Abstract
Introduction
Escherichia coli O103:H2 occurs as verotoxigenic E. coli (VTEC) carrying only vtx 1 or vtx 2 or both variants, but also as vtx-negative atypical enteropathogenic E. coli (aEPEC). The majority of E. coli O103:H2 identified from cases of human disease are caused by the VTEC form. If aEPEC strains frequently acquire verotoxin genes and become VTEC, they must be considered a significant public health concern. In this study, we have characterized and compared aEPEC and VTEC isolates of E. coli O103:H2 from Swedish cattle.
Methods
Fourteen isolates of E. coli O103:H2 with and without verotoxin genes were collected from samples of cattle feces taken during a nationwide cattle prevalence study 2011–2012. Isolates were sequenced with a 2×100 bp setup on a HiSeq2500 instrument producing >100× coverage per isolate. Single-nucleotide polymorphism (SNP) typing was performed using the genome analysis tool kit (GATK). Virulence genes and other regions of interest were detected. Susceptibility to transduction by two verotoxin-encoding phages was investigated for one representative aEPEC O103:H2 isolate.
Results and Discussion
This study shows that aEPEC O103:H2 is more commonly found (64%) than VTEC O103:H2 (36%) in the Swedish cattle reservoir. The only verotoxin gene variant identified was vtx 1a . Phylogenetic comparison by SNP analysis indicates that while certain subgroups of aEPEC and VTEC are closely related and have otherwise near identical virulence gene repertoires, they belong to separate lineages. This indicates that the uptake or loss of verotoxin genes is a rare event in the natural cattle environment of these bacteria. However, a representative of a VTEC-like aEPEC O103:H2 subgroup could be stably lysogenized by a vtx-encoding phage in vitro.
Pathogenic Escherichia coli have evolved by the acquisition of mobile genetic elements containing virulence and fitness factors through horizontal gene transfer (Citation1). Enteropathogenic E. coli (EPEC) are characterized by variants of the locus of enterocyte effacement (LEE), a genetic element that encodes intimin and components for a type III secretion system as well as several other virulence factors. These factors allow EPEC to cause attaching and effacing (A/E) lesions in the human intestinal epithelium (Citation2). Most human-pathogenic verotoxigenic E. coli (VTEC, also known as STEC) originate from populations of EPEC lysogenized by one or more phages carrying genes encoding verotoxin 1 and/or verotoxin 2 (vtx 1 , vtx 2 ) (Citation1). Although typical human-pathogenic EPEC carry the EPEC adherence factor plasmid, this plasmid is frequently absent in VTEC-like EPEC. Therefore, these strains are considered ‘atypical’ EPEC (aEPEC) (Citation2). Ruminants are the natural reservoir of many forms of aEPEC and VTEC (Citation2) and are colonized with a high prevalence and great diversity of these bacteria, but rarely develop symptoms as a result. Both VTEC and the different types of EPEC can cause diarrhea in infected humans, but VTEC can also cause more severe forms of bloody diarrhea and occasionally fatal complications like hemolytic uremic syndrome (HUS) (Citation3). The E. coli O157:H7 serotype is the most common cause of human VTEC infection in Sweden as well as in many countries worldwide, but non-O157 serogroups including O26, O103, and O121 also cause a significant number of cases every year (Citation4). A major outbreak of enteroaggregative VTEC O104:H4 in 2011 caused more than 4,000 cases in several European countries, including Sweden (Citation5),and led to an increased interest in the role of verotoxin-encoding phages in the emergence of new VTEC types.
