Abstract
Background: The healthy vascular endothelium, which forms the barrier between blood and the surrounding tissues, is known to efficiently respond to stress signals like hypoxia and inflammation by adaptation of cellular physiology and the secretion of (soluble) growth factors and cytokines. Exosomes are potent mediators of intercellular communication. Their content consists of RNA and proteins from the cell of origin, and thus depends on the condition of these cells at the time of exosome biogenesis. It has been suggested that exosomes protect their target cells from cellular stress through the transfer of RNA and proteins. We hypothesized that endothelium-derived exosomes are involved in the endothelial response to cellular stress, and that exosome RNA and protein content reflect the effects of cellular stress induced by hypoxia, inflammation or hyperglycemia. Methods: We exposed cultured endothelial cells to different types of cellular stress (hypoxia, TNF-α-induced activation, high glucose and mannose concentrations) and compared mRNA and protein content of exosomes produced by these cells by microarray analysis and a quantitative proteomics approach. Results: We identified 1,354 proteins and 1,992 mRNAs in endothelial cell-derived exosomes. Several proteins and mRNAs showed altered abundances after exposure of their producing cells to cellular stress, which were confirmed by immunoblot or qPCR analysis. Conclusion: Our data show that hypoxia and endothelial activation are reflected in RNA and protein exosome composition, and that exposure to high sugar concentrations alters exosome protein composition only to a minor extend, and does not affect exosome RNA composition.
To access the supplementary material to this article: Tables SI-SIV and Figures S1-2, please see Supplementary files under Article Tools online.
Exosomes are small, spherical vesicles that are secreted upon fusion of the limiting membrane of multivesicular bodies with the plasma membrane Citation1 Citation2. They are produced by many cell types and are involved in intercellular signalling Citation3 Citation4. As a result of inward budding during the formation of the intraluminal vesicles, exosomes contain cytosolic content, including proteins, mRNAs and miRNAs (Citation5–Citation7). Exosome-mediated signalling can occur through receptor-ligand interactions between membrane-bound proteins on the exosome and corresponding receptors on the cell surface Citation8. Exosome content can also be transferred into target cells, either through direct fusion of exosomes with the cell membrane or through active uptake, mediated by endocytosis Citation4 Citation9 Citation10. The latter 2 mechanisms would not only allow intercellular communication through protein interactions but through RNAs and cytoplasmic (signalling) proteins as well Citation6.
As mediators of intercellular communication, exosomes are involved in cellular stress responses. The secretion of stress-induced heat shock protein (Hsps) by several cells, including tumour cells, B lymphocytes, astrocytes and endothelial cells, has been described to be exosome associated (Citation11–Citation16). Exosomal Hsps are present on the surface of exosomes and can modulate immunological responses Citation16. Importantly, the presence of oxidative stress has been shown to alter RNA content of exosomes released by mouse mast cells, suggesting that stress-related signalling via exosomes does not only occur through protein transfer but also through shuttling of RNA Citation17.
The vascular endothelium, which forms the endothelial barrier between blood and the surrounding tissues, is known to efficiently respond to stress signals such as hypoxia and inflammation by adaptation of cellular physiology and secretion of (soluble) growth factors and cytokines that recruit endothelial progenitor cells or cells that play a role in the innate immune response Citation18. It has been demonstrated that endothelial cells also secrete exosomes Citation11 Citation19 and several reports show that endothelial cells can also be targeted by exosomes derived from different cell types Citation6 Citation20. Because of the important role of the vascular endothelium as a first line of defense against inflammatory, hypoxic and possibly also chronic stress (Citation21–Citation23), we hypothesized that endothelial cell-derived exosomes mediate communication of stress-related signals and that the content of these exosomes reflects the cellular stress in the cell of origin. We compared exosomes secreted by endothelial cells cultured under normal conditions, hypoxic conditions (model for ischemia), in the presence of TNF-α (model for inflammation and endothelial activation) and in the presence of high glucose (model for hyperglycemia) or mannose (osmotic control for glucose) concentrations. Exposure of endothelial cells to hypoxia or TNF-α modulated both protein and mRNA content of exosomes derived from these cells, whereas exposure to high glucose or mannose concentrations did not influence exosome protein or mRNA profiles. Our data indicate that protein and RNA content of endothelial cell-derived exosomes reflects signals of acute stress (hypoxic and endothelial activation) but not of short-term high sugar concentrations, present in the cells of origin.
Materials and methods
Cell culture
Human microvascular endothelial cells [HMEC-1; CDC; Atlanta, GA, USA Citation24] were maintained (up to passage number 27) in MCDB131 medium (Life Technologies, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin (all from Life Technologies, Grand Island, NY, USA), 50 nm Hydrocortisone, 10 mM L-glutamine and 10 ng/ml rhEGF at 37°C, 5% CO2 and ambient oxygen level (20%).
Exosome isolation
Exosomes were isolated from the medium by differential centrifugation as described previously Citation25 Citation26. Cells were grown to 70–75% confluence before exosome-free medium (prepared using FCS, which was centrifuged for at least 1 hour at 200,000g followed by 0.20 µm filter-sterilization) was added. During the 24-hour culturing period in exosome-free medium, cellular stress was induced by culturing in 2% O2 (hypoxia) or medium supplemented with 3.240 g/l glucose or mannose or 10 ng/ml TNF-α (all chemicals from Sigma, St Louis, MO, USA).
The culture medium was centrifuged sequentially for 15 min at 1,500g, 30 min at 10,000g and 60 min at 100,000g using a Beckman LE-80K centrifuge with SW32-Ti and SW60-Ti rotors (Beckman Instruments, Inc., Fullerton, CA, USA). Exosomes, pelleted in the 100,000g centrifugation step were washed twice by re-suspending in PBS and centrifugation at 100,000g.
