Abstract
Exosomes are a class of extracellular vesicles that are secreted by various cell types. Unlike other extracellular vesicles (ectosomes and apoptotic blebs), exosomes are of endocytic origin. The roles of exosomes in vaccine/drug delivery, intercellular communication and as a possible source of disease biomarkers have sparked immense interest in them, resulting in a plethora of studies. Whilst multidimensional datasets are continuously generated, it is difficult to harness the true potential of the data until they are compiled and made accessible to the biomedical researchers. Here, we describe ExoCarta (http://www.exocarta.org), a manually curated database of exosomal proteins, RNA and lipids. Datasets currently present in ExoCarta are integrated from both published and unpublished exosomal studies. Since its launch in 2009, ExoCarta has been accessed by more than 16,000 unique users. In this article, we discuss the utility of ExoCarta for exosomal research and urge biomedical researchers in the field to deposit their datasets directly to ExoCarta.
Exosomes, membranous vesicles of endocytic origin, are signalling organelles secreted by normal and disease cells (Citation1–Citation5). Originally described three decades ago Citation6 Citation7, exosomes contain a subproteome of the cells and are found in many bodily fluids Citation1 Citation8. Released upon fusion of multivesicular bodies (MVBs) with the plasma membrane (PM), exosomes are of 40–100 nm in diameter, are of endocytic origin, have a cup shaped appearance as visioned by electron microscopy, have a buoyant density in sucrose of 1.10–1.21 g/mL and sediment at 100,000 g Citation9 Citation10. They harbour proteins/RNA/lipids that reflect the functionality of the host cell and posses molecular signatures or footprints resembling the diseased cell from which they were secreted Citation11. Exosomes exhibit a typical lipid bilayer membrane and are high in phosphatidylserine (PS) residues on their surface Citation10. The field of exosomes has witnessed renewed interest in the past 5 years mainly due to the discovery of luminal RNAs including mRNA and miRNA in exosomes Citation12. The finding of exosome mediated non-selective transfer Citation12 of inactive forms of both mRNA and miRNAs to neighbouring cells has spurred numerous studies on exosomes (). This enormous interest in exosomal studies can be attributed to 3 main reasons: Citation1 purported role of exosomes in intercellular signalling; Citation2 use as delivery vehicles for vaccines and drugs and Citation3 as possible sources of disease biomarkers.
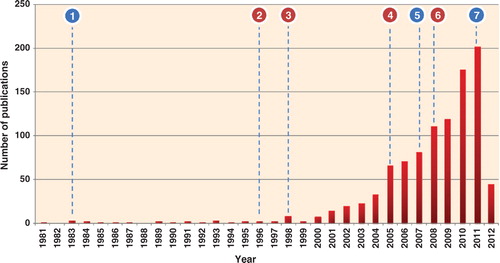
Fig. 1. Histogram of exosomal studies over the past 30 years. Immense interest in exosomes was seen during the last 7 years; more than 70% of the studies on exosomes were published (2005–2012). The statistics is generated based on PubMed indexed exosomal studies (keywords: exosomes or exosome-like) and adding the initial observations on exosomes. The resulting articles were manually verified by reading the title and abstract to ensure that the study referred exosomes, secreted vesicles of endocytic origin. Whilst each exosomal article improved our biological understanding, several articles contributed significantly. Important discoveries reported in the last 30 years are highlighted in the figure (blue – these findings moved exosomal research to a new next level; pink – trendsetting studies that formed the basis of current exosomal research): −1, discovery of exosomes Citation6 Citation47; −2, immune functions of B cell-secreted exosomes Citation48; −3, exosomes promote induction of antitumor immune responses in mice Citation49; −4, clinical trials of exosomes Citation50 Citation51; −5, discovery of exosomal RNA and transfer Citation12; −6, exosomes as a source of diagnostic biomarkers Citation52; −7, targeted delivery of siRNA through exosomes to brain Citation14.
Exosome-based drug delivery holds immense promise in the field of therapeutics including delivery of drugs across blood brain barrier and in the use of patient-derived microvesicles as a source of personalised drug delivery vehicle Citation13. The first proof-of-concept for the potential of exploiting these bioactive vesicles for targeted drug delivery was performed using dendritic cell-derived exosomes for siRNA delivery to the brain after systemic injection Citation14. The specificity of using exosomes as a drug carrying vehicles has created new opportunities for treatments of many diseases, most importantly, without significant side effects Citation15.
