Abstract
Introduction
Protein deimination, defined as the post-translational conversion of protein-bound arginine to citrulline, is carried out by a family of 5 calcium-dependent enzymes, the peptidylarginine deiminases (PADs) and has been linked to various cancers. Cellular microvesicle (MV) release, which is involved in cancer progression, and deimination have not been associated before. We hypothesize that elevated PAD expression, observed in cancers, causes increased MV release in cancer cells and contributes to cancer progression.
Background
We have previously reported that inhibition of MV release sensitizes cancer cells to chemotherapeutic drugs. PAD2 and PAD4, the isozymes expressed in patients with malignant tumours, can be inhibited with the pan-PAD-inhibitor chloramidine (Cl-am). We sought to investigate whether Cl-am can inhibit MV release and whether this pathway could be utilized to further increase the sensitivity of cancer cells to drug-directed treatment.
Methods
Prostate cancer cells (PC3) were induced to release high levels of MVs upon BzATP stimulation of P2X7 receptors. Western blotting with the pan-protein deimination antibody F95 was used to detect a range of deiminated proteins in cells stimulated to microvesiculate. Changes in deiminated proteins during microvesiculation were revealed by immunoprecipitation and immunoblotting, and mass spectrometry identified deiminated target proteins with putative roles in microvesiculation.
Conclusion
We report for the first time a novel function of PADs in the biogenesis of MVs in cancer cells. Our results reveal that during the stimulation of prostate cancer cells (PC3) to microvesiculate, PAD2 and PAD4 expression levels and the deimination of cytoskeletal actin are increased. Pharmacological inhibition of PAD enzyme activity using Cl-am significantly reduced MV release and abrogated the deimination of cytoskeletal actin. We demonstrated that combined Cl-am and methotrexate (MTX) treatment of prostate cancer cells increased the cytotoxic effect of MTX synergistically. Refined PAD inhibitors may form part of a novel combination therapy in cancer treatment.
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
Microvesicles (MVs) are defined as intact, submicron, phospholipid-rich vesicles that are ubiquitously released from the cell membrane of diverse cell types upon stimulation and/or apoptosis (Citation1). As cells are constantly challenged in the environment by various factors ranging from chemical to physical stress, a variety of vesicles are released that differ in size and composition depending on the status and origin of the cell (Citation2).
Cell activation and subsequent MV release have been shown to depend on an influx of calcium ions through pores made by sublytic complement, or released by the endoplasmic reticulum (ER) and through various calcium channels on activated cells (Citation1, Citation3). The rise in cytosolic calcium leads to the activation of enzymes such as calpain, gelsolin, scramblase as well as protein kinases. Simultaneously, enzymes such as translocase and phosphatases are inhibited, therefore resulting in cytoskeletal reorganisation, loss of membrane asymmetry, membrane blebbing and MV formation and release (Citation4).
Over the last decade, MVs have been implicated to play roles directly or indirectly in the pathogenesis of various diseases including cancer. The presence of MVs in cancer patients had already been noticed since the late 1970s (Citation5) and various investigators demonstrated elevated levels of MVs in the blood from cancer patients compared to healthy individuals (Citation6–Citation8). Physiologically, MVs have been identified as mediators of intercellular communication by transferring growth factors, micro RNAs and enzymes between cells and therefore influencing processes as diverse as differentiation, migration and angiogenesis (Citation9, Citation10). This mechanism has also been shown to aid tumour survival and spread as the various soluble proteins, pathological growth factor receptors and micro RNAs are transported via MVs (Citation9, Citation10).
MV release has also been identified as one of the mechanisms contributing to cancer drug resistance, as cancer cells can evade chemotherapeutic agents by increasing active drug efflux through MV shedding (Citation11, Citation12). In the same vein, recent work from our laboratory has shown that by inhibiting microvesiculation of cancer cells their resistance to anticancer drug treatment with methotrexate (MTX) is reduced (Citation12).
Protein deimination is a post-translational modification caused by a family of Ca2+-dependent enzymes, the peptidylarginine deiminases (PADs) (Citation13). Protein deimination is the irreversible post-translational conversion of protein-bound arginine and positively charged methylarginine residues to the neutral amino acid, citrulline. This leads to structural and functional changes in target proteins and has critical physiological and pathological consequences (Citation14–Citation16).
In mammals, 5 PAD isozymes (PADs 1–4 and 6) have been identified that exhibit tissue-specific expression patterns, vary in their subcellular localisation and selectively target proteins (Citation15, Citation17) (Citation18). In humans, the PAD genes are localised to a well-organized gene cluster at 1p36.13, which is also identified to be the cluster region for the tumour suppressor gene RUNX3 (Citation19).
While PAD-mediated protein deimination regulates numerous physiological functions, PAD dysregulation and resulting changes in protein deimination are implicated in central nervous system damage (Citation20, Citation21), neurodegenerative diseases (Citation22), various human autoimmune diseases (Citation23–Citation25) and cancer (Citation26). Both PAD2 and PAD4 isozymes have been linked with cancer and are reported to be overexpressed in blood and tissues of patients suffering from malignant tumours (Citation3, Citation27) (Citation28).
PAD2 is the most broadly expressed PAD isozyme in the body, detected in multiple organs including cells of the haematopoietic lineage, spleen, secretory glands and brain (Citation29–Citation31). Known PAD2 substrates include myelin basic protein in the nervous system (Citation32) and vimentin, an intermediate filament that provides support to cell organelles in conjunction with the cytoskeleton (Citation33, Citation34). Importantly, PAD2 has been shown to play a role in gene regulation in breast cancer cells (Citation28, Citation35).
PAD4 is the most characterised PAD isozyme and is expressed mainly in haematopoietic progenitor cells and cells of the immune system (granulocytes, monocytes and natural killer cells). It is the only isoform to contain a classic nuclear localisation signal and is commonly localised in the nucleus of the cell (Citation34, Citation36–Citation38) although recently PAD2 and PAD3 have also been reported in the nucleus (Citation21, Citation28) (Citation39). PAD4 has been shown to translocate to the nucleus in response to upregulated tumour necrosis factor alpha (TNFα) (Citation40), as well as to play a role in innate immunity by the formation of neutrophil extracellular traps during bacterial infection (Citation23, Citation36) (Citation41). PAD4 acts as a transcriptional co-regulator for a range of factors such as p53, p300, p21 and ELK1. This regulatory function of PAD4 is thought to be mediated via deimination of the N-terminal tails of various histone proteins (Citation42–Citation44). PAD4 also acts as a co-mediator of gene transcription and epigenetic cross talk with histone deacetylase 2 (HDAC2) to regulate p53 gene activity during DNA damage playing a role in apoptosis (Citation45).
PAD4 has been co-localised with cytokeratin (CK), an established tumour marker. Various isoforms of CK (CKs 8, 18, and 19) are deiminated, making them resistant to caspase-mediated cleavage, in turn, contributing to the disruption of apoptosis in cancer tumours (Citation46). PAD4 has also been linked with the regulation of oestrogen receptor target gene activity, mediated by oestrogen stimulation via histone tail deimination (Citation47). In addition, the PAD4 isozyme has been shown to act as a cofactor in epidermal growth factor mediated target gene activity activating the expression of the proto-oncogene c-fos (Citation44), interacting with p53 and influencing the expression of its target genes (Citation42, Citation43) (Citation48, Citation49).
As both microvesiculation and PAD enzyme activation are calcium-dependent events that have been shown to be elevated in certain human diseases including autoimmune diseases and cancer (Citation22, Citation23) (Citation26, Citation35) (Citation50, Citation51), we hypothesized that PAD enzyme activation and microvesiculation might play synergistic roles in cancer progression. Here, we demonstrate this association in the prostate cancer cell line, PC3.
Materials and methods
Cell culture
The highly metastatic prostate cancer cell line PC3 (Sigma–Aldrich, Gillingham, U.K.) and a control immortalised normal prostate cell line (PNT2; ECACC) were cultured in MV-free complete growth medium (CGM) consisting of EMV (exosome and MV)-free RPMI 1640 supplemented with 10% EMV-free foetal bovine serum (FBS; Hyclone, Thermo Scientific, Paisley, UK) in the absence of antibiotics. The CGM medium supplemented with 10% FBS was then centrifuged at 100,000g for 2 h to remove exosomes and MVs before using it in cell culture. EMV-free RPMI, phosphate-buffered saline (PBS), normal human serum (NHS) and FBS were prepared by centrifugation (100,000g/2 h) and filtering (0.1 µm pore size filter).