Quantitative proteomics
Exosomes were lysed in lysis buffer (2% SDS, 1% Triton-X100, 0.1 M Tris pH 7.4, 1 tablet Complete EDTA-free protease inhibitors (Roche, Indianapolis, IN, USA) and concentrations were determined using a BCA protein assay (Pierce, Rockford, IL, USA)). Samples from control and stress-exposed endothelial cells (100 µg each) were labelled with iTRAQ (isobaric tagging for relative and absolute quantitation) reagents (Life Technologies), purified and fractionated with strong cation exchange chromatography (SCX) and analysed on a Velos Orbitrap mass spectrometer. First, proteins were precipitated. Protein samples were mixed with 6 volumes of cold acetone (−20°C) and incubated for 60 min at −20°C. After 10 min of centrifugation at 13,000g at 4°C, the supernatant was removed and the rest of acetone was allowed to evaporate from the uncapped tubes at room temperature. Next, proteins were digested; for this, protein pellets were reconstituted with 20 µl of dissolution buffer (0.5 M triethylammonium bicarbonate), 1 µl denaturant (2% SDS) and 2 µl reducing reagent [50 mM tris-(2-carboxyethyl) phosphine]. The mixtures were mixed by vortexing, spun down and incubated at 60°C for 1 hour. Free cysteines were blocked by adding 1 µl of 200 mM Methyl methanethiosulfonate in isopropanol and incubated 10 min at room temperature. Trypsin (Promega V5111) was reconstituted with de-ionized water at 1 µg/µl concentration. Ten µl trypsin solution was added to each vial and incubated overnight at 37°C followed by iTRAQ labelling: 8-plex iTRAQ reagents were allowed to reach room temperature and then reconstituted with 50 µl of isopropanol. Each label reagent was mixed with the corresponding protein digest and incubated at room temperature for 2 hours. Samples were pooled into a new vial and dried using a centrifugal evaporator (Speedvac). After reconstituted with 0.1% formic acid (FA), the digest was desalted on an Oasis HLB 1 cc column (Waters, Milford, MA, USA) and eluted with 60% acetonitrile (ACN) 0.1% FA. Eluted peptide mixtures were dried by centrifugation evaporation, reconstituted with 100 µl SCX buffer A (10 mM KH2PO4, 20% ACN, pH 2.7) and separated on a PolyLC PolySULFOETHYL A column (200×2.1 m, 5 µm, 200 Å) with a linear 200 µl/min gradient of 0–70% buffer B (10 mM KH2PO4, 20% ACN, 500 mM KCl, pH 2.7) in 45 min on an Agilent 1200 LC device with Chemstation B.02.01 control software (Agilent, Santa Clara, CA, USA). Fractions were collected each minute and eventually pooled into 24 fractions. After vacuum centrifugation to evaporate the solute, fractions were desalted, eluted, dried as described above and reconstituted with 0.1% FA. Liquid chromatography was performed on an Eksigent nanoLC-Ultra 1D plus system (Eksigent, Dublin, CA, USA). Peptide digest was first loaded on a Zorbax 300SB-C18 trap (Agilent) at 6 µl/min for 5 min, then separated on a PicoFrit analytical column (100 mm long, ID 75 µm, tip ID 10 µm, packed with BetaBasic 5 µm 300 Å particles; New Objective, Woburn, MA, USA) using a 40-min linear gradient of 5–35% ACN in 0.1% FA at a flow rate of 250 nl/min. Mass analysis was carried out on an LTQ Orbitrap Velos (Thermo Fisher Scientific, San Jose, CA, USA) with data-dependent analysis mode, where MS1 scanned full MS mass range from m/z 300 to 2,000 at 30,000 mass resolution and 6 HCD MS2 scans were sequentially carried out at resolution of 7,500 with 45% collision energy, both in the Orbitrap.
Database search and quantitative data analysis
MS/MS spectra from 24 fractions were searched against the Swiss Prot (Swiss Institute of Bioinformatics, updated August 10, 2010, 21,241 entries) database, taxonomy Human using our 6-processor Mascot (Matrix Science, London, UK; version 2.3) cluster at NIH (http://biospec.nih.gov), with precursor mass tolerance at 20 ppm, fragment ion mass tolerance at 0.05 Da, trypsin enzyme with 2 miscleavages, methyl methanethiosulfonate of cysteine and iTRAQ 8plex of lysine and the N-terminus as fixed modifications, and deamidation of asparagine and glutamine, oxidation of methionine and iTRAQ 8plex of tyrosine as variable modifications.
The resulting.dat file was load into Scaffold Q+ (version Scaffold_3_00_04, Proteome Software Inc., Portland, OR, USA) to filter and quantitate peptides and proteins. Peptide identifications were accepted at 95.0% or higher probability as specified by the Peptide Prophet algorithm Citation27 and a false discovery rate (FDR) of less than 1%. Protein identifications were accepted at 99.0% or higher probability and contained at least 2 identified peptides with FDR less than 1%. Protein probabilities were assigned by the Protein Prophet algorithm Citation28. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Peptides were quantitated using the centroided reporter ion peak intensity, with minimum of 5% of the highest peak in the spectrum. Intrasample channels were normalised based on the median ratio for each channel across all proteins. The isobaric tagged samples were normalised by comparing the median protein ratios for the reference channel. Quantitative protein values were derived from only uniquely assigned peptides.
