Abstract
Background
Antibiotic-resistant bacteria pose challenges to healthcare delivery systems globally; however, limited information is available regarding the prevalence and spread of such bacteria in the environment. The aim of this study was to compare the prevalence of antibiotic-resistant bacteria in large-bodied gulls (Larus spp.) at urban and remote locations in Southcentral Alaska to gain inference into the association between antibiotic resistance in wildlife and anthropogenically influenced habitats.
Methods
Escherichia coli was cultured (n=115 isolates) from fecal samples of gulls (n=160) collected from a remote location, Middleton Island, and a more urban setting on the Kenai Peninsula.
Results
Screening of E. coli from fecal samples collected from glaucous-winged gulls (Larus glaucescens) at Middleton Island revealed 8% of isolates were resistant to one or more antibiotics and 2% of the isolates were resistant to three or more antibiotics. In contrast, 55% of E. coli isolates derived from fecal samples collected from large-bodied gulls (i.e. glaucous, herring [Larus argentatus], and potentially hybrid gulls) on the Kenai Peninsula were resistant to one or more antibiotics and 22% were resistant to three or more antibiotics. In addition, total of 16% of the gull samples from locations on the Kenai Peninsula harbored extended-spectrum cephalosporin-resistant E. coli isolates (extended-spectrum beta-lactamases [ESBL] and plasmid-encoded AmpC [pAmpC]), in contrast to Middleton Island where no ESBL- or pAmpC-producing isolates were detected.
Conclusion
Our findings indicate that increased prevalence of antibiotic resistance is associated with urban environments in Southcentral Alaska and presumably influenced by anthropogenic impacts. Further investigation is warranted to assess how migratory birds may maintain and spread antimicrobial-resistant bacteria of relevance to human and animal health.
Antibiotic-resistant bacteria are becoming increasingly problematic for healthcare systems globally (Citation1). Increased use of antibiotics in human and veterinary medicine, as well as food production systems, has resulted in the alteration of bacterial ecosystem dynamics and selection for resistance (Citation2). Antibiotic compounds and biocides in the environment as a consequence of anthropogenic activities are increasingly under scrutiny as playing an important role in the emergence of antibiotic resistance (Citation3). The amount of Escherichia coli in wild animals, relative abundance of integrons, and level of antibiotic resistance in the natural environment have all been correlated with proximity to human activity (Citation3–Citation6). However, even in places with relatively low human population density, antibiotic resistance has been identified (Citation7). It has therefore been suggested that migrating birds could play a role in the dispersal of antibiotic-resistant bacteria in the environment (Citation8, Citation9).
Numerous recent scientific investigations have focused on migratory birds, specifically gulls (family Laridae), as model species for understanding the occurrence and spread of antibiotic resistance in the environment for several reasons. First, gulls often thrive in anthropogenically influenced habitats where the level of antibiotic-resistant bacteria and antibiotic residues in the environment may be relatively high. As such, gulls may serve as reservoirs of E. coli carrying extended-spectrum beta-lactamases (ESBL, here including pAmpC) and other resistant strains, as in Portugal where 32% of gulls were carriers of ESBL, of which 98% were bla CTX-M, the most globally common type among humans (Citation10). Second, gulls often congregate and forage in response to human activities such as fishing, sewage discharge, and waste management, providing opportunities for transmission of antibiotic-resistant bacteria between distantly related taxa. For instance, in France, 9.4% of yellow-legged gulls (Larus michahellis) were found to carry ESBL-producing E. coli and transmission between humans and gulls was suggested, as isolates from both birds and humans carried ESBL genes of the same CTX-M type (Citation11). Third, gulls are often migratory and although they could potentially disperse antibiotic-resistant bacteria over long distances, E. coli cultured from Franklin's gull (Leucophaeus pipixcan) feces collected in Canada and Chile had genetically distinct ESBL genes, which does not support the dispersal of antibiotic-resistant bacteria between continents (Citation12, Citation13). Therefore, the role of migratory birds in the long-distance dispersal of antibiotic-resistant bacteria remains unresolved.