Verotoxin-encoding prophages can be stably maintained in the bacterial genome, but at least some strains of VTEC can lose the prophage and thereby the ability to produce the verotoxin, reverting to the aEPEC state (Citation6, Citation7). During VTEC outbreaks, strains that are highly similar to the outbreak strain but lack verotoxin genes can sometimes be isolated from patients or suspected sources (Citation6–Citation8). Conversely, several studies have described successful experimental infection by verotoxin-encoding phages of a broad range of E. coli strains as well as of Shigella sonnei, while in vitro production of stable lysogens appears to be more challenging (Citation9–Citation12). The E. coli serotype O103:H2 occurs as verotoxin-negative aEPEC as well as VTEC carrying only vtx 1 or vtx 2 or both variants (Citation13, Citation14). The verotoxin-negative variants have been shown to be the dominant O103:H2 population in ruminant reservoirs (Citation15, Citation16), but E. coli O103:H2 with vtx 1 is the most common cause of human VTEC illness (Citation17–Citation19).
The high prevalence of aEPEC O103:H2 in animals poses a potential threat for the human population. Assuming that frequent conversions by lysogeny of prophages occur in the animal host, in contaminated foodstuffs or even in the gastrointestinal tract of an infected human, the aEPEC strains must be considered a significant risk. In contrast, if the aEPEC and VTEC strains we find in ruminants represent distinct lineages, the conversions are likely to be rare events and the aEPEC strains less of a public health concern. In this study, we have compared aEPEC and VTEC isolates of E. coli O103:H2 from the Swedish cattle reservoir using high-throughput sequencing. We have also investigated the in vitro ability of strains of VTEC O103:H2 to revert to the aEPEC state and the ability of an aEPEC O103:H2 to acquire verotoxingenes by phage transduction.
Materials and methods
A nationwide VTEC O157:H7 prevalence study was performed on cattle at slaughter in Sweden during a one-year period, 2011–2012. One thousand fecal samples and 500 skin (ear) samples were collected from major slaughterhouses, with the number of samples proportional to the number of animals slaughtered at each slaughterhouse. From each collected sample, 10 g of feces or approximately 25 g of skin was pre-enriched in 90 ml of buffered peptone water (BPW) for 6–8 h at 37°C. Aliquots of 1.5 ml were stored at −70°C with 10–15% glycerol added. All samples were screened prior to freezing by real-time polymerase chain reaction (PCR) for the O103 wzx gene as follows. After enrichment, 10 µl of BPW was streaked on MacConkey agar and incubated overnight at 37°C. From the MacConkey plates, DNA extractions were prepared by the suspension of colony material in 500 µl nuclease-free water and heat lysis at 100°C for 10 min. The suspensions were then centrifuged at 13,000×g for 10 min and the supernatant was used as PCR template. PCR was performed according to Perelle et al. (Citation20), with the following modifications: PCR was performed in 15-µl reaction volumes containing 2X PerfeCTa qPCR Toughmix with Low ROX (Quanta Biosciences, Gaithersburg, MD), 500 nM of each primer, 100 nM of probe, and 2 µl template. The PCR was run in an ABI 7500 Fast thermal cycler (Applied Biosystems, Foster City, CA) with the following temperature profile: 95°C for 3 min followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. For this study, E. coli O103:H2 was isolated from a subset of the stored samples based on low C t value when screening for the O103 wzx gene. For the isolation, the frozen BPW was thawed and mixed with 9 ml fresh BPW and incubated for 4 h at 37°C. One milliliter of each sample was then transferred to 90 ml modified tryptic soy broth with 8 mg novobiocin added and incubated at 41.5°C for 16–20 h. Immunomagnetic separation of these enriched samples was performed using the Dynabeads EPEC/VTEC O103 kit (Life Technologies, Carlsbad, USA), according to the manufacturer's instructions. The beads were distributed on three washed sheep blood agar plates. The plates were supplemented with 8 mg/l vancomycin and 0.05 mg/l cefixime to inhibit unspecific growth, in particular, gram-positive bacteria and swarming Proteus spp., respectively. The plates were incubated at 37°C for 18–24 h. Hemolytic colonies were tested by slide agglutination using sera for the O103 antigen (SSI, Copenhagen, Denmark). Agglutination-positive colonies were confirmed by PCR for the presence of the intimin eae gene (Citation21), the O103 wzx gene (Citation20), and the H2 fliC gene (Citation22) as well as by E. coli O103:H2-specific CRISPR PCR (Citation23). A single isolate from each positive sample was included in the study. A total of 14 isolates were collected and verified in this way and available for sequencing. Isolates that did not fulfill the criteria for aEPEC (intimin) or VTEC (any verotoxin) of the serotype O103:H2 were excluded from further analysis.