Immunoblotting
Exosomes were lysed in lysis buffer, and concentrations were determined using a BCA protein assay (Pierce). Equal amounts of exosomes were subjected to 12% SDS-PAGE electrophoresis and transferred to PVDF membranes (Thermo Scientific). PVDF membranes were blocked in 5% low-fat dry milk powder (Campina, Amersfoort, The Netherlands) in TBS with 0.1% Tween-20 (blocking buffer). PVDF membranes were incubated with primary antibodies in blocking buffer, washed in TBS with 0.1% Tween-20 (TBST), followed by incubation with HRP-conjugated secondary antibodies in blocking buffer. Proteins were detected with Chemiluminescent Peroxidase Substrate (Sigma) and imaged on the Molecular Image ChemiDoc XRS system (Biorad, Hercules, CA, USA). After imaging, PVDF membranes were stripped from their antibodies using ReBlot Plus Mild Antibody Stripping Solution (Millipore, Bedford, NY, USA), blocked and re-probed for β-actin as a loading control. The primary antibodies used were β-actin (A5441, 1:15,000, Sigma), Flotillin-1 [sc-25506, 1:500, Santa Cruz Biotechnologies (Santa Cruz, CA, USA)], ICAM-I (SC-8439, 1:1,000, Santa Cruz Biotechnologies), LOXL2 [AF2639, 1:400, R&D Systems (Minneapolis, MN, USA)], PlexinA1 [PAB7879, 1:500, Abnova (Taipei City, Taiwan)], TNFAIP3 (MAB3716, 1:1000, Abnova), semenogelin-1 (SEMG1) [H00006406-B01P, 1:200, Novus Biologicals (Littleton, CO, USA)] and TUG (SC-101260, 1:200, Santa Cruz Biotechnologies). Secondary antibodies were HRP-conjugated swine anti-rabbit [P0399, 1:2,000, Dako (Glostrup, Denmark)], rabbit anti-Mouse (P0260, 1:2,000, Dako) and rabbit anti-goat (P0160, 1:2,000, Dako).
Gene expression analysis
Exosomes were pelleted and washed twice with PBS. Total RNA was isolated using a mirVana RNA isolation kit (Invitrogen), concentration was determined using a Nanodrop spectrophotometer and RNA integrity was verified using Bioanalyzer pico RNA chips (Agilent). Labelled cRNA was generated according to the Illumina “whole-genome gene expression direct hybridization assay” protocol and, after determining the concentration using a NanoDrop spectrophotometer, hybridized on Illumina HumanHT12-v4 Beadchips (Illumina, San Diego, CA, USA). Gene expression data obtained from Illumina Beadstudio was normalised using “R” bioconductor with the “lumi” package Citation29. Genes that passed the detection call in >1 sample were included in the analysis. To identify differentially expressed genes between the various conditions, we created a linear model on the basis of moderated t-statistics using the “limma” package Citation30. Genes with a FDR <0.05 were considered significant.
Quantitative reverse transcription-PCR (qRT-PCR)
cDNA was synthesized from same set of samples analysed by microarray analysis using the BioRad iScript cDNA Synthesis Kit. qRT-PCR analysis was performed on the BioRad CFX96 RT system, using the IQ Sybr Green Super Mix (Biorad). Primer sequences were taken from the Harvard Primer Databank Citation31 and synthesized by Sigma-Aldrich. The following primers were used: B2M (5′-CCAAGGAAGGCGTCTAAGGC-3′, 5′- TGCACTCCAGCAGTAGGTGT-3′), BNIP3 (5′-CAGGGCTCCTGGGTAGAACT-3′, 5′-CTACTCCGTCCAGACTCATGC-3′), CCL2 (5′-CAGCCAGATGCAATCAATGCC-3′, 5′-TGGAATCCTGAACCCACTTCT-3′), CIRBP (5′-AGGGCTGAGTTTTGACACCAA-3′, 5′-ACAAACCCAAATCCCCGAGAT-3′), GAPDH (5′-AAGGTGAAGGTCGGAGTCAAC-3′, 5′-GGGGTCATTGATGGCAACAATA-3′), IL-8 (5′-GAATGGGTTTGCTAGAATGTGATA-3′, 5′-CAGACTAGGGTTGCCAGATTTAAC-3′), IL-32 (5′-TGGCGGCTTATTATGAGGAGC-3′, 5′-CTCGGCACCGTAATCCATCTC-3′), NDRG1 (5′-TCGAGACTTTACATGGCTCTGT-3′, 5′-TCATGCCGATGTCATGGTAGG-3′), RPLP0 (5′-TCGACAATGGCAGCATCTAC-3′, 5′-ATCCGTCTCCACAGACAAGG-3′),SOD2 (5′-GCTCCGGTTTTGGGGTATCTG-3′, 5′-GCGTTGATGTGAGGTTCCAG-3′), TNIP1 (5′-CTAGTGTGACGGCAGGTAAGG-3′, 5′-GCTGCTTCATGGACCGGAA-3′) and VCAM (5′-GGGAAGATGGTCGTGATCCTT-3′, 5′-TCTGGGGTGGTCTCGATTTTA-3′). Ct values were normalised per gene and experiment. ddCt was calculated using the geometric mean of 3 common housekeeping genes, which did not show any variation in the genomic screening (RPLP0, B2M, and GAPDH). Statistical analysis was performed by 2-tail Student t-test. Differences were considered statistically significant at p < 0.05.
Gene ontology and pathway enrichment analysis
Gene Ontology Term for Term analysis was performed using Ontologizer Citation32 Citation33. The p-values for significant overrepresentation of gene ontology terms in the identified gene set as compared to human genes was calculated using a modified Fisher exact test (1-tailed) to which a Benjamini-Hochberg correction for multiple testing was applied with the significance threshold at p = 0.0100.