Purification of MVs
MVs were purified according to previously established protocols (Citation52). In brief, the supernatant was collected from the cell cultures following experiments and centrifuged once at 200g for 5 min to remove the cells. The supernatant was then centrifuged at 4,000g for 1 h to remove cell debris and further at 15,000g for 2 h to pellet MVs, which were then washed once by resuspending in sterile, EMV-free PBS and centrifuged again at 15,000g for 2 h. The MV pellet was resuspended in sterile, EMV-free, MV-free PBS and quantified [by nanoparticle tracking analysis (NTA), as described below], or analysed for phosphatidylserine exposure (Citation52) or quantified using the Guava EasyCyte microcapillary flow cytometer (10,000 events, 0.24 µl/s flow rate).
PAD isotype expression in cancer and non-cancerous cells
To determine the PAD isotype expressed in cancer and control cells, PC3 and PNT2 cells were labelled with PAD2 and PAD4 antibodies and analysed by flow cytometry, fluorescence microscopy and Western blotting.
Flow cytometry
In brief, 5×105 viable cells were placed in 1.5 ml Eppendorf tubes in triplicate and fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature (RT). The cells were then washed 3 times with cold EMV-free PBS at 400g for 5 min and resuspended in permeabilisation buffer (PB: 0.5% Triton-X 100 in PBS) for 5 min at RT. Permeabilised cells were washed 3 times and incubated with primary antibody (PAD2 or PAD4, 1:500 in 3% BSA/PBS) at 4°C for 1 h on a shaking platform. The cells were washed 3 times and incubated with FITC-conjugated anti-rabbit IgG (Sigma–Aldrich) in 3% BSA/PBS at 4°C for 1 h, washed 3 times with cold 1% BSA/PBS again and resuspended in 200 µl of PBS containing 3% BSA, 1% NaN3 and analysed by flow cytometry using the Guava EasyCyte microcapillary flow cytometer at a flow rate of 0.56 µl/s.
NTA protocol
The particles present in samples were measured by NTA, using the NTA (NS500; Nanosight, Amesbury, UK), equipped with an sCMOS camera and a 405 nm diode laser. Data acquisition and processing were performed using the NTA software version NTA 3.0 0068. Background extraction was applied together with automatic settings for the minimum expected particle size, minimum track length and blur and the ambient temperature was set at 23°C. Silica beads (100 nm diameter; Microspheres-Nanospheres, Cold Spring, NY, USA) were used to configure and calibrate the instrument. Samples were diluted 10- to 50-fold in particle-free PBS to maintain the number of particles in the field of view between approximately 20 and 40 particles. For each sample, 4 videos of 30 s duration were recorded generating replicate histograms that were averaged. Only measurements with at least 1,000 completed tracks were analysed (Citation53).
Fluorescence microscopy
PC3 and PNT2 cells (5×105 cells/well) were seeded on a cover slip in a 24-well plate in triplicate, incubated for 24 h, washed gently with prewarmed PBS, fixed with 4% PFA for 10 min at RT, washed 3 times with cold PBS and resuspended in PB for 5 min at RT. The buffer was then removed and the cells were washed 3 times as before.
After incubation with PAD2 or PAD4 primary antibodies (1:500 dilution in 3% BSA/PBS) for 1 h at 4°C on a shaking platform, the cells were washed 3 times with cold PBS and further incubated with AlexaFluor 488 conjugated anti-rabbit IgG secondary antibody (Invitrogen; 5 µg/ml in 3% BSA/PBS) at 4°C for 1 h on a shaking platform in the dark. The cells were then washed 3 times with cold 1% BSA/PBS and the cover slips mounted on to slides with DAPI-VECTASHIELD medium (Vector Laboratories, Inc., Burlingame, CA, USA). Images were acquired using a fluorescence microscope (1X81 motorized inverted fluorescence microscope, Olympus Corporation, Hamburg, Germany). The mean green fluorescence intensity of the fluorescence images was analysed as per the manufacturer's instructions using the Cell∧M imaging software (Olympus Corporation) provided with the Olympus 1X81 fluorescence microscope (Olympus Corporation).
SDS–PAGE
PC3 and PNT2 cells were lysed, diluted in 4× SDS sample buffer and incubated at 95°C for 4 min. Protein electrophoresis was performed as previously described (Citation54) using the Mini PROTEAN III Electrophoresis System (Bio-Rad, Hemel Hempstead, U.K.) and freshly prepared 8% polyacrylamide separating gels. Electrophoretic separation was performed at 100 V (constant voltage) and gels were either stained with Coomassie Brilliant Blue (BDH Ltd, Poole, England) or transferred onto nitrocellulose membrane for Western blotting analysis.
Western blotting
SDS–PAGE gels were transferred to a Hybond C nitrocellulose membrane for further analysis using a semi-dry transfer device (Bio-Rad Sartoblot System). Blotting paper, nitrocellulose membrane and the sandwich-blotting cassette were equilibrated in Sartoblot buffer. Electroblotting was carried out at 150 mA for 1 h.
The membranes were incubated in blocking buffer (6% non-fat milk/PBS-T) for 1 h at RT or at 4°C, overnight, on a shaker. Following blocking, the membranes were rinsed with PBS-T and incubated with the primary antibody for 1 h at RT on a shaker (PAD2, PAD4 1:500, Abcam, Cambridge, U.K., in 3% non-fat milk/PBS-T). Anti-β-actin (1:1,000, Santa Cruz, Biotechnology Inc., Heidelberg, Germany) was used as a loading control. Three 10 min washing steps with PBS-T were performed and the membranes were incubated with secondary HRP-labelled goat-anti-mouse-IgG antibodies (1:1,000; Sigma–Aldrich) for 1 h at RT. After washing 6 times in PBS-T, visualization was performed using the enhanced chemiluminescence reagent system (ECL, Amersham Pharmacia, Little Chalfont, U.K.). Chemiluminescence was detected using the UVP ChemiDoc-It system (UVP Systems, Cambridge, UK). Where necessary, band intensities on the Western blot were analysed using Image J software according to the manufacturer's instructions.
Nuclear protein preparation
TSSM buffer (20 mM Tris, pH 8; 100 mM NaCl; 300 mM sucrose; 2 mM MgCl2) with protease inhibitor cocktail was added to cells on ice for 10 min followed by centrifugation at 720g for 10 min/4°C. The pellet was resuspended in TSS buffer (20 mM Tris, pH 8; 100 mM NaCl; 2 mM EDTA, pH 8; which was rendered 300 mM NaCl) and then homogenised in a Dounce homogeniser (20 strokes on ice), left on ice for 20 min and then centrifuged at 24,000g for 20 min/4°C. The supernatant was then quantified for protein using the BCA assay kit (Life Technologies Ltd., Paisley, U.K.).
The effect of PAD inhibition on MV release
To determine the effect of PAD enzyme activity on cellular MV release, cell cultures were incubated with the irreversible pan-PAD-inhibitor chloramidine (Cl-am) (Citation20).
The cell preparations (quantified and tested for viability) were washed once with prewarmed EMV-free PBS (Supplementary Fig. 1), resuspended in prewarmed serum- and EMV-free RPMI 1640 (100,000g/2 h and filtered through 0.1 µm pore size membrane) supplemented with 2 mM CaCl2 at 5×105 cells and treated with BzATP (2′(3′)-O-(4-benzoylbenzoyl)adenosine-5′-trisphosphatetriethylammonium salt (300 µM)) to induce MV release (37°C for 30 min), serial concentrations of Cl-am (ranging from 5 to 50 µM) being included where necessary to inhibit microvesiculation. In some experiments, cells were incubated with 10% EMV-free NHS [from Human male AB plasma, Sigma–Aldrich; as a source of sublytic complement to stimulate MVs (Citation51)], rendered EMV-free by centrifugation at 100,000g for 2 h and filtering through a 0.1-µm pore size filter membrane, and further incubated at 37°C in a 12-well plate for 30 min with shaking. The plate was placed on ice for 1 min and the supernatant from each well was transferred to sterile 1.5 ml Eppendorf tubes on ice and centrifuged at 200 g for 5 min to remove the cell debris. The supernatants were treated as described earlier for isolation of MVs. The resulting MV pellet was suspended in 200 µl of sterile EMV-free PBS and analysed using the Nanosight (NS500; Nanosight, Amesbury, UK) or by flow cytometry (Millipore, Watford, U.K., microcapillary flow cytometer).