With the exponential increase in exosomal studies, the datasets generated are multidimensional originating from heterogeneous experimental platforms. Whilst most of the generated molecular (protein/RNA/lipid) data are mentioned in the inline text of the published article, a vast majority is often placed as supplementary information or not provided (especially with high throughput techniques) Citation16 Citation17. Importantly, whether in inline text or in supplementary tables, these exosomal molecular data in published articles are not easily queriable Citation16. In order to obtain novel biological insights, it is a perquisite to collate exosomal molecules in a centralized repository Citation18. For this reason, ExoCarta was created in 2009 as a free web-based resource that catalogs proteins and RNA identified in exosomes Citation19. ExoCarta is manually curated by expert scientists (http://www.exocarta.org) and contains molecular data on published and unpublished exosomal studies. It catalogs information on the exosomal isolation and purification procedures, samples used, investigator details and exosomal molecular components such as proteins, mRNA and miRNA as reported in the specific articles Citation20. ExoCarta can be queried using the gene symbol/name or browsed as a group based on the organism, molecular content type and the sample material. It is updated on a quarterly basis wherein newly published datasets are manually curated and appended to the existing repertoire of molecular data. Additionally, new features are added as per the demand of the exosomal field (for example, the importance of lipid molecules in exosomes prompted their inclusion in ExoCarta) Citation20.
Utility of ExoCarta
The current data and usage statistics of ExoCarta for the last 3 years are shown in and , respectively. Currently, ExoCarta contains 12,232 and 3,139 protein and RNA entries, respectively (). Few investigators have volunteered to submit their data directly to ExoCarta (http://www.exocarta.org/credits). In the following sections, the utility of ExoCarta is discussed:
As shown in , just over 16,000 unique users have visited ExoCarta over the last three years since its launch in 2009. The numbers are approximate and are based on unique IP addresses. A single user accessing ExoCarta from multiple computers will be counted multiple times. Also if multiple users from one educational institution access the database, they will be counted once. Regardless of the numbers, ExoCarta is a useful resource to the biological community.
ExoCarta has been routinely used by various groups for establishing exosomal markers (Citation21–Citation28).
Using the data downloaded from ExoCarta, 19 proteomic studies that identified at least 30 proteins were analysed to obtain the general protein composition of exosomes Citation9. Conversely, in addition to the conserved set of proteins, exosomes are also shown to contain a tissue-specific signature Citation11. The presence of tissue-specific signatures makes exosomes important in intercellular signalling and as possible source of disease biomarkers. Users can browse mRNA- and protein-based quantitative data to assess the tissue specificity of proteins and its presence in exosomes can be investigated through ExoCarta.
In ExoCarta, the isolation procedures along with the buoyant density are listed where available. ExoCarta allows researchers to download the entire dataset and filter them based on specific methods (e.g. sucrose density gradient centrifugation) for further analysis. Gyorgy et al. performed meta-analysis on the datasets downloaded from ExoCarta filtered based on density gradient centrifugation Citation29. The study observed the enrichment in the number of membrane proteins and depletion in nuclear proteins in exosomes.
Welton et al. performed a statistical over-representation analysis with datasets (limited to mass spectrometry) obtained from ExoCarta Citation30. The analysis highlighted a significant over-representation of proteins that are implicated in oncogenesis.
Prior to the release of ExoCarta, investigators spend considerable amount of time to collate previous exosomal studies and comparing them with protein/RNA/lipid identifications from their own studies. ExoCarta now allows researchers to download all studies as a single file and thereby aid in a quick comparison. The use of ExoCarta in reducing erroneous protein identifications from datasets obtained from plasma and urine Citation31 and in aiding quick comparison has been acknowledged (Citation31–Citation36).
From our preliminary analysis (Mathivanan and Simpson, unpublished observations), there is more than 60% overlap between proteins that are predicted to be non-classically secreted [based on SecretomeP Citation37] and those detected in exosomes. Gyorgy et al. reported that 30% of cytoplasmic proteins identified in exosomes were predicted to be secreted by non-classical secretory pathways [SecretomeP Citation37]. ExoCarta has been utilized to check for the presence of intracellular proteins that are detected in ovarian cancer ascites fluid Citation38 and wasp venom Citation39. Exosomal protein secretion as a possible mechanism of non-classical secretory pathway has prompted various groups to query ExoCarta for proteins lacking signal peptides Citation40 Citation41. Researchers can query ExoCarta to check whether their protein (not secreted by the classical secretory pathway) of interest is detected in any exosomal studies. Such additional information will prompt the investigators to design new studies to unravel the biological implications of exosome-based secretion.