Nanoparticle tracking analysis (NTA)
Size distribution of vesicles in 100,000g pellets were analysed using a Nanosight LM10-HS (NanoSight, Amesbury, UK) with a 532-nm laser-equipped sample chamber. The sample chamber was completely filled with 100,000g pellets resuspended in PBS. Shutter and gain were manually adjusted for optimal detection and were kept at this setting for all samples. A 1-min AVI file was recorded and analysed using NTA (version 2.2, build 0370, Nanosight) software to calculate size distributions and vesicle concentrations using the following settings: calibration: 166 nm/pixel; blur: auto; detection threshold: 14, minimum track length: auto, temperature: 22°C, viscosity: 0.95 cP. Differences in vesicle concentration and modal sizes were analysed by 1-way ANOVA using a Bonferroni-Holm post-hoc test to correct for multiple testing, assuming p-values <0.05 to be significant.
Electron microscopy
Transmission electron microscopy was performed as described by Slot and Geuze Citation34. Briefly, carbon-coated Formvar filmed grids were placed on exosome suspension for 20 min and washed with 0.15% glycine in PBS 3 times followed by a 0.1% BSA in PBS wash. Vesicles were fixed in 1% glutaraldehyde in PBS for 5 min and washed twice with PBS. For CD63 labelling, grids were placed on 5 µl 1% BSA in PBS containing 5 µg/ml anti-human CD63 (CLB, Amsterdam, The Netherlands) for 20 min, washed 4 times with 0.1% BSA in PBS and incubated on a drop of 1% BSA in PBS containing rabbit anti-mouse polyclonal antibody (Dako) for 20 min. Secondary antibodies were labelled by incubation on PBS containing 1% BSA and 10 nm gold particles coupled to protein A for 10 min. Specimen were fixed in 1% glutaraldehyde in PBS for 5 min and washed twice with PBS. After 4 washes on distilled water, grids were placed on ice-cold 1.8% methylcellulose (25 Ctp)/0.4% uranyl acetate (MC-AU) for 5 min, and after drying, vesicles were visualised using a FEI Tecnai 12 (FEI, Hillsboro, OR, USA) transmission electron microscope.
Sucrose gradient analysis
The 100,000g pellets were resuspended in 250 µl 2.5 M sucrose, 20 mM TRIS HCl pH 7.4 and floated in a SW60 tube for 16 hours at 190,000g using a linear sucrose gradient (2.0–0.25 M sucrose, 20 mM Tris-HCl, pH 7.4). Gradient fractions (250 µl) were collected from the top used for subsequent immunoblot and electron microscopy analyses.
Results
Sample characterization
Cultured endothelial cells produce exosomes and microvesicles, which can be isolated from the culture medium Citation11 Citation19 Citation35 Citation36. Vesicles present in the obtained 100,000g pellet (after 2 PBS washes) of cells grown under standard condition (no stress) were characterized with respect to their density, shape and size using sucrose density centrifugation, transmission electron microscopy and NTA.
Immunoblotting of sucrose density gradient fractions for Flotillin-1 reveals bands in fractions between 1.07 and 1.16 g/ml with peak fractions at a density of 1.10–1.11 g/ml (a). Transmission electron microscopy analysis of vesicles in the pooled 4 peak fractions (1.09–1.12 g/ml) shows vesicles at sizes around 100 nm, of which some contain CD63 protein. Also, larger vesicles could be detected (b), and NTA of the 100,000g pellet (c) confirmed the observed size distribution. Based on isolation procedure and vesicle characteristics, we term the vesicles present in the 100,000g pellet, which were used for subsequent analyses, exosomes, although we accept that our preparation also contains larger microvesicles.
Fig. 1. Characterization of endothelial cell-derived exosomes. Vesicles in the final, washed 100,000g pellets from endothelial cell culture supernatant cultured under standard conditions were analysed by sucrose density gradient and subsequent immunoblotting for Flotillin-1 (a), transmission electron microscopy (b; 10 nm gold: CD63) and by NTA (c). Size distributions of vesicles in washed 100,000g pellets from supernatants of stress-exposed endothelial cells were compared by NTA (d–g; conditions indicated, grey area represents SD).
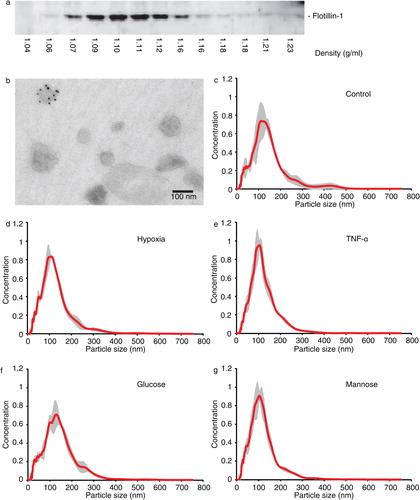
Subsequently, endothelial cells were grown under different stress conditions (hypoxia, TNF-α or high glucose) in exosome-depleted medium for 24 hours. Cells were grown in high mannose medium to serve as an osmotic control, allowing us to distinguish effects of high glucose from changes induced by osmotic stress. Washed 100,000g pellets were analysed by Nanosight analysis to investigate effects of cellular stress on vesicle size distribution, revealing that cellular stress did not significantly affect vesicle size or concentration (d–g; supplemental information Table SI).
Protein content confirms cellular origin of exosomes
To investigate whether the stress conditions under which our endothelial cells were cultured are reflected in exosome protein content, we performed iTRAQ-based quantitative proteomics analysis Citation37 Citation38. Exosomal proteins were digested, labelled and analysed using LC-MS/MS. Accepting protein identifications based on identification of at least 2 peptides with a probability higher than 95% and minimal total protein identification probability of 99% resulted in the identification of 1,354 proteins (Supplemental Information, Table SII). Analysis of enriched gene ontology (GO)-terms Citation32 representing cellular component classes (Supplemental Information, Fig. S1) reveals that proteins annotated to cytoplasmic content were abundantly present in the isolated exosomes (GO-terms “intracellular,” “cytosol,” “ribosome” and “ribonuclear complex”). Furthermore, GO-terms indicating that cellular vesicles are analysed (“vesicle,” “membrane-bounded vesicle” and “cytoplasmic membrane-bounded vesicle”) are significantly enriched. Clues on the origin of the protein sample are provided by the significantly enriched GO-terms “melanosome,” “pigment granule,” “endosomal part” and “endosome membrane,” reflecting lysosome-related organelles that have previously been identified as enriched in exosome protein samples Citation39.