In previous research to understand the presence and possible dissemination of antibiotic-resistant bacteria in remote regions of Alaska, sampling of glaucous gull (Larus hyperboreus) feces near the village of Barrow in 2005 did not provide evidence for antibiotic-resistant E. coli phenotypes; however, vancomycin-resistant enterococci were detected (Citation14). Antibiotic resistance was also identified in 8% of E. coli isolates from Russia and Greenland (Citation7). In contrast, 48% of E. coli isolates recovered from glaucous gulls using habitats adjacent to the Barrow landfill in 2010 were resistant to at least one antibiotic compound (Citation15). Therefore, investigations to assess spatiotemporal patterns of antibiotic resistance in E. coli infecting gulls, and presence in the environment within Alaska, are necessary to understand processes driving changes in prevalence.
In this study, we tested the hypothesis that the prevalence of antibiotic-resistant bacteria in gulls is associated with anthropogenic impacts to local habitats by sampling large-bodied gulls at both urban environments and more remote locations within the same geographic region of Southcentral Alaska. Our prediction was that the incidence of antibiotic-resistant E. coli phenotypes and genotypes and detection of multi-resistant strains would be higher in samples collected from gulls associated with urban environments as compared to those collected at a more remote location within the same general geographic region. Our results will be useful for informing future investigations of dispersal and maintenance of potential bacterial pathogens by migratory birds and for understanding patterns of antibiotic resistance in bacteria infecting wildlife in Alaska.
Material and methods
Sample collection
A total of 160 fecal samples were collected in June and July 2014 from large-bodied gulls at two sites from each of two general locations within Southcentral Alaska – Middleton Island and the Kenai Peninsula (). Middleton Island is a remote location in Southcentral Alaska, approximately 115 km offshore from the mainland in the Gulf of Alaska. Middleton Island was once the site of an active military installation (ca., 1958–1963) but is now uninhabited by humans with the exceptions of small field crews of scientists conducting research activities on the island during summer months and Federal Aviation Administration employees (typically two) that work on the island approximately half-time year round. Middleton Island is occupied by large numbers of gulls each summer including multiple breeding colonies of glaucous-winged gulls (Larus glaucescens). Fecal samples were obtained during 22–26 June 2014 from glaucous-winged gulls at sites on the northern (59.46°N, 146.29°W) and southern (59.41–59.42°N, 146.33–146.36°W) margins of Middleton Island, near nesting colonies prior to hatch. Individual fecal samples were collected by swirling a sterile swab in fresh feces and placing in bacteria freeze media in cryovials. Samples from Middleton Island were kept cool with icepacks for 4–8 days until frozen at −80°C.
Fig. 1 Map depicting sampling locations for large-bodied gulls (circles) in Southcentral Alaska. The approximate location of Anchorage, the most populous city in Alaska, is indicated with a star.
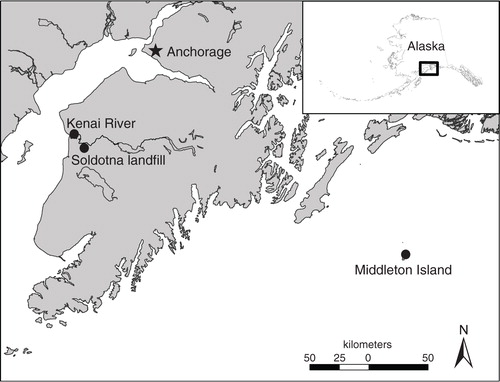
The Kenai Peninsula is a landmass extending south of Anchorage, Alaska, the most populous city in the state, and supports numerous communities and an extensive road system. Communities with the highest human population on the Kenai Peninsula are Soldotna and Kenai, both located adjacent to the Kenai River below Skilak Lake. The Kenai River is the largest waterway on the Kenai Peninsula and supports commercial, sport, and personal-use salmon fisheries during summer. In addition to a year-round population of approximately 58,000 residents as of 2015 (United States Census Bureau), the Kenai Peninsula also receives a considerable influx of tourists each summer as well as recreational users from the nearby city of Anchorage. Samples were collected from glaucous-winged gulls, herring gulls (Larus argentatus), and potentially, hybrids of these two species on the Kenai Peninsula at the tidal flats at the mouth of the Kenai River (60.54°N, 151.26°W) and at the Soldotna landfill (60.44°N, 151.11°W) on 24 July 2014. Fecal samples were collected as previously described and kept cool until frozen at −80°C within 12 h.