Immunoprecipitation (IP) of deiminated target proteins
To analyse deiminated proteins from experiments in which cells had been stimulated to microvesiculate, 500 µg of cell lysates (in RIPA buffer; 150 mM NaCl, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8 – Sigma–Aldrich) from PC3 and control cells treated with or without Cl-am were immunoprecipitated with the pan-citrulline F95 antibody (4 µg) that specifically recognizes protein bound citrullines (Citation55), using the Catch and Release v2.0 IP kit (Millipore), according to the manufacturer's instructions. Bound proteins were eluted using denaturing Elution Buffer containing 5% β-mercaptoethanol and the eluate stored at −20°C for further analysis.
Mass spectrometry analysis of deiminated proteins from PC3 cells stimulated for microvesiculation
To identify proteins that were deiminated during MV stimulation of PC3 cells, 2,000 µg of cell lysates from healthy PC3 cells only, or PC3 cells pretreated with or without 50 µM Cl-am and then stimulated with 300 µM BzATP, were immunoprecipitated with F95 antibody and eluted with 70 µl of non-denaturing buffer 4 times successively for maximum recovery of protein. The eluate was then subjected to mass spectrometry analysis.
Mass spectrometry analysis: in-solution trypsin digestion and MSE label-free quantitation
Protein samples in duplicate were denatured by the addition of 20 µl of 100 mM Tris-HCl buffer (pH 7.2) supplemented with 5 M dithioerythreitol and 6 M urea at RT for 60 min on a shaking platform. Free thiol groups in the samples were then carboamidomethylated by the addition of 6 µl of 100 mM Tris-HCl (pH 7.8) supplemented with 5 M iodoacetamide for 45 min. Next, the samples were incubated with 180 µl of 1 µg sequence grade trypsin dissolved in ddH2O for 12–16 h at 37°C. The samples were centrifuged and the supernatant was collected and analysed by mass spectrometry. Prior to analysis, each digest was spiked with 1 pmol of an enolase tryptic digestion (Saccharomyces cerevisiae), which acted as an internal standard for the quantitation of each protein.
The processed samples were then identified and quantified by direct analysis using a nanoAcquity UPLC (ultra performance liquid chromatography) and a QTOF (quadrupole time of flight) Premier mass spectrometer (Waters Corporation, Manchester, UK). Briefly, the samples were trapped and desalted using a Symmetry C18 5 µm, 5 mm×300 µm precolumn in 0.1% formic acid for a total time of 4 min at a rate of 4 µl/min and then eluted. The eluate was then separated using a 15 cm×75 µm C18 reverse-phase analytical column using a gradient of 3–40% acetonitrile containing 0.1% formic acid over a period of 120 min at a flow rate of 250 nl/min. The column was washed and regenerated at 300 nl/min for 10 min using 99% acetonitrile containing 0.1% formic acid. After the removal of all non-peptide and non-polar materials, the column was re-equilibrated under the initial starting conditions for 20 min. All columns were maintained at 35°C (Citation56). After purification, the samples were analysed in positive ion mode with the QTOF operated in v-mode with a typical resolving power of 10,000 fwhm after the calibration of the TOF analyser. Accurate mass using LC-MS was collected in a data-independent and alternating, low and collision energy mode. The data were then processed using the ProteinLynx GlobalServer version 2.4 (Waters Corporation) (Citation56).
Effect of PAD inhibition on the cytotoxic effect of methotrexate on cancer cells
To test whether Cl-am would enhance the cytotoxic effect of MTX, adherent PC3 cells were washed and seeded at 5×104 cells/well in 24-well plates 24 h before the experiment to allow the cells to attach and settle. On the day of the experiment, the cells were washed once and resuspended in EMV-free CGM with various concentrations of either MTX, or chloramidine, or MTX in combination with 100 µM chloramidine (chloramidine having been preincubated for 30 min prior to adding MTX) in triplicate and incubated for a further 48 h at 37°C (5% CO2, humidified conditions). Cells without any treatments served as controls.
After incubation, the supernatant was collected. The cells were trypsinised and centrifuged together with the supernatant at 500 g for 5 min. The wells were then washed once with prewarmed PBS, which was transferred to the individual tubes containing the cells. The cells were centrifuged once more at 500g for 5 min and resuspended in prewarmed RPMI 1640 and analysed for apoptosis by flow cytometry using the Guava ViaCount assay (Guava Millipore) as per the manufacturer's protocol.
Statistical tests
Statistical analysis was performed by using the Student's t-test, one-way analysis of variance (ANOVA) with Dunnett's multiple comparison test or two-way ANOVA with Bonferroni post hoc test where appropriate, using GraphPad Prism software, version 5.0 (GraphPad Software, San Diego, CA, USA). Differences giving a P-value<0.05 were considered statistically significant.
Results
PAD2 and PAD4 are expressed in both benign prostate PNT2 cells and PC3 cancer cells
To investigate the role of PAD activity in normal versus cancer cells, we began by evaluating the expression of PAD isozymes and found that by Western blotting that PC3 and PNT2 cells express both PAD2 and PAD4 isozymes, PAD2 and PAD4 being more highly expressed in PC3 than PNT2 cells (a and b). Fluorescent microscopy further suggested an intracellular, cytoplasmic and perinuclear location (Supplementary Fig. 2a and b).
Fig. 1. Immunoblotting and flow cytometry reveal higher detected PAD4 than PAD2 in PC3 and PNT2 cells and higher PAD2/4 expression in PC3 than PNT2 cells. (a and b) Western blot analysis and associated densitometry analysis (using Image J software) representing band intensity of healthy PC3 and PNT2 controls indicate higher detected PAD4 than PAD2 and of PAD2 and PAD4 in prostate cancer cells (PC3) compared to benign prostate cells (PNT2). Loading control: β-actin and Coomassie-stained SDS–PAGE. PC3 (c and d) and control PNT2 (c and e) cells were fixed, permeabilised and labelled with anti-PAD2 and anti-PAD4 antibody followed by FITC-IgG secondary antibody and analysed for PAD expression via flow cytometry. (c) Median fluorescence intensity (MFI) is a indicative of expression levels of PAD/cell. Significant differences were seen between PAD2 and PAD4 expression in both cell lines. Histograms in (d) represent% positive PC3/PNT2 cells for PAD2 and PAD4. Data represent the mean±SEM of 2 independent experiments performed in triplicate. *P<0.05, **P<0.001.
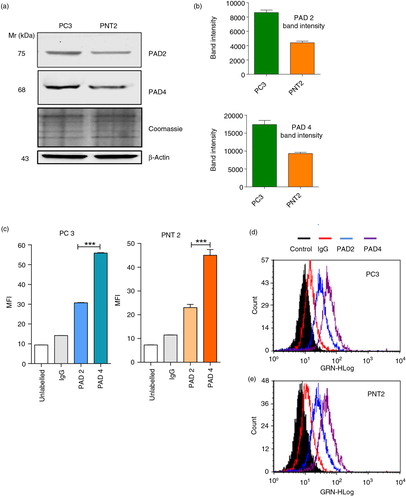
PAD2 and PAD4 expression by flow showed higher detected PAD4 than PAD2 in both cell lines (c), 83% positive cells for PAD4 compared to 38% for PAD2 in PC3 cells (d) and 70% for PAD4 and 30% for PAD2 in PNT2 cells (e).
Cl-am inhibits microvesiculation in PC3 and PNT2 cells and reduces nuclear PAD translocation during MV release
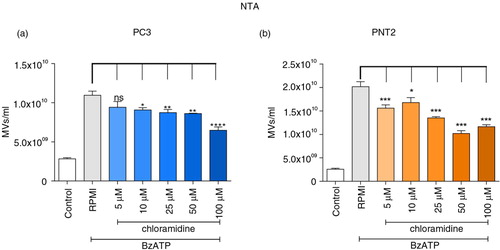
The effect of pharmacological inhibition of PAD on microvesiculation was monitored as the release of PS-rich vesicles showing a typical Forward/Side Scatter dot plot, a distinct population averaging at 200 nm in diameter by electron microscopy or just below 300 nm by NTA (Supplementary Fig. 3) and with weak expression of exosome markers TSG101 and Alix. PC3 prostate cancer cells were pretreated with serial concentrations of Cl-am and microvesiculation induced with 300 µM BzATP. MVs were then isolated and enumerated by NTA and flow cytometry.