Cellular stress conditions are reflected in exosome protein levels
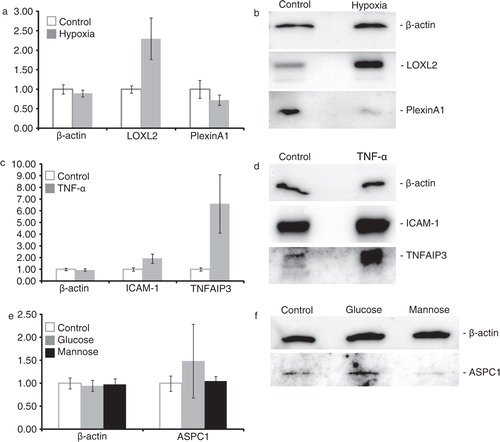
To compare the proteome of exosomes from cells grown under different conditions, all iTRAQ-labelled peptides were quantified. Two identical (control condition) exosome samples were included to assess reproducibility of quantitation between samples. All proteins for which the Log2 ratio of these 2 samples was between −0.2 and 0.2 and for which at least 2 peptides could be reliably quantified were further analysed (1,240 proteins, Supplemental Information, Table SIII). After manual inspection of quantified peptides, differentially abundant proteins could be identified for all stress conditions. Exposure to hypoxic conditions or TNF-α yielded several up- or downregulated proteins, whereas high glucose and mannose concentrations affected the abundance of only few proteins ( and Supplemental Information, Table SIII).
Table I. Proteins showing differential abundances: top up- and downregulated proteins, compared to control (>1.25-fold)
To confirm the quantitative proteomics data, differential abundances of a subset of proteins (based on availability of usable antibodies) were investigated using immunoblot analysis. β-Actin, a protein that was not affected in any condition, was used as a loading control. In agreement with the quantitative proteomics data (a), a decrease in Plexin A1 and an increase in lysyl oxidase like-2 (LOXL2) were observed in exosomes from endothelial cells cultured under hypoxic conditions (b). The increase of intercellular adhesion molecule 1 (ICAM-1) and tumor necrosis factor, alpha-induced protein 3 (TNFAIP3) in exosomes from endothelial cells cultured with 10 ng/ml TNF-α could also be confirmed (c,d). A minor increase of alveolar soft part sarcoma chromosomal region candidate gene 1 (ASPC1) was observed in exosomes from cells cultured in high glucose medium, but not in high mannose medium, in line with the quantitative proteomics analysis (e,f).
Gene expression analysis reveals stress-related exosomal mRNA abundances
To determine whether mRNA profiles in exosomes from stress-exposed endothelial cells vary between conditions, total RNA was isolated from washed 100,000g pellets followed by gene expression analysis on Illumina bead chip microarrays. After background subtraction, 10,533 mRNAs were confirmed to be present in endothelial cell-derived exosomes in more than 1 out of 24 samples, and 1,992 mRNAs were detected in all samples (Supplemental Information, Table SIV). We identified 18 genes that were significantly up- or downregulated in exosomes isolated from TNF-α treated cells and 3 genes in exosomes from cells cultured under hypoxia. No genes were significantly up- or downregulated in high-glucose conditions or in the isomolar mannose control (). We used RT-qPCR to verify significant differentially abundant RNAs observed in hypoxia (a) and 6 of the significantly upregulated mRNAs in exosomes from TNF-α treated cells (b) and confirmed differential abundance of all these mRNAs except for that encoding TNFAIP3-interacting protein 1 (TNIP1).
Table II. RNAs showing significant differential abundances (hypoxia, TNF-α): differentially abundant mRNAs
Fig. 3. Analysis and verification of exosome mRNA profiles. Quantitative PCR confirms differential abundance of mRNAs identified to be significantly increased or decreased in exosomes from hypoxia (a) or TNF-α exposed cells compared to those from control cells (b), error bars represent SEM, *p < 0.05. Clustering analysis based on the 100 most variable mRNAs distinguishes exosomes from endothelial cells exposed to hypoxia or TNF-α, but not to high glucose or mannose, from control cell-derived exosomes (c).
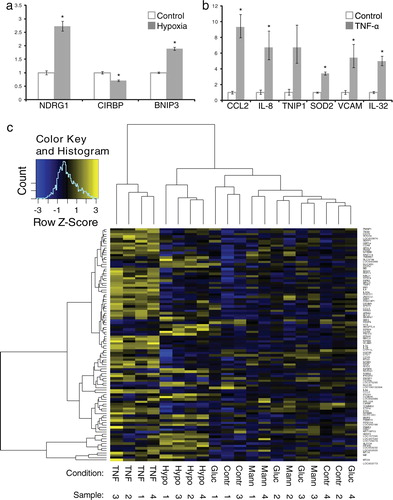
Using unsupervised hierarchical clustering of the top 100 genes that showed the highest intergroup variance (based on moderated t-statistics), we observed that mRNA profiles from exosomes isolated from TNF-α- and hypoxia-exposed cells consistently clustered together, whereas profiles of exosomes from glucose- and mannose-treated cells could not be distinguished from untreated controls (c).