For next-generation sequencing, DNA was extracted from colonies using the MasterPure DNA Purification kit (Epicentre, Madison, USA). Sequencing was performed using TruSeq libraries analyzed on a HiSeq2500 instrument (Illumina, San Diego, USA) with a 2×100 bp paired-end setup producing >100× average coverage per isolate. Read data sets were uploaded to the European Nucleotide Archive under project accession number PRJEB11451. For single-nucleotide polymorphism (SNP) typing, raw read data were trimmed and cleaned from duplicates using PrinSeq (Citation24). The trimmed reads were mapped against the complete genome of the E. coli O103:H2 12009 strain (GenBank Acc. AP010958) (Citation25) using Bowtie 2 (Citation26). Local realignment and SNP discovery were performed using the Genome analysis toolkit (GATK) (Citation27), with variants called using the Unified Genotyper algorithm. SNPs were filtered using the following parameters: QUAL (overall probability of polymorphism at position) > 1,000, QD (quality normalized by depth) > 2, FS (strand bias) < 60, ReadPosRankSumTest (alternative/reference allele positional bias) > − 8, MQ (overall mapping quality) > 40, and MQRankSum (alt/ref allele mapping quality bias) > − 12.5. Only SNPs in the chromosomal backbone of the reference were used in the SNP analysis, that is, prophages and integrative elements were excluded. The aligned SNP states in the different isolates were compared using the NeighborNet algorithm in SplitsTree4 (Citation28). Virulence genes were detected in the read data using the CGE VirulenceFinder web tool (Citation29), and the results were verified and manually corrected when necessary, using Bowtie 2 mapped reads against the virulence gene reference sequences.
To test the stability of the verotoxin-encoding prophages in vitro, aliquots of a fecal sample that had been tested and found to be free of any E. coli O103 were spiked with eight vtx 1 -positive E. coli O103:H2 strains. These fecal samples were then treated like the other project samples as previously described, that is, enriched, frozen, thawed, and enriched again for the re-isolation of the spiked-in isolates. Experimental transduction by verotoxin-encoding phages φ731 (Δvtx2::cat), originally derived from VTEC O103:H25 (Citation30, Citation31), and 933 W (Δvtx2::cat) (Citation32), originally derived from VTEC O157:H7, was attempted for a single isolate of verotoxin-negative E. coli O103:H2 as previously described (Citation11) with some modifications. Phage lysates were prepared from donor strains (E. coli C600 with phages 933 W, vtx2::cat or φ731, vtx2::cat). Donor strains were grown in LB-broth until the OD measured 0.3–0.5. Mitomycin C (Sigma-Aldrich, Saint Louis, USA) was added to a final concentration of 0.5 µg/ml and incubation was continued at 37°C overnight. Debris was removed by centrifugation at 3,000×g for 15 min and the supernatant was filtered (pore size 0.22 µM) to make a sterile phage lysate. The recipient strain was grown to OD 0.3–0.5 in LB-broth to attain a log-phase growing culture. To 4 ml of LB-broth, 500 µl phage lysate and 500 µl culture of recipient were added. After incubation for 4 h at 37°C, 100 µl was plated on LB-agar containing 25 µg/ml chloramphenicol. The plates were incubated overnight at 37°C before they were checked for resistant strains. Any growth was subsequently re-streaked and incubated on new chloramphenicol plates twice to isolate stably infected strains and remove any free phages that might interfere with PCR confirmation. To confirm that the resistant strains had been infected with the cat-carrying phage as opposed to becoming resistant to chloramphenicol by other means, the cat-gene was amplified using primers Cm-3 and Rho for φ731, and primers VCJ-F2 and VCJ-R2 for 933 W (). PCR was performed with a T100 thermal cycler (Bio-Rad, Hercules, USA) in 20 µl reactions containing 1 µl template DNA, 400 nM of each primer and 2X Multiplex PCR mastermix (Qiagen, Hilden, Germany). As expected, donor strains were positive and the recipient strain was negative for the presence of vtx2::cat. Four presumptive transductants were tested with PCR. The two isolates infected with φ731 (450 and 451) were PCR positive, confirming successful phage transduction. In contrast, the two isolates apparently infected with φ933 W (444 and 445) were PCR negative (), indicating that these isolates might have decreased sensitivity towards chloramphenicol resulting from mutations rather than from transduction with the 933 W phage.
