Abstract
Extracellular vesicles (EVs) have emerged as important mediators of intercellular communication in a diverse range of biological processes. For future therapeutic applications and for EV biology research in general, understanding the in vivo fate of EVs is of utmost importance. Here we studied biodistribution of EVs in mice after systemic delivery. EVs were isolated from 3 different mouse cell sources, including dendritic cells (DCs) derived from bone marrow, and labelled with a near-infrared lipophilic dye. Xenotransplantation of EVs was further carried out for cross-species comparison. The reliability of the labelling technique was confirmed by sucrose gradient fractionation, organ perfusion and further supported by immunohistochemical staining using CD63-EGFP probed vesicles. While vesicles accumulated mainly in liver, spleen, gastrointestinal tract and lungs, differences related to EV cell origin were detected. EVs accumulated in the tumour tissue of tumour-bearing mice and, after introduction of the rabies virus glycoprotein-targeting moiety, they were found more readily in acetylcholine-receptor-rich organs. In addition, the route of administration and the dose of injected EVs influenced the biodistribution pattern. This is the first extensive biodistribution investigation of EVs comparing the impact of several different variables, the results of which have implications for the design and feasibility of therapeutic studies using EVs.
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
Interest in the field of extracellular vesicles (EVs) has increased tremendously in the past decade. After being identified initially as secreted debris of platelets, EVs from many cell sources have now been recognized as important messengers in intercellular communication via horizontal transfer of proteins, RNA and bioactive lipids (Citation1–Citation5). EVs are found in all body fluids, including plasma, saliva, cerebrospinal fluid and urine (Citation6–Citation10).
EVs are generally divided into 3 subgroups depending on their biogenesis; (a) exosomes, with a diameter of 40–150 nm, which are released into the extracellular milieu when multi-vesicular bodies fuse with the cell membrane, (b) microvesicles, with a diameter of 150–1,000 nm, originating from direct budding of the plasma membrane and finally (c) apoptotic bodies, which display a broad size distribution (50–2,000 nm) (Citation1). Of these vesicles, exosomes have received the most attention and have been implicated in physiological functions, such as maintenance of stem cell niches and immune responses (Citation1, Citation2), as well as in pathological conditions, such as malignancies, metastasis, viral spread and neurological disorders (Citation3, Citation11) (Citation12).
Since various proteins and RNA associate with EVs, reflecting some of the contents and functions of their source cells, it has been investigated whether EVs could be employed as diagnostic or therapeutic agents. In addition to harnessing their innate therapeutic features, engineered EVs have been exploited as well (Citation13–Citation17). For example, dendritic cell (DC) exosomes are currently under assessment in clinical trials (e.g. NCT01159288) for cancer treatment and there have been several reports regarding enhanced tissue regeneration after treatment regimens with exosomes generated from mesenchymal stromal cells (MSCs) (Citation13, Citation14) (Citation15, Citation18) . Furthermore, Kordelas et al. recently reported successful treatment using MSC-derived EVs of a patient suffering from therapy-refractory graft-versus-host disease (Citation18). Hence, increasing evidence suggests that EVs can be applied to treat a range of different diseases, both in animal models and in patients.
Despite intense research in the field, only few studies (Citation19–Citation26) have analysed EV biodistribution and thus little is known about their in vivo trafficking. Herein, we aimed to assess the biodistribution profile of EVs smaller than 200 nm from a broad range of different cell types, including primary mouse DCs and human MSCs, using a previously described labelling method with a near-infrared dye. Furthermore, dose titration, time-lapse studies and different injections routes were examined.