Discussion
The data presented here demonstrate that protein and RNA levels in exosomes secreted by endothelial cells varies depending on culture condition and type of stress to which the cells are exposed to, suggesting that endothelial cells employ exosome-mediated cell communication to cope with conditions of cellular stress. Using sucrose gradient analysis and transmission electron microscopy, the isolated vesicles are demonstrated to contain characteristics that are typical for exosomes. Although some contamination with larger vesicles could be detected, the size distribution appears comparable to exosomes isolated from human umbilical vein endothelial cell (HUVEC)-derived exosomes Citation19. We subjected exosomal proteins and mRNAs to quantitative proteomics and gene expression analyses, identifying 1,354 proteins and 1,992 mRNAs, respectively. Analysis of enriched GO-terms provided confidence that our sample contained membrane-bounded vesicles with characteristics of endosomal vesicles and lysosome-related organelles, similar to the characteristics of B cell-derived exosomes Citation39. Furthermore, in agreement with prior exosome proteomics studies, components of the ESCRT complex and exosomal marker proteins as Flotillin-1 and -2, CD9, CD63, heat shock protein 70 (HSP70), MHC molecules, CD81, CD82 and proteins from the Rab family could be identified in our samples Citation3 Citation7 Citation39 Citation40. The tetraspanin protein CD63 was demonstrated to be present on a subset of exosomes, in line with the findings by van Niel et al. Citation41.
Nanoparticle tracking analysis was used to investigate whether cellular stress affects exosome size and concentration, and although minor differences could be detected, these differences did not appear significant. Using quantitative approaches to analyse exosomal protein and mRNA content, we demonstrate that exosomes from cells exposed to hypoxia or TNF-α could be clearly distinguished from control exosomes (and each other). Exposure to high glucose or mannose (osmotic control) concentrations resulted in minor differences in protein profiles and no significantly altered mRNA levels.
We verified the quantitative proteomics data by immunoblotting, and although changes in abundance were confirmed for proteins representing all stress conditions, some puzzling results were obtained. SEMG1, upregulated in exosomes from hypoxic cells, showed different band patterns in whole cell lysates and exosome samples (Supplemental Information, Fig. S2a). The lower band, detected in exosome sample, may represent a product of SEMG1 cleavage Citation42 Citation43, suggesting that this product is enriched in exosomes from endothelial cells grown in hypoxic conditions. The TNF-α induced increase in TNFAIP3 was confirmed by immunoblot analysis. However, the observed signal corresponds to a protein (-complex) of >170 kDa instead of the predicted size of 90 kDa (Supplemental Information, Fig. S2b), potentially reflecting protein aggregates containing TNFAIP3 due to the non-denaturing conditions used in this analysis.
We used RT-qPCR to validate a selection of differentially abundant mRNAs. Since no common housekeeping genes for RT-qPCR in exosomes are available, the relative changes were determined by comparing mRNA expression of several affected genes with the geometric mean of 3 commonly used housekeeping genes, which were unaffected by all conditions in the gene expression analysis. The magnitudes of changes differed from the changes observed in the gene expression analysis but all changes investigated in this manner could be verified, confirming that indeed exosomal RNA levels reflect cellular stress.
Treatment of HMEC-1 cells with high concentrations of glucose or mannose affected only few proteins and had no significant effect on any of the mRNAs found in exosomes. Potentially, our experimental setup does not induce sufficient stress to be reflected in exosomes from these cell, and exposure time Citation44 and sugar concentrations may need to be increased. A minor increase of ASPC1 was observed in exosomes from endothelial cells cultured in high glucose medium but not for a similar concentration of mannose (in line with the quantitative proteomics analysis). In contrast, complement component 4A (CO4A) is equally downregulated in exosomes from cells exposed to glucose and mannose, indicating an effect of osmotic stress.
The major changes observed in exosomes from TNF-α-exposed cells involved proteins that are commonly affected by TNF-α in endothelial cells, such as ICAM-1 and TNFAIP3 (Citation45–Citation47), and interestingly, our data are consistent with the observed upregulation of ICAM-1 in endothelial microparticles from TNF-α-treated endothelial cells in a qualitative proteomics study Citation48. Our gene expression analysis revealed 18 genes significantly up- or downregulated in exosomes from TNF-α treated cells. The genes and proteins affected by TNF-α display a variety of functions, including superoxide protection (superoxide dismutase 2, SOD2), immune responses (chemokine (C-C) ligand 2, interleukin-8 and -32, vascular adhesion molecule 1) and the NF-κB pathway (NF-kappa B, NF-kappa BIA, TNIP1). These changes represent the typical protein and gene expression profiles of activated endothelial cells (Citation49–Citation54), clearly reflecting the stress condition of exosome producing cells.
Proteins found upregulated in exosomes from cells grown under hypoxia suggest that cytoskeletal and extracellular matrix rearrangements can be induced by these exosomes, involving proteins such as LOXL2, fibronectin and collagen. Induction of such processes by hypoxia in endothelial cells is common Citation55, and because of their secretion into the environment, exosomes may especially contribute to extracellular matrix remodelling. In contrast, changes identified in the microarray analysis concern genes involved in stress response (N-myc downstream regulated 1, cold inducible RNA binding protein) and apoptosis (BCL2/adenovirus E1B 19 kDa interacting protein 3) (Citation56–Citation59). Taken together, both protein and mRNA content of exosomes from endothelial cells cultured under hypoxic conditions are affected and distinguish them from control cell-derived exosomes.
Although it remains to be determined whether exosomes from stressed endothelial cells also serve different functional goals, our data underscore that the content of exosomes can be used to assess the physiological condition of their producing cells. This principle has increased the interest in exosomes as a source for disease biomarkers, for example, for tumours and kidney diseases (Citation60–Citation64).
Altogether, our study demonstrates that cellular stress conditions are reflected in exosome protein and RNA content and suggest a role for endothelial exosomes in the transfer of stress signals to target cells in case of stress, activation and damage, as well as disease.