Table 1 Primers used for confirmation of phage-infected isolates
Table 2 Strains and isolates used in this study
Results and discussion
Among cattle fecal samples originally collected for a nationwide O157:H7 prevalence study, we found vtx-negative (64%) and vtx 1a -positive (36%) isolates of intimin-positive E. coli O103:H2 in cattle, but no O103:H2 isolates with vtx 2 . In agreement, Sekse et al. found that less than 20% of intimin-positive E. coli O103:H2 isolates from Norwegian sheep encoded vtx 1 genes, while vtx 2 was completely absent (Citation15). Furthermore, a recent study in Australia identified only intimin-positive E. coli O103:H2 isolates from ruminants and none that were verotoxin positive (Citation16). Thus, the most common VTEC-like O103:H2 population in ruminants appears to be intimin positive, but without verotoxin genes. A possible alternative explanation would be that the verotoxin genes are lost during the isolation procedure. To test this, we added eight vtx 1 -positive isolates to portions of an O103-negative cattle fecal sample and re-isolated them, with all isolates remaining verotoxin positive.
The primary goal of this study was to characterize and compare E. coli O103:H2 isolates. Therefore, the isolation of E. coli O103:H2 was not attempted from all samples in the prevalence study, and no prevalence of VTEC/aEPEC O103:H2 in Swedish cattle can be reliably calculated from the data. In addition to the VTEC and aEPEC described here, we found an abundance of E. coli O103:H2 with a hemolysis phenotype that lacked both verotoxin and intimin. For many of these avirulent isolates, the O103:H2 CRISPR PCR (Citation23) for specific detection of E. coli O103:H2 was positive, suggesting these isolates are phylogenetically related to aEPEC and VTEC O103:H2. Similar findings were reported from a prevalence study of E. coli O103:H2 in Norwegian sheep (Citation15). These incidental findings can be a problem when trying to find a particular strain of O103:H2 in ruminants, for example, for investigating suspected cases of infection from animals to humans.
SNP typing of the 14 E. coli aEPEC and VTEC O103:H2 isolates separated them into five major groups, two of which consisted only of VTEC and the remaining three only of aEPEC (). The fact that VTEC and aEPEC form separate lineages in this limited set of isolates might indicate that the uptake or loss of verotoxin-encoding phages is a rare event in the natural cattle environment of E. coli O103:H2. aEPEC O103:H2 may consequently pose less of a risk for human health than we could have assumed if any lineage contained both verotoxin states. As more E. coli O103:H2 genome sequences become available in the next few years, it will be evident whether this observation also holds for lineages of E. coli O103:H2, which are absent in the Swedish cattle reservoir, for example, those carrying vtx 2 .
Fig. 1 Virulence gene repertoire (left) and single-nucleotide polymorphism variation network (right), inferred from next-generation sequencing of Swedish cattle isolates of E. coli O103:H2. Black squares indicate present genes, white absent. For efa1, a ‘T’ indicates the presence of a truncated gene.