Isolation of E. coli and antibiotic susceptibility testing
Eighty samples from each Middleton Island (remote location) and the Kenai Peninsula (urban location) were screened for E. coli and the resultant isolates were tested for antibiotic susceptibility. An aliquot of 100 µl from each fecal sample was incubated overnight at 37°C in 4 ml of Luria-Bertani broth (Department of Clinical microbiology, Uppsala University Hospital). Cotton swabs soaked in the broth were streaked on cystein lactose electrolyte deficient agar plates (Department of Clinical Microbiology, Uppsala University Hospital) and incubated overnight at 37°C. One presumptive E. coli isolate from each sample was collected and confirmed using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF; Bruker Corporation, Germany) as previously described (Citation16). E. coli isolates were spread evenly on Muller-Hinton agar (Department of Clinical Microbiology, Uppsala University Hospital) and 10 antibiotic discs – ampicillin 10 µg, cefadroxil 30 µg, chloramphenicol 30 µg, nalidixic acid 30 µg, nitrofurantoin 100 µg, mecillinam 10 µg, tetracycline 30 µg, tigecycline 15 µg, streptomycin 10 µg, and sulfamethoxazole trimethoprim 25 µg (Oxoid Ltd, Hants, UK) – were placed on the plate. Inhibition zone diameters were compared to clinical breakpoints for disc diffusion provided by European Committee on Antibiotic Susceptibility (EUCAST). Currently, streptomycin and tetracycline lack EUCAST clinical breakpoints for disc susceptibility testing, therefore breakpoints for these antibiotics were defined according to the Normalized Resistance Interpretation (NRI) method (Citation17).
For statistical analyses, samples collected from two urban sites on the Kenai Peninsula and two remote sites on Middleton Island were pooled by location. The prevalence of antibiotic-resistant isolates and proportion of isolates resistant to three or more antibiotic compounds were compared using Fischer's exact test (GraphPad Prism version 6). A P value ≤0.05 was considered significant.
ESBL and carbapenem resistance screening
Fifty randomly selected samples from each of the Kenai Peninsula and Middleton Island collections were screened for extended-spectrum cephalosporin (ESBL- and pAmpC) and carbapenem-resistant E. coli. Fecal swabs were placed in tubes with 1 mL phosphate-buffered saline and vortexed. Aliquots from each tube were transferred to two vials containing 4 mL Luria-Bertani broth with either a cefpodoxime 10-µg disc (vial 1) or a ertapeneme 10-µg disc (vial 2) and incubated overnight at 37°C. Cotton swabs soaked in the broth from vial 1 were streaked on MacConkey agar plates with cefotaxime 5-µg disc and ceftazidime 10-µg disc and from vial 2 on MacConkey agar plates with cefpodoxime 10-µg disc and ertapenem 10-µg disc (Oxoid Ltd). All plates were incubated overnight at 37°C. Colonies growing close to an antibiotic disc were identified as E. coli using MALDI-TOF. To identify E. coli isolates with ESBL- or AmpC phenotype, isolates from the previous step were prepared and spread on Muller-Hinton agar (Department of Clinical Microbiology, Uppsala University Hospital) according to EUCAST disc diffusion method for antimicrobial susceptibility testing and five antibiotic discs – amoxicillin/clavulanic acid 30/1 µg, cefotaxime 5 µg, ceftazidime 10 µg, cefepime 30 µg, and cefoxitin 30 µg – were placed on the plate according to standard protocol at the Department of Clinical Microbiology in Uppsala University Hospital. Specific inhibition of bacterial growth around the antibiotic discs was used to identify ESBL/pAmpC-phenotypes (Citation18, Citation19). Isolates that displayed ESBL or AmpC phenotype were subjected to either two multiplex-PCR detecting bla CTX-M, bla SHV, bla TEM, bla OXA-1 or one multiplex-PCR detecting pAmpC genes and the specific gene variants were determined by sequencing as previously described (Citation20). The ESBL/pAmpC-producing E. coli isolates were further tested for resistance to additional antibiotic compounds. The chosen antibiotics are either frequently used in veterinary and human medicine or are saved for treatment of infections caused by ESBL/pAmpC/carbapenemase-producing E.coli. Isolates were spread on Muller-Hinton agar (Department of Clinical Microbiology, Uppsala University Hospital) and five antibiotic discs – gentamicin 10 µg, ciprofloxacin 5 µg, meropenem 10 µg, tetracycline 30 µg, and sulfamethoxazole trimethoprim 25 µg (Oxoid Ltd) – were placed on the plate. Inhibition zone diameters were compared to clinical breakpoints for disc diffusion provided by EUCAST and the NRI method (Citation16), as described above. In addition, all isolates were tested for colistin susceptibility using ETEST (bioMérieux, Marcy l’Etoile, France) according to manufacturer’s guidelines. However, due to that the recently described plasmid encoded mcr-1 have been described to show MICs below EUCAST breakpoint and the unreliability of colistin susceptibility results with E-test all ESBL-isolates were also tested for the presences of mcr-1 (Citation21, Citation22).