Conflict of interest and funding
The authors declare no conflicts of interest.
Supplementary Figure S1
Download PDF (46 KB)Supplementary Figure S2
Download PDF (357.4 KB)Supplementary Table SI
Download MS Excel (16.5 KB)Supplmentary Table SII
Download MS Excel (102.9 KB)Supplementary Table SIII
Download MS Excel (1.4 MB)Supplementary Table SIV
Download MS Excel (45.9 KB)Supplemental figure, uncropped gel images
Download PDF (531.4 KB)Acknowledgements
B.W.M.v.B. is supported by the Netherlands Foundation for Cardiovascular excellence (Grant No. 003/08) and The Netherlands Organization for Scientific Research (NGI/ZonMW Horizon project 935190280) and a UMC Utrecht Focus and Massa grant (Grant No. DIGD-DGK-DHL); M.C.V. is supported by The Netherlands Organization for Scientific Research (ZonMW VIDI grant), the Netherlands Institute for Regenerative Medicine and a UMC Utrecht Focus and Massa grant (Grant No. DIGD-DGK-DHL); O.G.d.J. is supported by the Netherlands Institute for Regenerative Medicine. Y.C. and M.G. are supported by the National Heart, Lung and Blood intramural program of the National Institutes of Health. P.V. and R.M.S. are supported by The European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement (Grant No. 260627). We thank J. Brummelman and P.M. de Bree for excellent technical assistance.
References
- Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. 2002, 3: 321-30. 10.3402/jev.v1i0.18396.
- Poliakov A, Spilman M, Dokland T, Amling CL, Mobley JA. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. 2009, 69: 159-167. 10.3402/jev.v1i0.18396.
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. 2002, 2: 569-610.
- Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. 2004, 16: 415-21. 10.3402/jev.v1i0.18396.
- Ohshima K, Inoue K, Fujiwara A, Hatekeyama K, Kanto K, Watanabe Yet al.Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. 2010, 5: e13247. 10.3402/jev.v1i0.18396.
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. 2007, 9: 654-9. 10.3402/jev.v1i0.18396.
- Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze H. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. 1998, 273: 20121-7. 10.3402/jev.v1i0.18396.
- Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. 2008, 180: 7249-58.
- Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. 2007, 1: 156-8. 10.3402/jev.v1i0.18396.
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JLet al.Functional delivery of viral miRNAs via exosomes. 2010, 107: 6328-33. 10.3402/jev.v1i0.18396.
- Zhan R, Leng X, Liu X, Wang X, Gong J, Yan Let al.Heat shock protein 70 is secreted from endothelial cells by a non-classical pathway involving exosomes. 2009, 387: 229-33. 10.3402/jev.v1i0.18396.
- Taylor AR, Robinson MB, Gifondorwa DJ, Tytell M, Milligan CE. Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. 2007, 67: 1815-29. 10.3402/jev.v1i0.18396.
- Lancaster GI, Febbraio MA. Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. 2005, 280: 23349-55. 10.3402/jev.v1i0.18396.
- Clayton A, Turkes A, Navabi H, Mason MD, Tabi Z. Induction of heat shock proteins in B-cell exosomes. 2005, 118: 3631-8. 10.3402/jev.v1i0.18396.
- Gupta S, Knowlton AA. HSP60 trafficking in adult cardiac myocytes: role of the exosomal pathway. 2007, 292: H3052-6. 10.3402/jev.v1i0.18396.
- Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JAet al.Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. 2005, 65: 5238-47. 10.3402/jev.v1i0.18396.
- Eldh M, Ekstrom K, Valadi H, Sjostrand M, Olsson B, Jernas Met al.Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. 2010, 5: e15353. 10.3402/jev.v1i0.18396.
- Goligorsky MS, Kuo MC, Patschan D, Verhaar MC. Review article: endothelial progenitor cells in renal disease. 2009, 14: 291-7. 10.3402/jev.v1i0.18396.
- Sheldon H, Heikamp E, Turley H, Dragovic R, Thomas P, Oon CEet al.New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. 2010, 116: 2385-94. 10.3402/jev.v1i0.18396.
- Al-Nedawi K, Szemraj J, Cierniewski CS. Mast cell-derived exosomes activate endothelial cells to secrete plasminogen activator inhibitor type 1. 2005, 25: 1744-9. 10.3402/jev.v1i0.18396.
- Basile DP. The endothelial cell in ischemic acute kidney injury: implications for acute and chronic function. 2007, 72: 151-6. 10.3402/jev.v1i0.18396.
- Fagan SC, Hess DC, Hohnadel EJ, Pollock DM, Ergul A. Targets for vascular protection after acute ischemic stroke. 2004, 35: 2220-5. 10.3402/jev.v1i0.18396.
- Fisher M. Injuries to the vascular endothelium: vascular wall and endothelial dysfunction. 2008, 5Suppl 1S4-11.
- Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DCet al.HMEC-1: establishment of an immortalized human microvascular endothelial cell line. 1992, 99: 683-90. 10.3402/jev.v1i0.18396.
- Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJet al.B lymphocytes secrete antigen-presenting vesicles. 1996, 183: 1161-72. 10.3402/jev.v1i0.18396.
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. . 2006; Chapter 3: 3.22.1–3.22.29.
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. 2002, 74: 5383-92. 10.3402/jev.v1i0.18396.
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. 2003, 75: 4646-58. 10.3402/jev.v1i0.18396.
- Huber W, von HA, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. 2002, 18Suppl 1S96-104. 10.3402/jev.v1i0.18396.
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. 2004, 3: Article 3.
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. 2003, 31: e154. 10.3402/jev.v1i0.18396.
- Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0 – a multifunctional tool for GO term enrichment analysis and data exploration. 2008, 24: 1650-1. 10.3402/jev.v1i0.18396.