Koppen et al. used ExoCarta to identify orthologs of exosomal markers to characterize exosomes secreted by drosophila cells Citation42.
It has been reported that immunoaffinity capture yields high quality exosomes, provided antibodies exist for the exosomal membrane protein of choice Citation43. Epithelial cell adhesion molecule (EPCAM), glycoprotein A33 (GPA33), HER2 (ERBB2) and CD63 molecule (CD63) are some of the previously used proteins to isolate exosomes from cell culture media or bodily fluids. As ExoCarta catalogs exosomal protein data from a wide range of cell types and tissues, it allows the investigators to choose the membrane protein that can be used for the exosomal immunoaffinity capture in their sample of interest. For example, GPA33 is not expressed in neuronal cells, which preclude it from being used for isolating exosomes that are of neuronal origin. However, neuronal membrane protein L1 cell adhesion molecule (L1CAM) or common proteins found in exosomes (CD63) can be used for isolating exosomes to homogeneity.
As more functional roles of exosomes are uncovered, the implications of exosomal membrane and luminal proteins in signalling cannot be ignored. ExoCarta provides users with protein interactors for their molecule of interest [the protein-protein interaction data is obtained from Biological General Repositories for Interaction Datasets (BioGRID) Citation44 and Human Protein Reference Database (HPRD) Citation45]. In the graphical display of protein interaction network present in the molecule page of ExoCarta, proteins detected in exosomes are highlighted in pink (). It can be speculated that, after the non-selective transfer of the luminal contents to the target cells, the transferred luminal proteins may interact with target cell proteins triggering downstream responses. Even though the molecular interactors vary depending on the cell type and the function performed, ExoCarta can provide information on the known protein interactors of the exosomal protein of interest ().
Fig. 2. A snapshot of ExoCarta and interaction network of CD9. Snapshot of ExoCarta homepage is displayed. Protein interaction network of CD9 (tetraspanin family) shows its protein interactors that are identified in exosomes (pink) and not (blue). Each individual tetraspanin protein performs differently via actions through their respective interactors; for instance, CD9 and CD82 mediate metastasis inhibition by several mechanisms whereas CD151 supports tumour progression by activating MMPs Citation53 Citation54.
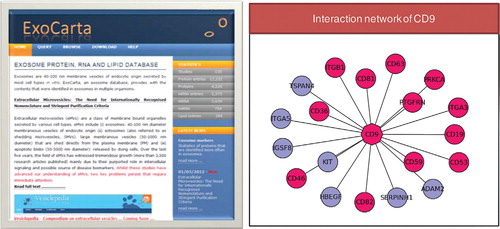
Table I. ExoCarta data statistics
Table II. ExoCarta usage statistics
The need for the involvement of exosomal community
With the explosion of exosomal studies, to maintain and update ExoCarta is a difficult task. However, with the active participation of the exosomal community, the database can be updated regularly. To facilitate active involvement of the exosomal research community, data contributions are acknowledged in the credits section of ExoCarta. The submitted datasets can be highlighted as private (only be accessed by the authors or their collaborators) or public (accessible by everyone).
ExoCarta is not a gold standard
Isolation and purification protocols employed in any exosomal study are of paramount interest as high quality exosomes are needed for any biological study. As data compiled in ExoCarta are derived directly from published articles, the quality of the datasets in ExoCarta is as good as the published articles. We emphasise caution when using datasets from ExoCarta as many published studies have purified exosomes by the far simpler differential centrifugation approach often ending with a crude preparation of exosomes with contaminants including ectosomes, apoptotic blebs and protein aggregates Citation30 Citation43 Citation46 . Whilst the exosomal community is addressing this issue, it has to be accepted that much of the published information has been obtained from impure preparations.
