed to identify traits associated with bacterial adaptation to the avian intestinal environment, and to improve the biological and taxonomic understanding of this complex group of spirochaetes.
Conflict of interest and funding
The authors declare that there are no conflicts of interest.
References
- Bonnedahl J, Broman T, Waldenström J, Palmgren H, Niskanen T, Olsen B. In search of human-associated bacterial pathogens in Antarctic wildlife: report from six penguin colonies regularly visited by tourists. Ambio. 2005; 34: 430–2. doi: http://dx.doi.org/10.1639/0044-7447(2005)034[0430:ISOHBP]2.0.CO;2 [PubMed Abstract].
- Hernández J, Stedt J, Bonnedahl J, Molin Y, Drobni M, Calisto-Ulloa N, etal. Human-associated extended-spectrum β-lactamase in the Antarctic. Appl Environ Microbiol. 2012; 78: 2056–8. doi: http://dx.doi.org/10.1128/AEM.07320-11.
- Oxberry SL, Trott DJ, Hampson DJ. Serpulina pilosicoli, waterbirds and water: potential sources of infection for humans and other animals. Epidemiol Infect. 1998; 121: 219–25. doi: http://dx.doi.org/10.1017/S0950268898008863 [PubMed Abstract] [PubMed CentralFull Text].
- Jansson DS, Johansson K-E, Olofsson T, Råsbäck T, Vågsholm I, Pettersson B, etal. Brachyspira hyodysenteriae and other strongly beta-haemolytic and indole-positive spirochaetes isolated from mallards (Anas platyrhynchos). J Med Microbiol. 2004; 53: 293–300. doi: http://dx.doi.org/10.1099/jmm.0.05488-0 [PubMed Abstract].
- Jansson DS, Persson M, Zimmerman U, Johansson K-E. Phenotypic and genetic diversity among intestinal spirochaetes (genus Brachyspira) in free-living wild mallards (Anas platyrhynchos) sampled in southern Sweden. Syst Appl Microbiol. 2011; 34: 566–75. doi: http://dx.doi.org/10.1016/j.syapm.2011.10.001 [PubMed Abstract].
- Rubin JE, Harms NJ, Fernando C, Soos C, Detmer SE, Harding JC, etal. Isolation and characterization of Brachyspira spp. including “Brachyspira hampsonii” from lesser snow geese (Chen caerulescens caerulescens) in the Canadian Arctic. Microb Ecol. 2013b; 66: 813–22. doi: http://dx.doi.org/10.1007/s00248-013-0273-5.
- Hampson D. Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW. Brachyspiral colitis. Diseases of swine . 2012; Ames, IA: Wiley Blackwell. 680–96. 10th ed.
- Mappley LJ, La Regione RM, Woodward MJ. Brachyspira and its role in avian intestinal spirochaetosis. Vet Microbiol. 2014; 168: 245–60. doi: http://dx.doi.org/10.1016/j.vetmic.2013.11.019 [PubMed Abstract].
- Pettersson B, Wang M, Fellström C, Uhlén M, Molin G, Jeppsson B, etal. Phylogenetic evidence for novel and genetically different intestinal spirochetes resembling Brachyspira aalborgi in the mucosa of the human colon as revealed by 16S rDNA analysis. Syst Appl Microbiol. 2000; 23: 355–363.
- Westerman LJ, Stel HV, Schipper MEI, Bakker LJ, Neefjes-Borst EA, van den Brande JHM, etal. Development of a real-time PCR for identification of Brachyspira species in human colonic biopsies. PLoS One. 2012; 7: e52281. doi: http://dx.doi.org/10.1371/journal.pone.0052281 [PubMed Abstract] [PubMed CentralFull Text].
- Walker MM, Talley NJ, Inganäs L, Engstrand L, Jones MP, Nyhlin H, etal. Colonic spirochetosis is associated with colonic eosinophilia and irritable bowel syndrome in a general population in Sweden. Hum Pathol. 2015; 46: 277–83. doi: http://dx.doi.org/10.1016/j.humpath.2014.10.026 [PubMed Abstract].