Whilst the same trends in MV numbers were noted, compared to NTA () flow underestimated levels by 103-fold (Supplementary Fig. 4). Incubating PC3 and PNT2 cells with BzATP significantly increased the number of released MVs, whilst preincubating the cells with the pan-PAD inhibitor, Cl-am (5–100 µM), caused a significant reduction of microvesiculation (with no concomitant reduction in cell viability, Supplementary Fig. 5), which in the case of the NTA measurements was shown to be dose dependent (). This asserts a functional role of PAD enzymes in the microvesiculation of both normal and cancerous prostate cells.
Fig. 2. PAD inhibition reduces microvesiculation in PC3 prostate cancer and PNT2 control cells. PC3 and PNT2 cells pretreated with or without various concentrations of PAD-inhibitor chloramidine (Cl-am) were stimulated with BzATP and incubated at 37°C for 30 min. MVs were then isolated and counted by NTA (a and b). Significant reductions in microvesicle release were detected upon treatment with Cl-am. The data are represented as the mean±SEM of 3 experiments performed in triplicates. **P<0.005 and ***P<0.001 were considered statistically significant (one-way ANOVA).
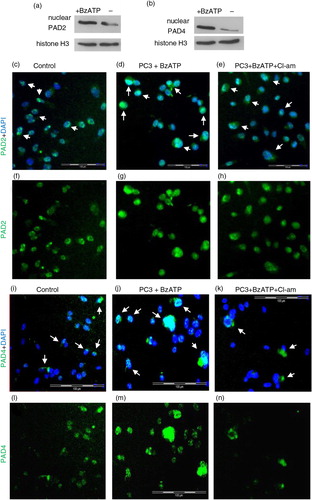
To further identify the localisation of PAD2 and PAD4 enzymes during BzATP stimulation and Cl-am inhibition, PC3 cells were pretreated with 10 µM Cl-am (30 min at 37°C), stimulated with BzATP and labelled with PAD2 or PAD4 antibody followed by isotype-matched AlexaFluor 488 antibodies and mounted onto slides for fluorescent microscopy analysis.
Compared to cells stimulated with BzATP to release MVs (a, d, g and b, j, m), there was a reduced expression of PAD2 and PAD4 in cells preincubated with Cl-am (a, e, h and b, k, n) (and in control, untreated cells, a, c, f and b, i, l). When stimulated to microvesiculate with BzATP, Western blotting showed an increased nuclear localisation of PAD2 and PAD4 (a and b, respectively) and fluorescence microscopy revealed a nuclear translocation, as reflected by the increase in fluorescence intensity from the cytoplasm in unstimulated cells (c, f, i, l, white arrows) to the nucleus (d, g, j, m, white arrows) although for PAD2 the fluorescence was relatively high around the nucleus, suggesting only a partial transfer. This increase in fluorescence intensity was reduced, however, upon preincubating cells with Cl-am prior to BzATP stimulation (e, h, k, n, white arrows).
Fig. 3. Increased expression of PAD2 and PAD4 upon microvesiculation is inhibited by chloramidine. Mean green fluorescence intensity [for PAD2 (a) and PAD4 (b)], as determined using Cell∧M imaging software on fluorescent microscopy images [panels (c–e) and (i–k)], respectively, indicates increased expression of PAD2 and 4 during BzATP stimulation of MV release. These values were obtained from PC3 cells preseeded on coverslips and pretreated with 10 µM Cl-am (e, h, k, n) or without Cl-am (d, g, j, m) for 30 min at 37°C. The cells were then washed and incubated at 37°C for 30 min in the presence (d, e, g, h and j, k, m, n) or absence (c, f, i, l) of 300 µM BzATP. (d, g, j, m) PAD2 and PAD4 (green) expression in PC3 cells are increased in response to stimulation of MV release (BzATP treatment) and reduced upon Cl-am treatment (e, h, k, n). (f–h) and (l–n) represent merged images of (c–e) and (i–k) respectively with nuclear (DAPI) staining. Scale bars – 100 µm, white bar, 20 µm.
![Fig. 3. Increased expression of PAD2 and PAD4 upon microvesiculation is inhibited by chloramidine. Mean green fluorescence intensity [for PAD2 (a) and PAD4 (b)], as determined using Cell∧M imaging software on fluorescent microscopy images [panels (c–e) and (i–k)], respectively, indicates increased expression of PAD2 and 4 during BzATP stimulation of MV release. These values were obtained from PC3 cells preseeded on coverslips and pretreated with 10 µM Cl-am (e, h, k, n) or without Cl-am (d, g, j, m) for 30 min at 37°C. The cells were then washed and incubated at 37°C for 30 min in the presence (d, e, g, h and j, k, m, n) or absence (c, f, i, l) of 300 µM BzATP. (d, g, j, m) PAD2 and PAD4 (green) expression in PC3 cells are increased in response to stimulation of MV release (BzATP treatment) and reduced upon Cl-am treatment (e, h, k, n). (f–h) and (l–n) represent merged images of (c–e) and (i–k) respectively with nuclear (DAPI) staining. Scale bars – 100 µm, white bar, 20 µm.](/cms/asset/c760d6cb-7bc5-4a03-9f25-09b9d0c74df3/zjev_a_11815560_f0003_ob.jpg)
Fig. 4. Nuclear PAD2 and PAD4 translocation during microvesiculation of PC3 cancer cells is reduced upon PAD inhibition. (a and b) Nuclear preparations indicate increased expression of PAD2 and PAD4, by Western blot, during BzATP-induced MV release. Cl-am treatment appears to reduce nuclear translocation of PAD2 and PAD4 (e, h, k, n). Panels in c–n represent magnified images of white squares in (nuclear PAD2 and PAD4 expression indicated by white arrows). Scale bars: 100 µm.
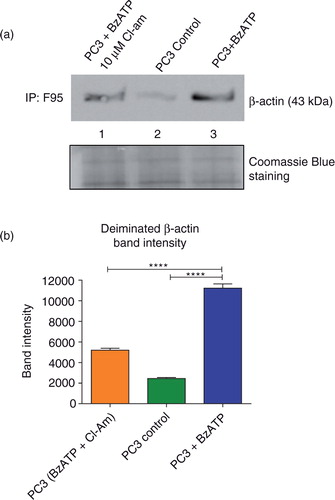
The cytoskeletal protein β-actin is deiminated by PADs during MV release
Having shown that PAD inhibition using Cl-am significantly reduced microvesiculation in both cancer and control cells, we hypothesized that actin filaments, which form part of the cytoskeleton and undergo considerable rearrangement allowing the shedding of MVs (Citation1), might be affected by changes in deimination, leading to reduced MV release.
PC3 cells (1×107) pretreated with 10 µM Cl-am were stimulated to microvesiculate as before. PC3 cells without any treatment served as controls. After incubation, the cells were washed and lysed with RIPA buffer and 500 µg of protein lysates from each treatment group immunoprecipitated using the pan-deimination F95 antibody and immunoblotted with anti-β-actin. Untreated PC3 cells had a very low expression of deiminated β-actin (a, L2 and b), whereas cells stimulated to microvesiculate had approximately 4 times greater levels (a, L3 and b). Cells that were preincubated with Cl-am and then stimulated to microvesiculate displayed a band intensity of β-actin twice as high as the control, but almost twice as low as BzATP-stimulated cells untreated with the PAD inhibitor (a, L1).
Fig. 5. MV stimulation of PC3 cells leads to increased β-actin deimination that is reduced upon PAD inhibition. PC3 cells were stimulated with BzATP to microvesiculate and total deiminated proteins immunoprecipitated using the F95 pan-deimination protein antibody. (a) Increased deiminated β-actin was observed in PC3 cells stimulated to microvesiculate with BzATP (Lane 3) compared to untreated, control PC3 cells (Lane 2). Upon treatment with 10 µM of the pan-PAD-inhibitor Cl-am prior to MV stimulation, a significant decrease was observed in the levels of deiminated β-actin (Lane 1). Equal amounts of protein extract immunoprecipitated with F95 were loaded as shown by Coomassie blue staining. (b) A bar chart representing deiminated β-actin band intensity as measured by protein densitometry and normalised intensity (calculated in Image J) from each of the treatments immunoblotted in (a).