Future directions
A manually curated compendium of extracellular vesicles (apoptotic blebs, exosomes, large dense core vesicles, microparticles, microvesicles and synaptic vesicles) called Vesiclepedia has recently been completed and will be launched soon. ExoCarta will be continuously maintained after the release of Vesiclepedia and will become a primary resource with high quality exosomal datasets. The quality control is critical, and inputs from the exosomal community through International Society of Extracellular Vesicles (ISEV) will aid in filtering the existing data and in the addition of new high quality datasets. One of the current problems with extracellular vesicles is the nomenclature used in naming the vesicles. The confusion in the nomenclature has led to typical exosome preparations sometimes being referred to as microvesicles and vice versa Citation46. With the involvement of the ISEV, the nomenclature of vesicles can be standardised and emphasis can be made on employing stringent purification protocols to isolate exosomes.
*Suresh Mathivanan Department of Biochemistry La Trobe Institute for Molecular Science La Trobe University Bundoora, Victoria 3086, Australia Tel: +61 03 9479 2565 Fax: +61 03 9479 1226 Email: [email protected]
Conflict of interest and funding
None declared.
Acknowledgements
This work was supported by the National Health and Medical Research Council, Australia (Program Grant No. 487922) to R.J.S. and a NH&MRC fellowship (1016599) to S.M.
References
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. 2002, 2: 569-79.
- Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. 2002, 3: 321-30. 10.3402/jev.v1i0.18374.
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. 2006, 140: 13-21. 10.3402/jev.v1i0.18374.
- Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. 2007, 1: 156-8. 10.3402/jev.v1i0.18374.
- Johnstone RM. Exosomes biological significance: a concise review. 2006, 36: 315-21. 10.3402/jev.v1i0.18374.
- Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. 1983, 33: 967-78. 10.3402/jev.v1i0.18374.
- Johnstone RM. Revisiting the road to the discovery of exosomes. 2005, 34: 214-9. 10.3402/jev.v1i0.18374.
- Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. 2009, 6: 267-83. 10.3402/jev.v1i0.18374.
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. 2010, 73: 1907-20. 10.3402/jev.v1i0.18374.
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. 2009, 9: 581-93. 10.3402/jev.v1i0.18374.
- Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. 2010, 9: 197-208. 10.3402/jev.v1i0.18374.
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. 2007, 9: 654-9. 10.3402/jev.v1i0.18374.
- Lakhal S, Wood MJ. Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. 2010, 33: 737-41. 10.3402/jev.v1i0.18374.
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. 2011, 29: 341-5. 10.3402/jev.v1i0.18374.
- Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, Barnes S, Grizzle W, Miller D, Zhang HG. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. 2010, 18: 1606-14. 10.3402/jev.v1i0.18374.
- Santos C, Blake J, States DJ. Supplementary data need to be kept in public repositories. 2005, 438: 738. 10.3402/jev.v1i0.18374.
- Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PCet al.Human Proteinpedia enables sharing of human protein data. 2008, 26: 164-7. 10.3402/jev.v1i0.18374.
- Vizcaino JA, Foster JM, Martens L. Proteomics data repositories: providing a safe haven for your data and acting as a springboard for further research. 2010, 73: 2136-46. 10.3402/jev.v1i0.18374.
- Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. 2009, 21: 4997-5000. 10.3402/jev.v1i0.18374.
- Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. 2012, 40: D1241-4. 10.3402/jev.v1i0.18374.
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. 2011, 13: 423-33. 10.3402/jev.v1i0.18374.
- Isern J, Fraser ST, He Z, Zhang H, Baron MH. Dose-dependent regulation of primitive erythroid maturation and identity by the transcription factor Eklf. 2010, 116: 3972-80. 10.3402/jev.v1i0.18374.
- Koumangoye RB, Sakwe AM, Goodwin JS, Patel T, Ochieng J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. 2011, 6: e24234. 10.3402/jev.v1i0.18374.
- Aalberts M, van Dissel-Emiliani FM, van Adrichem NP, van Wijnen M, Wauben MH, Stout TAet al.Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1 and GLIPR2 in humans. 2011, 86: 82. 10.3402/jev.v1i0.18374.
- Reinhardt TA, Lippolis JD, Nonnecke BJ, Sacco RE. Bovine milk exosome proteome. 2012, 75: 1486-92. 10.3402/jev.v1i0.18374.
- Carayon K, Chaoui K, Ronzier E, Lazar I, Bertrand-Michel J, Roques Vet al.Proteolipidic composition of exosomes changes during reticulocyte maturation. 2011, 286: 34426-39. 10.3402/jev.v1i0.18374.
- Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon Get al.Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. 2011, 46: 409-18. 10.3402/jev.v1i0.18374.