- Hampson DJ, Oxberry SL, La T. Potential for zoonotic transmission of Brachyspira pilosicoli. Emerg Infect Dis. 2006; 12: 869–70. doi: http://dx.doi.org/10.3201/eid1205.051180 [PubMed Abstract] [PubMed CentralFull Text].
- Råsbäck T, Jansson DS, Johansson K-E, Fellström C. A novel enteropathogenic, strongly haemolytic spirochaete isolated from pig and mallard, provisionally designated ‘Brachyspira suanatina’ sp. nov. Environ Microbiol. 2007; 9: 983–91. doi: http://dx.doi.org/10.1111/j.1462-2920.2006.01220.x.
- Rubin JE, Costa MO, Hill JE, Kittrell HE, Fernando C, Huang Y, etal. Reproduction of mucohaemorrhagic diarrhea and colitis indistinguishable from swine dysentery following experimental inoculation with “Brachyspira hampsonii” strain 30446. PLoS One. 2013a; 8: e57146. doi: http://dx.doi.org/10.1371/journal.pone.0057146.
- Costa MO, Hill JE, Fernando C, Lemieux HD, Detmer SE, Rubin JE, etal. Confirmation that “Brachyspira hampsonii” clade I (Canadian strain 30599) causes mucohemorrhagic diarrhea and colitis in experimentally infected pigs. BMC Vet Res. 2014; 10: 129. doi: http://dx.doi.org/10.1186/1746-6148-10-129 [PubMed Abstract] [PubMed CentralFull Text].
- Fellström C, Gunnarsson A. Phenotypical characterization of intestinal spirochaetes isolated from pigs. Res Vet Sci. 1995; 59: 1–4. doi: http://dx.doi.org/10.1016/0034-5288(95)90021-7.
- Kraaz W, Pettersson B, Thunberg U, Engstrand L, Fellström C. Brachyspira aalborgi infection diagnosed by culture and 16S ribosomal DNA sequencing using human colonic biopsy specimens. J Clin Microbiol. 2000; 38: 3555–60. [PubMed Abstract] [PubMed CentralFull Text].
- Fellström C, Karlsson M, Pettersson B, Zimmerman U, Gunnarsson A, Aspán A. Emended descriptions of indole negative and indole positive isolates of Brachyspira (Serpulina) hyodysenteriae. Vet Microbiol. 1999; 70: 225–38. doi: http://dx.doi.org/10.1016/S0378-1135(99)00146-7.
- Karlsson M, Fellström C, Gunnarsson A, Landén A, Franklin A. Antimicrobial susceptibility testing of porcine Brachyspira (Serpulina) species isolates. J Clin Microbiol. 2003; 41: 2596–2604. doi: http://dx.doi.org/10.1128/JCM.41.6.2596-2604.2003 [PubMed Abstract] [PubMed CentralFull Text].
- Råsbäck T, Fellström C, Gunnarsson A, Aspán A. Comparison of culture and biochemical tests with PCR for detection of Brachyspira hyodysenteriae and Brachyspira pilosicoli. J Microbiol Methods. 2006; 66: 347–53. doi: http://dx.doi.org/10.1016/j.animet.2005.12.008.
- Phillips ND, La T, Hampson DJ. A cross-sectional study to investigate the occurrence and distribution of intestinal spirochaetes (Brachyspira spp.) in three flocks of laying hens. Vet Microbiol. 2005; 105: 189–98. doi: http://dx.doi.org/10.1016/j.vetmic.2004.10.016 [PubMed Abstract].
- Jansson DS, Fellström C, Råsbäck T, Vågsholm I, Gunnarsson A, Ingermaa F, etal. Phenotypic and molecular characterization of Brachyspira spp. isolated from laying hens in different housing systems. Vet Microbiol. 2008; 130: 348–62. doi: http://dx.doi.org/10.1016/j.vetmic.2008.02.010 [PubMed Abstract].