Mass spectrometry analysis of deiminated protein candidates from PC3 cells stimulated for microvesiculation
Further to the likely involvement of deiminated β-actin in MV release, we repeatedly observed increased levels of deimination, comparing Western blots for deiminated proteins in stimulated versus resting cells (not shown). Whether such increases were of particular proteins or a global increase, we could not be sure, but with the aim to understand the involvement of PAD-mediated protein deimination in microvesiculation, we set out to identify further target proteins. Cell lysates from the 3 treatment groups: (a) untreated PC3 cells, (b) PC3 cells stimulated with BzATP and (c) PC3 cells treated with Cl-am prior to BzATP stimulation were immunoprecipitated with F95 antibody for the isolation of deiminated proteins. The eluates were then subjected to mass spectrometry analysis to identify differences in deiminated proteins that were present in each of the 3 samples.
Using ProteinLynx Global SERVER version 2.4 to process the data obtained from the analysis, deiminated proteins were identified through the human proteome UniProt database. As only duplicate samples were available, this was very much an exploratory experiment to indicate likely deiminated candidate proteins for future study. Altogether, 6 proteins were identified to be deiminated in PC3 cells (untreated and BzATP stimulated) excluding Yeast Enolase 1 (P00924), which is a protein that was not part of the sample, but was added manually as part of the analysis process. Of the 5 proteins found to be deiminated in PC3 cells without any stimulation or PAD inhibition (Table ), 4 had a confidence score of 2 (95% confidence – low probability of false positive; see “OK” column in Table ) and one protein had a confidence score of 1 (50% confidence – high probability of false positive).
Table I. Protein identities of deiminated protein candidates from PC3 cell lysates stimulated for microvesiculation
Actin alpha skeletal muscle was identified to be deiminated in untreated and BzATP-stimulated PC3 cells although with a reduced relative abundance upon Cl-am treatment. It is a highly conserved protein and plays a role in cell motility and microvesiculation. Glyceraldehyde-3-phosphate dehydrogenase deiminated in untreated PC3 cells is a multifunctional enzyme involved in glycolysis, nuclear functions such as transcription and DNA replication, as well as apoptosis (Citation57) and has also been implicated to play a role in the organization and assembly of the cytoskeleton (Citation58).
Deiminated nucleoside diphosphate kinase B was identified with high confidence only in untreated PC3 cells. It is involved in the synthesis of nucleoside triphosphates apart from ATP and has been identified as a candidate tumour metastasis suppressor, has histidine protein kinase activity and is a transcriptional activator of the regulatory myc gene that has been associated with cancer formation (Citation59, Citation60). Putative elongation factor 1 alpha like 3 as a deiminated protein was identified in untreated PC3 cells only. This protein has been reported to promote GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein synthesis (Citation61). Deiminated splicing regulatory glutamine lysine–rich protein 1 was identified with high confidence level only in PC3 cells stimulated with BzATP. Its function is to regulate alternative splicing by modulating the activity of other splice factors during RNA splicing (Citation61).
The PAD and microvesiculation inhibitor Cl-am sensitizes PC3 cells to methotrexate-mediated cytotoxicity
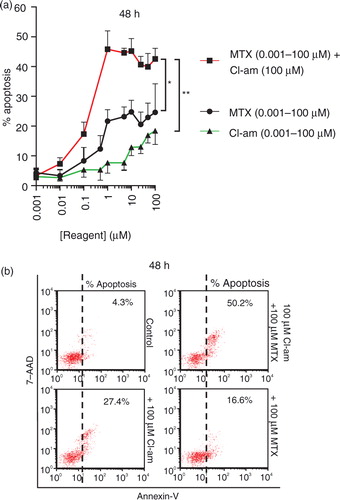
As Cl-am inhibits microvesiculation in PC3 cells, we sought to investigate whether treating cells with Cl-am in combination with MTX would affect cell survival. PC3 cells pretreated with Cl-am were given MTX in the range 0.001–100 µM (in the presence of CaCl2). After 24 h, the cells displayed a healthy morphology but over a 48-h period, increases in apoptosis were observed. PC3 cells treated with 1 µM MTX alone showed a significant increase in apoptosis to 22% over controls but when pretreated with 1 µM Cl-am, this increased to 47% (a). Cl-am (1 µM) alone only increased apoptosis to 6% compared to untreated control (4% apoptosis). Representative dot plots at 48 h using the highest concentrations of Cl-am and MTX (100 µM) are presented in b. Cl-am combined with MTX treatment thus had a synergistic effect on PC3 cancer cell viability over a 48-h period.
Fig. 6. The pharmacological PAD-inhibitor Cl-am synergistically increases the cytotoxic effects of the cancer drug methotrexate in PC3 prostate cancer cells. (a) PC3 cells were treated with Cl-am alone (0.001–100 µM) or with Cl-am fixed at 100 µM in combination with MTX (0.001–100 µM); MTX (0.001–100 µM) without any Cl-am was also used. Over a 48-h period, cells treated with 1 µM Cl-am alone had a slightly reduced viability showing average apoptosis levels of 6% compared to the highly viable, untreated cells (4% apoptosis). PC3 cells treated with 1 µM MTX alone showed a significant reduction in viability (apoptosis at 22%) compared to control cells. Combined treatment of 1 µM Cl-am with 1 µM MTX increased apoptosis to around 47%, indicating a synergistic effect of Cl-am and MTX treatment on PC3 cancer cell viability over a 48-h period. (b) Dot plot presentation of apoptosis (Annexin V) of PC3 cells after the 48-h incubation period with maximum concentrations of MTX, Cl-am (100 µM) or Cl-am/MTX from (a). *P<0.05, ***P<0.001.
Discussion
The role of MVs in helping maintain the delicate balance between health and disease has become a burgeoning field within biomedical research (Citation62). Apart from playing roles in various physiological functions including thrombosis, inflammation, angiogenesis, cellular cross talk and vasoconstriction (Citation63–Citation66), MVs have been linked with a wide range of clinical conditions such as diabetes, and cancer as well as cardiovascular, inflammatory and autoimmune disease (Citation50, Citation51) (Citation67–Citation69). Similarly, the PAD group of enzymes, which catalyses post-translational deimination of various target proteins, is receiving increasing interest in the medical and pharmacological fields due to implications in various autoimmune and neurodegenerative diseases, neuronal damage (Citation20, Citation21) as well as in the biology of cancer (Citation26).
However, to date no studies have aimed at elucidating any synergistic link between these 2 calcium-dependent mechanisms in cancer biology. Also, MV release being implicated in cancer progression and a significant expression of PAD4 has been reported in many tumour tissues including various forms of adenocarcinomas, breast carcinoma and malignant tumours as well as in the blood of cancer patients (Citation3, Citation46). In addition, various functions of PAD4 in cancer survival and progression have also been postulated. For example, CK, part of the cytoskeleton, has been identified as a deiminated target protein of PAD4. CK also plays a role in caspase-mediated apoptosis and is a potential tumour marker. When deiminated, CK is prevented from being cleaved by caspase, resulting in the inhibition of apoptosis of tumour cells (Citation46); this is intriguing in the context of MV release which is a feature of cells in early apoptosis. Another deiminated protein target of PAD4 is anti-thrombin, which loses its ability to inhibit thrombin and has various pro-cancer functions such as cell proliferation, angiogenesis and fibrin formation (Citation46).
PAD4 function has also been associated with cancer epigenetics (Citation70) through deimination of histone proteins that play a role in the regulation of various genes, including the tumour suppressor gene p53 (Citation45). Previously, elevated PAD4 levels have been detected in prostate adenocarcinoma (Citation46). We now show, similarly, that PAD isotypes 4 (and 2) are present at significantly elevated levels in metastatic prostate cancer cells (PC3) compared to the benign (PNT2) prostate cell line.