Results
General resistance of E. coli in gulls sampled in Southcentral Alaska
A total of 55 E. coli isolates were recovered from 80 samples screened from the Kenai Peninsula and 60 isolates were from 80 samples screened from Middleton Island, yielding isolation rates of 69 and 75% per location, respectively (). Of the E.coli isolates recovered from the urban Kenai Peninsula, 55% (30/55) were resistant to at least one antibiotic compound, whereas only 8% (5/60) of the isolates from the remote site Middleton Island displayed antibiotic resistance (). The difference in the prevalence of isolates resistant to at least one antibiotic between the two locations was statistically significant (Kenai Peninsula vs Middleton Island, two-tailed P value <0.0001, Fisher's exact test). Additionally, 22% (12/55) of E. coli isolates from the Kenai Peninsula were resistant to three or more compounds as compared to 2% (1/60) of isolates derived from samples at Middleton Island, supporting a statistical difference in the prevalence of multi-resistant E. coli between locations (Kenai Peninsula vs Middleton Island, two-tailed Fisher's exact test, P value <0.001; ). Considering antimicrobial activity in all E.coli isolates from the two areas, resistance to eight different antibiotic compounds could be found in Kenai Peninsula samples as compared to five different antibiotic compounds in Middleton Island isolates (). All of the E. coli isolates from both sample locations were susceptible to tigecycline.
Table 1 Isolation of E. coli from large-bodied gull samples collected at urban and remote locations in Southcentral Alaska, 2014
Table 2 Antibiotic resistance of E. coli from large-bodied gulls sampled in Southcentral Alaska relative to number of compounds
Table 3 Resistance of E. coli isolated from large-bodied gulls in Southcentral Alaska to specific antibiotic compounds
Phenotypic and genotypic testing of extended-spectrum cephalosporin-resistant E. coli (ESBL and/or pAmpC)
Out of 50 samples screened from the Kenai Peninsula (urban location), three E. coli isolates that displayed ESBL phenotype and five E. coli isolates that displayed AmpC phenotype were found. The ESBL isolates harbored bla CTX-M-15 and bla TEM-1 and the pAmpC isolates harbored bla CMY-2 (). Three ESBL/pAmpC-producing E. coli isolates were resistant to at least one other antibiotic compound and all were negative for mcr-1 via PCR (). On the Kenai Peninsula, all eight ESBL/pAmpC isolates were found among the gull samples from the Soldotna landfill site. No ESBL/pAmpC isolates were found at the Kenai River site or on Middleton Island (). No suspected carbapenem-resistant E. coli isolates were found.
Table 4 Phenotype, genotype, and antibiotic susceptibility of eight extended-spectrum cephalosporin-resistant E. coli; (ESBL/pAmpC) isolates from gulls sampled on the Kenai Peninsula, Alaska. Susceptibility (S) and resistance (R) to specific compounds are indicated
Discussion
Through our investigation of antibiotic-resistant E. coli derived from large-bodied gulls at two locations in Southcentral Alaska with different levels of anthropogenic influence, we observed differences in the prevalence of antibiotic-resistant and ESBL/pAmpC-producing E.coli. Thus, although our study design is correlative in nature, our results support the hypothesis that anthropogenic inputs may be a factor in determining the level of antibiotic resistance among E. coli infecting large-bodied gulls in Southcentral Alaska.