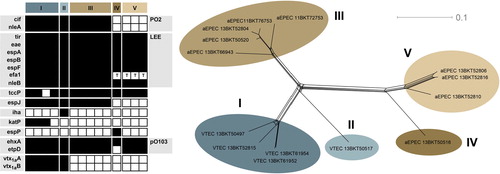
The results of the virulence gene typing are presented in . All isolates had the epsilon intimin variant. All isolates lacked the bfpA gene characteristic of typical EPEC, confirming their status as aEPEC or VTEC. Genes encoding verotoxin 1a were found in all isolates in SNP clusters I and II. Virulence genes in the LEE were conserved, apart from a truncated version of efa1 found in aEPEC SNP clusters IV and V. The same two clusters also appear to lack espJ and the prophage-associated O103:H2 PO2 region, which encodes several putative virulence determinants (Citation34). The large virulence plasmid pO103 was present and intact in all isolates except the only representative of cluster IV, which lacked approximately half the E. coli O103:H2 12009 pO103 plasmid sequence, including the etpD gene. Apart from the presence of verotoxin genes, there were no consistent differences in virulence gene repertoires of the VTEC clusters and the most closely related aEPEC cluster III. However, katP was found only in VTEC cluster I and has been shown to be significantly associated with E. coli O103:H2 isolates from human infections compared with isolates from cattle (Citation17). Thus, the possible existence of distinct lineages of VTEC E. coli O103:H2 that differ in their virulence for the human host warrants further investigation.
The actual benefit for EHEC strains carrying verotoxin genes is controversial (Citation35), but the verotoxin-negative lineages of E. coli O103:H2 are obviously at least as successful as their verotoxin-positive counterparts in the ruminant reservoirs that are the natural habitat of these bacteria. As the local ruminant reservoir is probably the source for a significant portion of cases of E. coli O103:H2 infection in humans, the scarcity of E. coli O103:H2 lineages that carry verotoxin 2, in particular, seems fortunate. Most domestic cases of E. coli O103:H2 infection in Sweden are caused by strains carrying only vtx 1 and produce comparatively mild symptoms (Citation36). In general, the risk of HUS appears to be higher for those infected by VTEC strains carrying vtx 2 , in particular, the subtype vtx 2a , while vtx 1 rarely causes severe disease (Citation37, Citation38). However, E. coli O103:H2 with only vtx 1 is a known cause of HUS (Citation18, Citation19).
A representative of a VTEC-like aEPEC O103:H2 subgroup III (11-BKT072753) was successfully lysogenized by φ731 (Δvtx2::cat) phage transduction in vitro, while transduction by the 933W (Δvtx2::cat) phage failed. Further studies will be needed to explain why only one of the phages was able to lysogenize this particular strain. The authors anticipate that the two phages are able to use the same insertion site in the host genome, so this might be due to other unknown host specificity factors or experimental differences such as different concentrations of the induced phages. The presence of the φ731 phage was persistent after the subculturing of the strain, suggesting stable insertion in the host genome. This shows the ability of Swedish aEPEC E. coli O103:H2 to convert to verotoxin 2-positive status, at least under laboratory conditions. As previous studies have found verotoxin-producing E. coli to be abundant and diverse in the gastrointestinal tract of ruminants (reviewed by Mauro et al. (Citation39)), lack of compatible donor strains is unlikely to explain the rarity or absence of verotoxin 2-positive O103:H2 strains in the cattle reservoir. Historically, VTEC research has, for obvious reasons, mostly been focused on strains directly responsible for causing severe illness in humans, while the extensive diversity of VTEC and VTEC-like bacteria occupying niches in animal reservoirs or the environment has been neglected. This study highlights the importance of collecting and characterizing both VTEC and aEPEC in order to increase our understanding of the role of verotoxin-encoding phages in the evolution and ecology of human-pathogenic VTEC.
Conflict of interest and funding
The authors declare no conflict of interest. This study was financed by the Swedish Civil Contingencies Agency. EB-R was partially supported by the EU FP7 project ALLBIO, project reference: 289452, funded under FP7-KBBE.
Acknowledgements
The authors acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax/Uppnex for providing assistance with massive parallel sequencing and computational infrastructure. The authors also thank the diagnostics staff at the molecular biology and VTEC labs at the SVA for assistance with bacteriology and PCR assays.