We report for the first time, that, upon stimulating cells to microvesiculate, nuclear translocation of PAD4 and a partial nuclear translocation of PAD2 was observed in PC3 cancer cells. Upon pharmacological inhibition of PAD activation prior to stimulating MV release, this nuclear PAD translocation was prevented. Furthermore, a dose-dependent inhibitory effect of PAD-inhibitor Cl-am on MV release was revealed in both tumorigenic and non-tumorigenic cell lines. Our data indicate that ATP stimulation appears to be associated with elevated MV release and translocation of PAD, especially PAD4. However, at this stage we cannot say whether this translocation per se is directly involved in MV release or conversely that MV release causes translocation. Specific PAD isozymes may yet be found to play a crucial role in the shedding of MVs and this is on-going work. A similar effect of PAD-inhibitor Cl-am was also observed with the normal prostate (PNT2) epithelial cells, indicating that PAD activity could have a significant role in the release of MVs in both cancerous and non-cancerous cells. As Cl-am acts as a pan-PAD inhibitor, it cannot be stipulated at this stage which particular PAD isozyme(s) (PAD2 or PAD4 ) is the main one in this process (or indeed whether they are equally important).
The mechanism of MV release may be regulated by influx of Ca2+ released by the ER or Ca2+ entering the cell through pores made by sublytic complement (Citation62) or through various calcium channels (Citation71, Citation72) on activated cells that bring about calpain-mediated cleavage of various cytoskeletal actin filaments leading to reorganisation of the cytoskeleton and facilitating MV release (Citation73). Actins have been shown to play an important role during microvesiculation, and both β-actin and F-actin stress fibres are involved in the redistribution of the actin-cytoskeleton during MV formation through the activation of Rho/Rho-associated kinase (ROCK) pathways during apoptosis and thrombin stimulation (Citation74). As both β- and γ-actins have been identified as deiminated target proteins in sera and synovial fluid from patients with rheumatoid arthritis (Citation75), we sought to establish whether there is a link between deiminated β-actin and microvesiculation. Our data show that deiminated β-actin is increased upon stimulating PC3 and PNT2 cells with BzATP, and markedly decreased when pretreating the cells with PAD inhibitor prior to stimulation with BzATP. Although some deiminated proteins were identified in PC3 cells pretreated with the PAD-inhibitor, none had a confidence score of 95% (Table , C).
Our work implies the involvement of PAD enzymes in MV release through deimination of target proteins involved in cytoskeletal rearrangement, which is an essential part of MV biogenesis. Further support comes from identification of actin alpha 1 as a deiminated protein target. Its confidence level was reduced to 50% in PC3 cells when pretreated with Cl-am prior to stimulation, suggesting that deimination of actin alpha-1, in addition to β-actin, may play an important role in the shedding of MVs. PAD upregulation and the resulting increase in deimination of cytoskeletal actin filaments would likely have an effect on the cytoskeletal reorganisation for the MV release process () but will need further testing. Indeed, other proteins found to be deiminated such as glyceraldehyde-3-phosphate dehydrogenase also play likely roles in cytoskeleton organization and assembly and warrant further investigation. While the effects of PAD2/4 activity on the deimination of actin and their inhibition by Cl-am have been largely measured here on MV release, we certainly cannot exclude that some of the vesicles measured were exosomes. Since the process of multivesicular body recruitment to the plasma membrane to release exosomal cargo likely involves actin and microtubular elements of the cytoskeleton (Citation76), it may well be that PADs and deimination of actin are also involved in exosomal release.
Fig. 7. The proposed role of PADs in microvesiculation and the potential therapeutic application of PAD inhibitors in anticancer therapy. PAD2 and PAD4 are cytosolic enzymes, which during the course of cells being stimulated, through raised intracellular calcium levels, to microvesiculate (e.g. using BzATP stimulation of P2X channels or NHS as a source of sublytic complement), translocate to the nucleus (PAD2 and PAD4) or remain partially in the cytosol (PAD2). These PAD isozymes play a potential novel role in the biogenesis of MV release. This may happen by influencing actin-cytoskeleton cleavage and actin rearrangement and/or by deimination of genes in the nucleus through still unknown pathways involved in microvesicle biogenesis . The pharmacological inhibition of PADs with the pan-PAD-inhibitor chloramidine (Cl-am) abrogates the release of MVs and when combined with the anticancer drug methotrexate (MTX) works synergistically to induce increased cytotoxic effects and apoptosis of PC3 tumour cells.
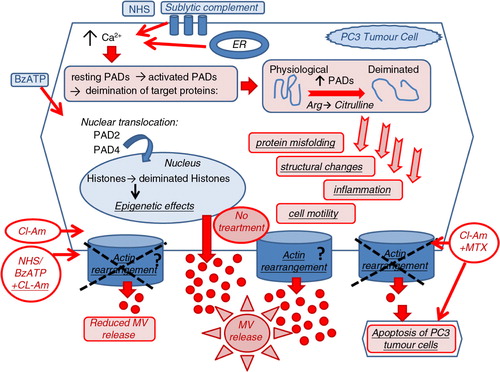
Several recent studies have reported the cytotoxic effect of PAD inhibitors on cancer cells without affecting normal cells (Citation20, Citation77) (Citation78). For instance, the potent PAD4 inhibitors fluoramidine (F-am) and chloramidine (Cl-am) were shown to exhibit cytotoxic effects at micromolar concentrations on several cancer cell lines including HL60, MCF-7 and HT-29, whereas no effect was observed on non-cancerous cells. Differentiation of HL60 and HT29 cells was also observed (Citation70). Furthermore, a synergistic effect has been shown whereby cell cytotoxicity was increased when combining PAD inhibitors with the cancer drug doxorubicin (Citation70). We now report a similar finding, whereby PC3 prostate cancer cells treated with PAD-inhibitor Cl-am and MTX exerted a synergistic cytotoxic effect compared to MTX alone. We can now postulate a novel mechanism for the synergistic effect of Cl-am/F-am with doxorubicin, reported previously (Citation70) and of Cl-am with MTX as described here. Essentially, Cl-am, which inhibits MV release through the inhibition of actin deimination may thus sensitize the cancer cells to other chemotherapeutic drug(s) (). Our present findings are in accordance with our previous observations and other studies (Citation12) reporting that neoplastic cells are rendered more sensitive to cancer drugs when their microvesiculation processes are inhibited, as this may prevent these cells from removing the drugs through MV release [Citation12 and on-going studies at CMIRC].
Various studies have shown that MVs play a role in cellular cross talk and act as carriers of active molecules that have an effect at distal sites from the origin of MV release. The presence of PAD4 has been reported in the plasma of cancer patients, although how it reaches this location remains unknown (Citation46). It is thus tempting to postulate that tumour cells may increase the production of MVs through a PAD4-mediated mechanism, which would also allow the PAD enzyme to be packaged in the MVs and carried into the plasma where it could deiminate various proteins such as antithrombin thus aiding the spread of tumourigenesis indirectly.
In summary, our findings suggest that PAD isozymes 2 and 4 play a significant role in cellular microvesiculation. This can happen either directly through the deimination of cellular actins, which affects actin-cytoskeletal rearrangement, or indirectly through the deimination of histone proteins in the nucleus. This is supported by the observed changes in nuclear PAD translocation in concert with PAD inhibition and changes in MV release, a mechanism that still needs further detailed investigation. Refined and isozyme-selective PAD inhibitors pose as promising therapeutic agents for current therapy regimes and may form part of novel future combination therapies in various forms of cancers.
Conflict of interest and funding
This work benefitted from HEFCE QR funding (RAE2008) and in part through funding from the Royal Society, the Sir Richard Stapely Educational Trust and NIH grant GM 079357. SK was a recipient of a VC scholarship from LMU.
JEV Suppl Figs1.pdf
Download PDF (502.1 KB)Acknowledgements
The authors thank the members of CMIRC for critically reading the manuscript and Maria McCrossan for assisting with the electron microscopy.
Notes
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
References
- Piccin A, Murphy WG, Smith OP. Circulating microparticles: pathophysiology and clinical implications. Blood Rev. 2007; 21: 157–71. [PubMed Abstract].
- Diamant M, Tushuizen ME, Sturk A, Nieuwland R. Cellular microparticles: new players in the field of vascular disease?. Eur J Clin Invest. 2004; 34: 392–401. [PubMed Abstract].