The higher incidence of antibiotic resistance detected in E. coli isolated from large-bodied gulls on the Kenai Peninsula relative to Middleton Island is likely, at least partially, a function of inputs from inhabitants and visitors of the towns of Soldotna and Kenai, urban areas in close proximity to sampling sites at the Soldotna landfill and mouth of the Kenai River. Although neither Soldotna nor Kenai are major metropolitan areas, direct inputs of antibiotic- resistant bacteria and biocides/antibiotic residues, which may lead to the emergence of antibiotic- resistant strains, are likely released into the environment via sewage treatment and the local landfill at higher rates as compared to areas lacking large permanent human settlements. Accordingly, the likelihood that gulls may encounter antibiotic-resistant E. coli or plasmids conferring resistant genes from anthropogenic sources via waste management practices may be higher in these more urban environments as compared to more remote sites. Food animal production occurs at low levels in Alaska and therefore is less likely a major contributor to antibiotic resistance in the area (Citation23).
The prevalence of antimicrobial-resistant E. coli detected in gulls at Kenai Peninsula sites (55%) was similar to that found in yellow-legged gulls in France (47%) (Citation11) and the most recent assessment of gulls at Barrow, Alaska (48%) (Citation15), areas in close proximity to human settlements providing anthropogenic inputs into the environment. Additionally, the finding that all ESBL/pAmpC-harboring E. coli isolated in this study were detected among gull samples collected at Kenai Peninsula, including globally disseminated bla CTX-M-15, is similar to the recent detection of bla CTX-M in Barrow, Alaska at a sampling site in close proximity to the local landfill (Citation15). As bla CTX-M-15 and bla CMY-2 are relatively common in humans and livestock in the USA (Citation24, Citation25), it is unsurprising to find these enzymes in gulls inhabiting Alaska landfills. Further studies to assess the occurrence of antibiotic resistance and antibiotic compounds in the soil and water around the Soldotna landfill may help to provide inference regarding the acquisition of antibiotic-resistant bacteria by gulls in this area.
The relatively low level of antibiotic resistance detected in E. coli isolated from glaucous-winged gulls at Middleton Island is likely a reflection of the geographic isolation of this location from mainland North America and a lack of anthropogenic input of antibiotic-resistant bacteria and/or antibiotic compounds. In a survey from the remote Commander Islands (Russia) in the Bering Sea, Hernandez et al. found 4% (6/145 isolates) of E. coli isolated from seabirds, including glaucous-winged gulls, were resistant to one or more antibiotic compounds (Citation26), comparable to the level found in gulls at Middleton Island (8%). Thus, it appears that glaucous-winged gulls sampled at remote breeding sites have relatively little exposure to antibiotic-resistant bacteria, or compounds driving their emergence, during the breeding season. The detection of some level, albeit low, of antimicrobial resistance among E. coli cultured from glaucous-winged gull samples collected at Middleton Island suggests that either low levels of resistance may be maintained among bacteria infecting wild birds in the absence of anthropogenic inputs or that resistant bacterial strains arrive at breeding grounds via migrating gulls returning from wintering and staging areas. It should be noted, however, that levels of antibiotic resistance in E. coli among avian populations are not static. The observed prevalence of ESBL-producing E. coli in gulls sampled at Barrow, Alaska has increased between 2005 and 2010 (Citation15). Thus, Middleton Island could be an informative site for future assessments aiming to address the regional and global dissemination of antibiotic-resistant genes through time.
Although differences of antibiotic resistance levels among E. coli infecting gulls at two Southcentral Alaska locations are likely directly influenced by bacteria and plasmids from human sources, the role of antibiotic residues in the environment driving the development of resistance in wild birds needs to be further investigated. Furthermore, whether birds can maintain antibiotic-resistant E. coli for long durations in the absence of human impact and disperse such bacteria via migration remains unclear and is worthy of further study. Finally, given the relatively high prevalence of antibiotic resistance detected in large-bodied gulls on the Kenai Peninsula (i.e. 55% of isolates tested) and the geographic and temporal proximity of our sampling site at the mouth of the Kenai River to a popular personal-use sockeye salmon (Oncorhynchus nerka) fishery, additional research may be warranted to understand the risk for human bacterial E. coli infections via environmental transmission at this site during summer.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
Acknowledgements
We appreciate field help provided by Brianna Williams and Morgan Walker and laboratory support provided by Jenny Isaksson, Badrul Hasan, and Henrik Reutherborg. We appreciate manuscript reviews provided by Christina Ahlstrom and John Pearce. We are grateful to John Reed for assistance with the preparation of . We thank Scott Hatch for logistical support while sampling on Middleton Island. This work was funded, in part, by the US Geological Survey through the Contaminants Biology Program of the Environmental Health Mission Area. None of the authors have any financial interests or conflict of interest with this article. Any use of trade names is for descriptive purposes only and does not imply endorsement by the US government.