- Grossmann S, Bauer S, Robinson PN, Vingron M. Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. 2007, 23: 3024-31. 10.3402/jev.v1i0.18396.
- Slot JW, Geuze HJ. Cryosectioning and immunolabeling. 2007, 2: 2480-91. 10.3402/jev.v1i0.18396.
- Brodsky SV, Zhang F, Nasjletti A, Goligorsky MS. Endothelium-derived microparticles impair endothelial function in vitro. 2004, 286: H1910-5. 10.3402/jev.v1i0.18396.
- Mezentsev A, Merks RM, O'Riordan E, Chen J, Mendelev N, Goligorsky MSet al.Endothelial microparticles affect angiogenesis in vitro: role of oxidative stress. 2005, 289: H1106-14. 10.3402/jev.v1i0.18396.
- Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. 2007, 7: 340-50. 10.3402/jev.v1i0.18396.
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan Set al.Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. 2004, 3: 1154-69. 10.3402/jev.v1i0.18396.
- Buschow SI, van Balkom BW, Aalberts M, Heck AJ, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. 2010, 88: 851-6. 10.3402/jev.v1i0.18396.
- Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. 2008, 8: 4083-99. 10.3402/jev.v1i0.18396.
- van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan Net al.Intestinal epithelial cells secrete exosome-like vesicles. 2001, 121: 337-49. 10.3402/jev.v1i0.18396.
- Brillard-Bourdet M, Rehault S, Juliano L, Ferrer M, Moreau T, Gauthier F. Amidolytic activity of prostatic acid phosphatase on human semenogelins and semenogelin-derived synthetic substrates. 2002, 269: 390-5. 10.3402/jev.v1i0.18396.
- Jonsson M, Lundwall A, Linse S, Frohm B, Malm J. Truncated semenogelin I binds zinc and is cleaved by prostate-specific antigen. 2006, 27: 542-7. 10.3402/jev.v1i0.18396.
- Wang XL, Fu A, Spiro C, Lee HC. Proteomic analysis of vascular endothelial cells-effects of laminar shear stress and high glucose. 2009, 2: 445. 10.3402/jev.v1i0.18396.
- Cooper JT, Stroka DM, Brostjan C, Palmetshofer A, Bach FH, Ferran C. A20 blocks endothelial cell activation through a NF-kappaB-dependent mechanism. 1996, 271: 18068-73. 10.3402/jev.v1i0.18396.
- Opipari AW Jr, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. 1990, 265: 14705-8.
- Sluiter W, Pietersma A, Lamers JM, Koster JF. Leukocyte adhesion molecules on the vascular endothelium: their role in the pathogenesis of cardiovascular disease and the mechanisms underlying their expression. 1993, 22Suppl 4S37-44. 10.3402/jev.v1i0.18396.
- Peterson DB, Sander T, Kaul S, Wakim BT, Halligan B, Twigger Set al.Comparative proteomic analysis of PAI-1 and TNF-alpha-derived endothelial microparticles. 2008, 8: 2430-46. 10.3402/jev.v1i0.18396.
- Gu L, Tseng SC, Rollins BJ. Monocyte chemoattractant protein-1. 1999, 72: 7-29.
- Gerszten RE, Garcia-Zepeda EA, Lim YC, Yoshida M, Ding HA, Gimbrone Jr MAet al.MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. 1999, 398: 718-23. 10.3402/jev.v1i0.18396.
- Kim SH, Han SY, Azam T, Yoon DY, Dinarello CA. Interleukin-32: a cytokine and inducer of TNFalpha. 2005, 22: 131-42.
- Carlos T, Kovach N, Schwartz B, Rosa M, Newman B, Wayner Eet al.Human monocytes bind to two cytokine-induced adhesive ligands on cultured human endothelial cells: endothelial-leukocyte adhesion molecule-1 and vascular cell adhesion molecule-1. 1991, 77: 2266-71.
- Curran JE, Weinstein SR, Griffiths LR. Polymorphic variants of NFKB1 and its inhibitory protein NFKBIA, and their involvement in sporadic breast cancer. 2002, 188: 103-7. 10.3402/jev.v1i0.18396.
- Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. 2003, 536: 135-40. 10.3402/jev.v1i0.18396.
- Ostergaard L, Simonsen U, Eskildsen-Helmond Y, Vorum H, Uldbjerg N, Honore Bet al.Proteomics reveals lowering oxygen alters cytoskeletal and endoplasmatic stress proteins in human endothelial cells. 2009, 9: 4457-67. 10.3402/jev.v1i0.18396.
- Kovacevic Z, Sivagurunathan S, Mangs H, Chikhani S, Zhang D, Richardson DR. The metastasis suppressor, N-myc downstream regulated gene 1 (NDRG1), upregulates p21 via p53-independent mechanisms. 2011, 32: 732-40. 10.3402/jev.v1i0.18396.
- De LF, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. 2007, 313: 4130-44. 10.3402/jev.v1i0.18396.
- Zeng Y, Kulkarni P, Inoue T, Getzenberg RH. Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. 2009, 107: 179-88. 10.3402/jev.v1i0.18396.
- Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. 2001, 61: 6669-73.
- van Balkom BW, Pisitkun T, Verhaar MC, Knepper MA. Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. 2011, 80: 1138-45. 10.3402/jev.v1i0.18396.
- Zhou H, Yuen PS, Pisitkun T, Gonzales PA, Yasuda H, Dear JWet al.Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. 2006, 69: 1471-6.
- Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. 2009, 10: 42-6. 10.3402/jev.v1i0.18396.
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves Met al.Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. 2008, 10: 1470-6. 10.3402/jev.v1i0.18396.
- Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. 2009, 6: 267-83. 10.3402/jev.v1i0.18396.