- Inal JM, Kosgodage U, Azam S, Stratton D, Antwi-Baffour S, Lange S. Blood/plasma secretome and microvesicles. Biochim Biophys Acta. 2013; 1834: 2317–25. [PubMed Abstract].
- Lynch SF, Ludlam CA. Plasma microparticles and vascular disorders. Br J Haematol. 2007; 137: 36–48. [PubMed Abstract].
- Friend C, Marovitz W, Henie G, Henie W, Tsuei D, Hirschhorn K et al. Observations on cell lines derived from a patient with Hodgkin's disease. Cancer Res. 1978; 38: 2581–91. [PubMed Abstract].
- Ginestra A, La P, Saladino F, Cassara D, Nagase H, Vittorelli ML. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res. 1998; 18: 3433–7. [PubMed Abstract].
- Kim HK, Song KS, Park YS, Kang YH, Lee YJ, Lee KR et al. Elevated levels of circulating platelet microparticles, VEGF, IL-6 and RANTES in patients with gastric cancer: possible role of a metastasis predictor. Eur J Cancer. 2003; 39: 184–91. [PubMed Abstract].
- Zwicker JI, Liebman HA, Neuberg D, Lacroix R, Bauer KA, Furie BC et al. Tumor-derived tissue factor-bearing microparticles are associated with venous thromboembolic events in malignancy. Clin Cancer Res. 2009; 15: 6830–40. [PubMed Abstract] [PubMed CentralFull Text].
- Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010; 123: 1603–11. [PubMed Abstract] [PubMed CentralFull Text].
- Roos MA, Gennero L, Denysenko T, Reguzzi S, Cavallo G, Pescarmona GP et al. Microparticles in physiological and in pathological conditions. Cell Biochem Funct. 2010; 28: 539–48. [PubMed Abstract].
- Bebawy M, Combes V, Lee E, Jaiswal R, Gong J, Bonhoure A. Membrane microparticles mediate transfer of P-glycoprotein to drug sensitive cancer cells. Leukemia. 2009; 23: 1643–9. [PubMed Abstract].
- Jorfi S, Inal JM. The role of microvesicles in cancer progression and drug resistance. Biochem Soc Trans. 2013; 41: 293–8. [PubMed Abstract].
- Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC et al. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010; 49: 4852–63. [PubMed Abstract] [PubMed CentralFull Text].
- Rogers GE. Occurrence of citrulline in proteins. Nature. 1962; 194: 1149–51. [PubMed Abstract].
- Shirai H, Blundell TL, Mizuguchi K. A novel superfamily of enzymes that catalyze the modification of guanidino groups. Trends Biochem Sci. 2001; 26: 465–8. [PubMed Abstract].
- Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem. 1996; 271: 30709–16. [PubMed Abstract].
- Chavanas S, Mechin MC, Takahara H, Kawada A, Nachat R, Serre G et al. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene. 2004; 330: 19–27. [PubMed Abstract].
- Vossenaar ER, Nijenhuis S, Helsen MM, van der Heijden A, Senshu T, van den Berg WB et al. Citrullination of synovial proteins in murine models of rheumatoid arthritis. Arthritis Rheum. 2003; 48: 2489–500. [PubMed Abstract].
- Ellsworth RE, Vertrees A, Love B, Hooke JA, Ellsworth DL, Shriver CD. Chromosomal alterations associated with the transition from in situ to invasive breast cancer. Ann Surg Oncol. 2008; 15: 2519–25. [PubMed Abstract].
- Lange S, Rocha-Ferreira E, Thei L, Mawjee P, Bennett K, Thompson PR et al. Peptidylarginine deiminases: novel drug targets for prevention of neuronal damage following hypoxic ischemic insult (HI) in neonates. J Neurochem. 2014; 130: 555–62. [PubMed Abstract] [PubMed CentralFull Text].
- Lange S, Gögel S, Leung KY, Vernay B, Nicholas AP, Causey CP et al. Protein deiminases: new players in the developmentally regulated loss of neural regenerative ability. Dev Biol. 2011; 355: 205–14. [PubMed Abstract].
- Mohanan S, Cherrington BD, Horibata S, McElwee JL, Thompson PR, Coonrod SA. Potential role of peptidylarginine deiminase enzymes and protein citrullination in cancer pathogenesis. Biochem Res Int. 2012; 2012: 895343. [PubMed Abstract] [PubMed CentralFull Text].
- Anzilotti C, Pratesi F, Tommasi C, Migliorini P. Peptidylarginine deiminase 4 and citrullination in health and disease. Autoimmun Rev. 2010; 9: 158–60. [PubMed Abstract].
- Anzilotti C, Merlini G, Pratesi F, Tommasi C, Chimenti D, Migliorini P. Antibodies to viral citrullinated peptide in rheumatoid arthritis. J Rheumatol. 2006; 33: 647–51. [PubMed Abstract].
- Migliorini P, Pratesi F, Tommasi C, Anzilotti C. The immune response to citrullinated antigens in autoimmune diseases. Autoimmun Rev. 2005; 4: 561–4. [PubMed Abstract].
- Nicholas AP, Lu L, Heaven M, Kadish I, van Groen T, Accavitti-Loper MA, Nicholas AP, Bhattacharya SK. Ongoing studies of deimination in neurodegenerative diseases using the F95 antibody. Protein deimination in human health and disease. 2014; New York, NY: Springer Science+Business Media. 257–80.
- Wang L, Chang X, Yuan G, Zhao Y, Wang P. Expression of peptidylarginine deiminase type 4 in ovarian tumors. Int J Biol Sci. 2010; 6: 454–64. [PubMed Abstract] [PubMed CentralFull Text].
- Cherrington BD, Zhang X, McElwee JL, Morency E, Anguish LJ, Coonrod SA. Potential role for PAD2 in gene regulation in breast cancer cells. PLoS One. 2012; 7: e41242. [PubMed Abstract] [PubMed CentralFull Text].
- Senshu T, Akiyama K, Ishigami A, Nomura K. Studies on specificity of peptidylarginine deiminase reactions using an immunochemical probe that recognizes an enzymatically deiminated partial sequence of mouse keratin K1. J Dermatol Sci. 1999; 21: 113–26. [PubMed Abstract].
- Watanabe K, Akiyama K, Hikichi K, Ohtsuka R, Okuyama A, Senshu T. Combined biochemical and immunochemical comparison of peptidylarginine deiminases present in various tissues. Biochim Biophys Acta. 1988; 966: 375–83. [PubMed Abstract].
- Urano Y, Watanabe K, Sakaki A, Arase S, Watanabe Y, Shigemi F et al. Immunohistochemical demonstration of peptidylarginine deiminase in human sweat glands. Am J Dermatopathol. 1990; 12: 249–55. [PubMed Abstract].
- Pritzker LB, Joshi S, Harauz G, Moscarello MA. Deimination of myelin basic protein. 2. Effect of methylation of MBP on its deimination by peptidylarginine deiminase. Biochemistry. 2000; 39: 5382–8. [PubMed Abstract].
- Inagaki M, Nishi Y, Nishizawa K, Matsuyama M, Sato C. Site-specific phosphorylation induces disassembly of vimentin filaments in vitro. Nature. 1987; 328: 649–52. [PubMed Abstract].
- Vossenaar ER, Radstake TR, van der Heijden A, van Mansum MA, Dieteren C, de Rooij DJ et al. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann Rheum Dis. 2004; 63: 373–81. [PubMed Abstract] [PubMed CentralFull Text].
- McElwee JL, Mohanan S, Griffith OL, Breuer HC, Anguish LJ, Cherrington BD. Identification of PADI2 as a potential breast cancer biomarker and therapeutic target. BMC Cancer. 2012; 12: 500. [PubMed Abstract] [PubMed CentralFull Text].
- Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002; 277: 49562–8. [PubMed Abstract].
- Asaga H, Nakashima K, Senshu T, Ishigami A, Yamada M. Immunocytochemical localization of peptidylarginine deiminase in human eosinophils and neutrophils. J Leukoc Biol. 2001; 70: 46–51. [PubMed Abstract].
- Nakashima K, Hagiwara T, Ishigami A, Nagata S, Asaga H, Kuramoto M et al. Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 1999; 274: 27786–92. [PubMed Abstract].