- Atyeo RF, Stanton TB, Jensen NS, Suriyaarachichi DS, Hampson DJ. Differentiation of Serpulina species by NADH oxidase gene (nox) sequence comparisons and nox-based polymerase chain reaction tests. Vet Microbiol. 1999; 67: 47–60. doi: http://dx.doi.org/10.1016/S0378-1135(99)00030-9 [PubMed Abstract].
- Trott DJ, Stanton TB, Jensen NS, Duhamel GE, Johnson JL, Hampson DJ. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int J Syst Bacteriol. 1996; 46: 206–15. doi: http://dx.doi.org/10.1099/00207713-46-1-206 [PubMed Abstract].
- Rodhe J, Rothkamp A, Gerlach GF. Differentiation of porcine Brachyspira species by a novel nox PCR-based restriction fragment length polymorphism analysis. J Clin Microbiol. 2002; 40: 2598–600. doi: http://dx.doi.org/10.1128/JCM.40.7.2598-2600.2002.
- Johansson K-E, Duhamel GE, Bergsjö B, Olsson Engvall E, Persson M, Pettersson B, etal. Identification of three clusters of canine intestinal spirochaetes by biochemical and 16S rDNA sequence analysis. J Med Microbiol. 2004; 53: 345–50. doi: http://dx.doi.org/10.1099/jmm.0.05479-0 [PubMed Abstract].
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32: 1792–7. doi: http://dx.doi.org/10.1093/nar/gkh34 [PubMed Abstract] [PubMed CentralFull Text].
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000; 17: 540–52.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–9. doi: http://dx.doi.org/10.1093/molbev/mst197 [PubMed Abstract] [PubMed CentralFull Text].
- Jansson DS, Fellström C, Johansson K-E. Intestinal spirochetes isolated from wild-living jackdaws, hooded crows and rooks (genus Corvus): provisionally designated “Brachyspira corvi” sp. nov. Anaerobe. 2008; 14: 287–95. doi: http://dx.doi.org/10.1016/j.anaerobe.2008.09.002 [PubMed Abstract].
- Martínez-Lobo FJ, Hidalgo Á, García M, Argüello H, Naharro G, Carvajal A, etal. First identification of “Brachyspira hampsonii” in wild European waterfowl. PLoS One. 2013; 8: e82626. doi: http://dx.doi.org/10.1371/journal.pone.0082626.
- Carboneras C., Order ANSERIFORMES . del Hoyo J, Elliott A, Sargatal J. Family ANATIDAE (ducks, geese and swans). Handbook of the birds of the world, Vol. 1 (Ostrich to ducks) . 1996; Barcelona: Lynx Edicions. 546–55. 1st ed.
- Burger AE , del Hoyo J, Elliott A, Sargatal J . Order CHARADRIIFORMES. Family CHIONIDAE (sheathbills). Handbook of the birds of the world, Vol. 3 (Hoatzin to Auks) . 1996; Barcelona: Lynx Edicions. 546–55. 1st ed.
- Furness RW. del Hoyo J, Elliott A, Sargatal J. Order CHARADRIIFORMES. Family STERCORARIIDAE (skuas). Handbook of the birds of the world, Vol. 3 (Hoatzin to Auks) . 1996; Barcelona: Lynx Edicions. 556–71. 1st ed.
- Fellström C, Zimmerman U, Gunnarsson A. The use of culture, pooled samples and PCR for identification of herds infected with Brachyspira hyodysenteriae. Anim Health Res Rev. 2001; 2: 37–43.
- Hovind-Hougen K, Birch-Andersen A, Henrik-Nielsen R, Orholm M, Pedersen JO, Teglbjærg PS, etal. Intestinal spirochetosis: morphological characterization of the spirochete Brachyspira aalborgi gen. nov., sp. nov. J Clin Microbiol. 1982; 16: 1127–6. [PubMed Abstract] [PubMed CentralFull Text].