References
- Pitout JD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008; 8: 159–66.
- Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008; 321: 365–7.
- Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, etal. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis. 2013; 13: 155–65.
- Gordon DM. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology. 2001; 147: 1079–85.
- Rolland RM, Hausfater G, Marshall B, Levy SB. Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl Environ Microbiol. 1985; 49: 791–4.
- Skurnik D, Ruimy R, Andremont A, Amorin C, Rouquet P, Picard B, etal. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli . J Antimicrob Chemother. 2006; 57: 1215–19.
- Sjolund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, Pinhassi J, etal. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg Infect Dis. 2008; 14: 70–2.
- Khan AU. Worldwide spread of NDM-1: are migratory birds culprits?. J Infect Dev Ctries. 2015; 9: 120–1.
- Guenther S, Aschenbrenner K, Stamm I, Bethe A, Semmler T, Stubbe A, etal. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS One. 2012; 7: e53039.
- Simoes RR, Poirel L, Da Costa PM, Nordmann P. Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli . Emerg Infect Dis. 2010; 16: 110–12.
- Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, etal. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One. 2009; 4: e5958.
- Bonnedahl J, Stedt J, Waldenstrom J, Svensson L, Drobni M, Olsen B. Comparison of extended-spectrum beta-lactamase (ESBL) CTX-M genotypes in Franklin gulls from Canada and Chile. PLoS One. 2015; 10: e0141315.
- Hernandez J, Johansson A, Stedt J, Bengtsson S, Porczak A, Granholm S, etal. Characterization and comparison of extended-spectrum beta-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin's gulls (Leucophaeus pipixcan) and humans in Chile. PLoS One. 2013; 8: e76150.
- Drobni M, Bonnedahl J, Hernandez J, Haemig P, Olsen B. Vancomycin-resistant enterococci, Point Barrow, Alaska, USA. Emerg Infect Dis. 2009; 15: 838–9.
- Bonnedahl J, Hernandez J, Stedt J, Waldenstrom J, Olsen B, Drobni M. Extended-spectrum beta-lactamases in Escherichia coli and Klebsiella pneumoniae in gulls, Alaska, USA. Emerg Infect Dis. 2014; 20: 897–9.
- Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, etal. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009; 49: 543–51.
- Kronvall G, Kahlmeter G, Myhre E, Galas MF. A new method for normalized interpretation of antimicrobial resistance from disk test results for comparative purposes. Clin Microbiol Infect. 2003; 9: 120–32.
- Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988; 10: 867–78.
- Polsfuss S, Bloemberg GV, Giger J, Meyer V, Bottger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2011; 49: 2798–803.
- Egervärn M, Börjesson S, Byfors S, Finn M, Kaipe C, Englund S, etal. Escherichia coli with extended-spectrum beta-lactamases or transferable AmpC beta-lactamases and Salmonella on meat imported into Sweden. Int J Food Microbiol. 2014; 171: 8–14.
- Hindler JA, Humphries RM. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol. 2013; 51: 1678–84.
- Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, etal. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016; 16: 161–8.
- 2015 State Agriculture overview: US Department of Agriculture (2016). Available from: https://www.nass.usda.gov/Quick_Stats/Ag_Overview/stateOverview.php?state=ALASKA [cited 18 August 2016]..
- Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009; 64(Suppl 1): i3–10.
- Paterson D, Egea P, Pascual A, López-Cerero L, Navarro M, Adams-Haduch J, etal. Extended-spectrum and CMY-type β-lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clin Microbiol Infect. 2010; 16: 33–8.
- Hernandez J, Bonnedahl J, Eliasson I, Wallensten A, Comstedt P, Johansson A, etal. Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX-M-15, found in Glaucous-winged gull at remote Commander Islands, Russia. Environ Microbiol Rep. 2010; 2: 329–32.