References
- Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013; 26: 822–80. [PubMed Abstract] [PubMed CentralFull Text].
- Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli . Emerg Infect Dis. 8: 508–13.
- Nguyen Y, Sperandio V. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect Microbiol. 2012; 2: 90. [PubMed Abstract] [PubMed CentralFull Text].
- The National Board of Health and Welfare. Infektion med EHEC/VTEC – Ett nationellt strategidokument. 2014. ISBN 978-91-7555-249-1.
- Karch H, Denamur E, Dobrindt U, Finlay BB, Hengge R, Johannes L, etal. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol Med. 2012; 4: 841–48. [PubMed Abstract] [PubMed CentralFull Text].
- Bielaszewska M, Prager R, Kock R, Mellmann A, Zhang W, Tschape H, etal. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl Environ Microbiol. 2007; 73: 3144–50. [PubMed Abstract] [PubMed CentralFull Text].
- Mellmann A, Lu S, Karch H, Xu JG, Harmsen D, Schmidt MA, etal. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol. 2008; 74: 67–72. [PubMed Abstract] [PubMed CentralFull Text].
- L'Abee-Lund TM, Jorgensen HJ, O'Sullivan K, Bohlin J, Ligard G, Granum PE, etal. The highly virulent 2006 Norwegian EHEC O103:H25 outbreak strain is related to the 2011 German O104:H4 outbreak strain. PloS One. 2012; 7: e31413. [PubMed Abstract] [PubMed CentralFull Text].
- Tozzoli R, Grande L, Michelacci V, Ranieri P, Maugliani A, Caprioli A, etal. Shiga toxin-converting phages and the emergence of new pathogenic Escherichia coli a world in motion. Front Cell Infect Microbiol. 2014; 4: 80. [PubMed Abstract] [PubMed CentralFull Text].
- Muniesa M, Serra-Moreno R, Jofre J. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx genes appeared conserved. Environ Microbiol. 2004; 6: 716–25. [PubMed Abstract].
- Schmidt H, Bielaszewska M, Karch H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage phi3538 isolated from Escherichia coli O157:H7. Appl Environm Microbiol. 1999; 65: 3855–61.
- Muniesa M, Blanco JE, De Simon M, Serra-Moreno R, Blanch AR, Jofre J. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology. 2004; 150: 2959–71. [PubMed Abstract].
- Beutin L, Kaulfuss S, Herold S, Oswald E, Schmidt H. Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J Clin Microbiol. 2005; 43: 1552–63. [PubMed Abstract] [PubMed CentralFull Text].
- Kappeli U, Hachler H, Giezendanner N, Beutin L, Stephan R. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000–2009. Emerg Infect Dis. 2011; 17: 180–5. [PubMed Abstract] [PubMed CentralFull Text].
- Sekse C, Sunde M, Hopp P, Bruheim T, Cudjoe KS, Kvitle B, etal. Occurrence of potentially human-pathogenic Escherichia coli O103 in Norwegian sheep. Appl Environ Microbiol. 2013; 79: 7502–09. [PubMed Abstract] [PubMed CentralFull Text].
- Moore SC, Pettazzoni S, McMillan KE, Mellor GE, Barlow RS, Fegan N. Characterization of E. coli O103 isolates from Australian ruminants. 2015; Boston, MA: VTEC.
- Karama M, Johnson RP, Holtslander R, Gyles CL. Phenotypic and genotypic characterization of verotoxin-producing Escherichia coli O103:H2 isolates from cattle and humans. J Clin Microbiol. 2008; 46: 3569–75. [PubMed Abstract] [PubMed CentralFull Text].
- Prager R, Liesegang A, Voigt W, Rabsch W, Fruth A, Tschape H. Clonal diversity of Shiga toxin-producing Escherichia coli O103:H2/H(-) in Germany. Infect Genet Evol. 2002; 1: 265–75. [PubMed Abstract].