- U KP Subramanian V, Nicholas AP, Thompson PR, Ferretti P. Modulation of calcium-induced cell death in human neural stem cells by the novel peptidylarginine deiminase-AIF pathway. Biochim Biophys Acta. 2014; 1843: 1162–71. [PubMed Abstract] [PubMed CentralFull Text].
- Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM et al. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006; 26: 11387–96. [PubMed Abstract].
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M et al. Histone deimination antagonizes arginine methylation. Cell. 2004; 118: 545–53. [PubMed Abstract].
- Tanikawa C, Ueda K, Nakagawa H, Yoshida N, Nakamura Y, Matsuda K. Regulation of protein Citrullination through p53/PADI4 network in DNA damage response. Cancer Res. 2009; 69: 8761–9. [PubMed Abstract].
- Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. 2008; 28: 4745–58. [PubMed Abstract] [PubMed CentralFull Text].
- Zhang X, Gamble MJ, Stadler S, Cherrington BD, Causey CP, Thompson PR et al. Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate c-Fos expression in breast cancer cells. PLoS Genet. 2011; 7: e1002112. [PubMed Abstract] [PubMed CentralFull Text].
- Li P, Wang D, Yao H, Doret P, Hao G, Shen Q et al. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene. 2010; 29: 3153–62. [PubMed Abstract] [PubMed CentralFull Text].
- Chang X, Han J. Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol Carcinog. 2006; 45: 183–96. [PubMed Abstract].
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004; 306: 279–83. [PubMed Abstract].
- Guo Q, Fast W. Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J Biol Chem. 2011; 286: 17069–78. [PubMed Abstract] [PubMed CentralFull Text].
- Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF et al. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008; 283: 20060–8. [PubMed Abstract] [PubMed CentralFull Text].
- Nomura S, Ozaki Y, Ikeda Y. Function and role of microparticles in various clinical settings. Thromb. Res. 2008; 123: 8–23. [PubMed Abstract].
- Castellana D, Toti F, Freyssinet JM. Membrane microvesicles: macromessengers in cancer disease and progression. Thromb Res. 2010; 125: S84–8. [PubMed Abstract].
- Ansa-Addo EA, Lange S, Stratton D, Antwi-Baffour S, Cestari I, Ramirez MI et al. Human plasma membrane-derived vesicles halt proliferation and induce differentiation of THP-1 acute monocytic leukemia cells. J Immunol. 2010; 185: 5236–46. [PubMed Abstract].
- Gardiner C, Ferreira YJ, Dragovic RA, Redman CW, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013; 2: 19671. doi: http://dx.doi.org/10.3402/jev.v2i0.19671.
- Inal JM, Schifferli JA. Complement C2 receptor inhibitor trispanning and the beta-chain of C4 share a binding site for complement C2. J Immunol. 2001; 168: 5213–21. [PubMed Abstract].
- Nicholas AP, Nicholas JN. Preparation of a monoclonal antibody to citrullinated epitopes: its characterization and some applications to immunohistochemistry in human brain. Glia. 2002; 37: 328–36.
- Manwaring V, Heywood WE, Clayton R, Lachmann RH, Keutzer J, Hindmarsh P. The identification of new biomarkers for identifying and monitoring kidney disease and their translation into a rapid mass spectrometry-based test: evidence of presymptomatic kidney disease in pediatric Fabry and type-I diabetic patients. J Proteome Res. 2013; 12: 2013–21. [PubMed Abstract].
- Ercolani L, Florence B, Denaro M, Alexander M. Isolation and complete sequence of a functional human glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1988; 263: 15335–41. [PubMed Abstract].
- Tisdale EJ. Glyceraldehyde-3-phosphate dehydrogenase is required for vesicular transport in the early secretory pathway. J Biol Chem. 2001; 276: 2480–6. [PubMed Abstract].
- Wieland T, Hippe HJ, Ludwig K, Zhou XB, Korth M, Klumpp S. Reversible histidine phosphorylation in mammalian cells: a teeter-totter formed by nucleoside diphosphate kinase and protein histidine phosphatase 1. Methods Enzymol. 2010; 471: 379–402. [PubMed Abstract].
- Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993; 261: 478–80. [PubMed Abstract].
- UniProt Consortium. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014; 42: 191–8.
- Inal JM, Ansa-Addo EA, Stratton D, Kholia S, Antwi-Baffour SS, Jorfi S et al. Microvesicles in health and disease. Arch Immunol Ther Exp. 2012; 60: 107–21. [PubMed Abstract].
- Toth B, Lok CA, Boing A, Diamant M, van der Post JA, Friese K et al. Microparticles and exosomes: impact on normal and complicated pregnancy. Am J Reprod Immunol. 2007; 58: 389–402.
- Berckmans RJ, Nieuwland R, Kraan MC, Schaap MC, Pots D, Smeets TJ et al. Synovial microparticles from arthritic patients modulate chemokine and cytokine release by synoviocytes. Arthritis Res Ther. 2005; 7: R536–44. [PubMed Abstract] [PubMed CentralFull Text].
- Kim HK, Song KS, Chung JH, Lee KR, Lee SN. Platelet microparticles induce angiogenesis in vitro. Br J Haematol. 2004; 124: 376–84. [PubMed Abstract].
- Boulanger CM, Scoazec A, Ebrahimian T, Henry P, Mathieu E, Tedgui A et al. Circulating microparticles from patients with myocardial infarction cause endothelial dysfunction. Circulation. 2001; 104: 2649–52. [PubMed Abstract].
- Chironi G, Simon A, Hugel B, Del PM, Gariepy J, Freyssinet JM et al. Circulating leukocyte-derived microparticles predict subclinical atherosclerosis burden in asymptomatic subjects. Arterioscler Thromb Vasc Biol. 2006; 26: 2775–80. [PubMed Abstract].
- Mallat Z, Hugel B, Ohan J, Leseche G, Freyssinet JM, Tedgui A. Shed membrane microparticles with procoagulant potential in human atherosclerotic plaques: a role for apoptosis in plaque thrombogenicity. Circulation. 1999; 99: 348–53. [PubMed Abstract].
- Antwi-Baffour S, Kholia S, Aryee YK, Ansa-Addo EA, Stratton D, Lange S et al. Human plasma membrane-derived vesicles inhibit the phagocytosis of apoptotic cells – possible role in SLE. Biochem Biophys Res Commun. 2010; 398: 278–83. [PubMed Abstract].
- Slack JL, Causey CP, Thompson PR. Protein arginine deiminase 4: a target for an epigenetic cancer therapy. Cell Mol Life Sci. 2011; 68: 709–20. [PubMed Abstract].
- Salzer U, Hinterdorfer P, Hunger U, Borken C, Prohaska R. Ca(++)-dependent vesicle release from erythrocytes involves stomatin-specific lipid rafts, synexin (annexin VII), and sorcin. Blood. 2002; 99: 2569–77. [PubMed Abstract].
- Pizzirani C, Ferrari D, Chiozzi P, Adinolfi E, Sandona D, Savaglio E et al. Stimulation of P2 receptors causes release of IL-1beta-loaded microvesicles from human dendritic cells. Blood. 2007; 109: 3856–64. [PubMed Abstract].
- Lemoinne S, Thabut D, Housset C, Moreau R, Valla D, Boulanger CM4 et al. The emerging roles of microvesicles in liver diseases. Nat Rev Gastroenterol Hepatol. 2014; 11: 350–61. [PubMed Abstract].
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001; 3: 339–45. [PubMed Abstract].
- van Beers JJ, Schwarte CM, Stammen-Vogelzangs J, Oosterink E, Bozic B, Pruijn GJ. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and beta-actin. Arthritis Rheum. 2013; 65: 69–80. [PubMed Abstract].
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles and friends. J Cell Biol. 2013; 200: 373–83. [PubMed Abstract] [PubMed CentralFull Text].
- Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. A fluoroacetamidine-based inactivator of protein arginine deiminase 4: design, synthesis, and in vitro and in vivo evaluation. J Am Chem Soc. 2006; 128: 1092–3. [PubMed Abstract] [PubMed CentralFull Text].
- Knuckley B, Luo Y, Thompson PR. Profiling Protein Arginine Deiminase 4 (PAD4): a novel screen to identify PAD4 inhibitors. Bioorg Med Chem. 2008; 16: 739–45. [PubMed Abstract] [PubMed CentralFull Text].