- van den Boorn JG, Picavet DI, van Swieten PF, van Veen HA, Konijnenberg D, van Veelen PAet al.Skin-depigmenting agent monobenzone induces potent T-cell autoimmunity toward pigmented cells by tyrosinase haptenation and melanosome autophagy. 2011, 131: 1240-51. 10.3402/jev.v1i0.18374.
- Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi Bet al.Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. 2011, 68: 2667-88. 10.3402/jev.v1i0.18374.
- Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth Jet al.Proteomics analysis of bladder cancer exosomes. 2010, 9: 1324-38. 10.3402/jev.v1i0.18374.
- Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. 2011, 11: 709-20. 10.3402/jev.v1i0.18374.
- Wang Z, Hill S, Luther JM, Hachey DL, Schey KL. Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT). 2012, 12: 329-38. 10.3402/jev.v1i0.18374.
- Stamer WD, Hoffman EA, Luther JM, Hachey DL, Schey KL. Protein profile of exosomes from trabecular meshwork cells. 2011, 74: 796-804. 10.3402/jev.v1i0.18374.
- da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. 2011, 86: 71. 10.3402/jev.v1i0.18374.
- Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2011. In press DOI: 10.1016/j.bcp.2011.12.037.
- Choi DS, Park JO, Jang SC, Yoon YJ, Jung JW, Choi DYet al.Proteomic analysis of microvesicles derived from human colorectal cancer ascites. 2011, 11: 2745-51. 10.3402/jev.v1i0.18374.
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. 2004, 17: 349-56. 10.3402/jev.v1i0.18374.
- Elschenbroich S, Ignatchenko V, Clarke B, Kalloger SE, Boutros PC, Gramolini AOet al.In-depth proteomics of ovarian cancer ascites: combining shotgun proteomics and selected reaction monitoring mass spectrometry. 2011, 10: 2286-99. 10.3402/jev.v1i0.18374.
- Baek JH, Lee SH. Identification and characterization of venom proteins of two solitary wasps, Eumenes pomiformis and Orancistrocerus drewseni. 2010, 56: 554-62. 10.3402/jev.v1i0.18374.
- Loei H, Tan HT, Lim TK, Lim KH, So JB, Yeoh KGet al.Mining the gastric cancer secretome: identification of GRN as a potential diagnostic marker for early gastric cancer. 2012, 11: 1759-72. 10.3402/jev.v1i0.18374.
- Ji H, Goode RJ, Vaillant F, Mathivanan S, Kapp EA, Mathias RAet al.Proteomic profiling of secretome and adherent plasma membranes from distinct mammary epithelial cell subpopulations. 2011, 11: 4029-39. 10.3402/jev.v1i0.18374.
- Koppen T, Weckmann A, Muller S, Staubach S, Bloch W, Dohmen RJet al.Proteomics analyses of microvesicles released by Drosophila Kc167 and S2 cells. 2011, 11: 4397-410. 10.3402/jev.v1i0.18374.
- Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AMet al.Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. 2012, 56: 293-304. 10.3402/jev.v1i0.18374.
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MSet al.The BioGRID interaction database: 2011 update. 2011, 39: D698-704. 10.3402/jev.v1i0.18374.
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala Pet al.Human protein reference database – 2006 update. 2006, 34: D411-4. 10.3402/jev.v1i0.18374.
- Simpson RJ, Mathivanan S. Extracellular microvesicles: the need for internationally recognised nomenclature and stringent purification criteria. 2012, 5: ii-ii.
- Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. 1983, 97: 329-39. 10.3402/jev.v1i0.18374.
- Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJet al.B lymphocytes secrete antigen-presenting vesicles. 1996, 183: 1161-72. 10.3402/jev.v1i0.18374.
- Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza Det al.Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. 1998, 4: 594-600. 10.3402/jev.v1i0.18374.
- Escudier B, Dorval T, Chaput N, Andre F, Caby MP, Novault Set al.Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. 2005, 3: 10. 10.3402/jev.v1i0.18374.
- Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TMet al.A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. 2005, 3: 9. 10.3402/jev.v1i0.18374.
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves Met al.XO Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. 2008, 10: 1470-6. 10.3402/jev.v1i0.18374.
- Shiomi T, Okada Y. MT1-MMP and MMP-7 in invasion and metastasis of human cancers. 2003, 22: 145-52. 10.3402/jev.v1i0.18374.
- Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. 2009, 9: 40-55. 10.3402/jev.v1i0.18374.