- Mariani-Kurkdjian P, Denamur E, Milon A, Picard B, Cave H, Lambert-Zechovsky N, etal. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J Clin Microbiol. 1993; 31: 296–301. [PubMed Abstract] [PubMed CentralFull Text].
- Perelle S, Dilasser F, Grout J, Fach P. Detection of Escherichia coli serogroup O103 by real-time polymerase chain reaction. J Appl Microbiol. 2005; 98: 1162–68. [PubMed Abstract].
- Nielsen EM, Andersen MT. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5’ nuclease PCR assay. J Clin Microbiol. 2003; 41: 2884–93. [PubMed Abstract] [PubMed CentralFull Text].
- Madic J, Peytavin de Garam C, Vingadassalon N, Oswald E, Fach P, Jamet E, etal. Simplex and multiplex real-time PCR assays for the detection of flagellar (H-antigen) fliC alleles and intimin (eae) variants associated with enterohaemorrhagic Escherichia coli (EHEC) serotypes O26:H11, O103:H2, O111:H8, O145:H28 and O157:H7. J Appl Microbiol. 2010; 109: 1696–705. [PubMed Abstract].
- Delannoy S, Beutin L, Fach P. Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR. J Clin Microbiol. 2012; 50: 4035–40. [PubMed Abstract] [PubMed CentralFull Text].
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011; 27: 863–64. [PubMed Abstract] [PubMed CentralFull Text].
- Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, etal. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci USA. 2009; 106: 17939–44. [PubMed Abstract] [PubMed CentralFull Text].
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Br J Pharmacol. 2012; 9: 357–9.
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, etal. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43: 491–8. [PubMed Abstract] [PubMed CentralFull Text].
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006; 23: 254–67. [PubMed Abstract].
- Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, etal. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014; 52: 1501–10. [PubMed Abstract] [PubMed CentralFull Text].
- Solheim HT, Sekse C, Urdahl AM, Wasteson Y, Nesse LL. Biofilm as an environment for dissemination of stx genes by transduction. Appl Environ Microbiol. 2013; 79: 896–900. [PubMed Abstract] [PubMed CentralFull Text].
- Sekse C, Muniesa M, Wasteson Y. Conserved Stx2 phages from Escherichia coli O103:H25 isolated from patients suffering from hemolytic uremic syndrome. Foodborne Path Dis. 2008; 5: 801–10. [PubMed Abstract].
- Serra-Moreno R, Acosta S, Hernalsteens JP, Jofre J, Muniesa M. Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol Biol. 2006; 7: 31. [PubMed Abstract] [PubMed CentralFull Text].
- Allison HE, Sergeant MJ, James CE, Saunders JR, Smith DL, Sharp RJ, etal. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect Immun. 2003; 71: 3409–18. [PubMed Abstract] [PubMed CentralFull Text].
- Leopold SR, Magrini V, Holt NJ, Shaikh N, Mardis ER, Cagno J, etal. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc Natl Acad Sci USA. 2009; 106: 8713–18. [PubMed Abstract] [PubMed CentralFull Text].
- Los JM, Los M, Wegrzyn A, Wegrzyn G. Altruism of Shiga toxin-producing Escherichia coli: recent hypothesis versus experimental results. Front Cell Infect Microbiol. 2012; 2: 166. [PubMed Abstract] [PubMed CentralFull Text].
- Public Health Agency of Sweden. Epidemiologisk årsrapport 2011. 2012. ISBN 978-91-86723-26-2.
- Orth D, Grif K, Khan AB, Naim A, Dierich MP, Wurzner R. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn Microbiol Infect Dis. 2007; 59: 235–42. [PubMed Abstract].
- Persson S, Olsen KE, Ethelberg S, Scheutz F. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol. 2007; 45: 2020–24. [PubMed Abstract] [PubMed CentralFull Text].
- Mauro SA, Koudelka GB. Shiga toxin: expression, distribution, and its role in the environment. Toxins. 2011; 3: 608–25. [PubMed Abstract] [PubMed CentralFull Text].