Material and methods
Cells
Dendritic cells (DCs): Bone marrow was flushed from the bone cavity of tibias and femurs of 8- to 10-week-old male C57BL/6 mice using a G27 syringe into DMEM GlutaMAX (Life Technologies, Carlsbad, CA, USA). Clumps were dissociated by re-suspension and samples were centrifuged for 10 minutes at 280g. The supernatant was removed and the pellet resuspended in red blood cell lysing buffer (Sigma-Aldrich, St Louis, MO, USA). Following a 5-minute incubation, the suspension was neutralized with DMEM and spun at 280g for 10 minutes. Cells were then plated at a concentration of 1×106 cells/ml in DMEM, 10% foetal bovine serum (FBS) and antibiotics (Penicillin and Streptomycin 5 mg/ml) and supplemented with 10 ng/ml murine granulocyte macrophage colony stimulating factor (GM-CSF) (Sigma-Aldrich). DCs were cultured for 4 days. On day 4, DCs were transfected with plasmids containing a Lamp2b construct fused to Rabies virus glycoprotein (RVG) [as described in (Citation17)] or mock transfected using TransIT LT1 transfection reagent (Mirus Bio, Madison, WI, USA) according to manufacturer's instructions. DCs were then cultured for a further 3 days before the medium was replaced with medium containing FBS, which had been spun at 120,000g for 70 minutes prior to medium preparation. Twenty four hours later the culture medium was collected, spun at 1,000g for 15 minutes and passed through a 0.22 µm filter. Thereafter, EVs were purified and labelled with DiR as described below.
MSCs were isolated from bone marrow aspirates of 2 healthy human volunteers as described previously (Citation27, Citation28). MSC donors gave informed consent and the study was approved by the Regional Ethics Review Board. Mononuclear cells separated by density centrifugation were cultured in DMEM (Life Technologies) supplemented with 10% heat-inactivated FBS. Purity was confirmed by flow cytometry (CD73+, CD105+, CD90+, HLA-I+, CD45−, CD34−, CD14−, HLA-DR−). Before EV isolation, MSCs (at passage 4) were cultured in OptiMEM (Life Technologies) for 48 hours. Thereafter, the EV labelling and isolation were performed as described below.
Cell lines: HEK293T, C2C12, B16-F10 and OLN-93 cells were grown in DMEM supplemented with 10% (HEK293T, B16-F10 and OLN-93) or 20% (C2C12) FBS.
Twenty four hours after cells were plated, the growth media was changed to OptiMEM or DMEM supplemented with 10% FBS depleted of serum EVs. Forty eight hours later, cells had reached 90% confluence. The supernatant was then collected for purification and labelling of EVs as described below.
EV purification with differential ultracentrifugation
Conditioned media (CM) was harvested and spun first at 300g for 5 minutes to remove cells, followed by 1,500g for 10 minutes to remove cell debris and thereafter filtrated through a 0.22-µm filter to remove any larger particles. The CM was then ultracentrifuged at 110,000g for 70 minutes to pellet the EVs. A second washing step was performed by resuspending the EV pellet in 25 ml of phosphate buffer saline (PBS) and ultracentrifuged at 110,000g for another 70 minutes. The pellet was then resuspended in PBS. All ultracentrifugation (UC) steps were performed using the Beckman Coulter (Brea, CA, USA) Type 70 Ti rotor at 4°C.
EGFP-positive EVs
To generate EGFP-positive EVs, HEK293T cells were transfected with CD63-EGFP plasmid using polyethylenimine (PEI) at a 1:4 (DNA/PEI) ratio. Four hours post-transfection, the cells were washed with PBS and the media changed to OptiMEM. The EVs were isolated 48 hours after media change, as described above.
DiR-labelled EVs
The filtered CM was incubated with 1 µM fluorescent lipophilic tracer DiR (1,1-dioctadecyl-3,3,3,3-tetramethylindotricarbocyanine iodide) (D12731, Invitrogen, Life Technologies) at room temperature (RT) for 15 minutes prior to EV isolation by the UC method as described above.
Nanoparticle analysis
Nanoparticle tracking analysis (NTA) was performed with NS500 nanoparticle analyser (NanoSight, Malvern, Worchestershire, UK) to measure the size distribution of DiR-labelled particles. NTA determines the size of particles based on Brownian motion and is commonly used for analysis of nanometre-sized particles (Citation29). For all our recordings, we used a camera level of 15–16 and automatic functions for all post-acquisition settings except for the detection threshold, which was fixed at 6. Samples were diluted in PBS between 1:100 and 1:20,000 to achieve a particle count of between 2×108 and 2×109 per ml. The camera focus was adjusted to make the particles appear as sharp dots. Using the script control function, five 30- or 60-second videos for each sample were recorded, incorporating a sample advance and a 5-second delay between each recording. For GFP-positive EVs, the NTA's 488 nm laser was used to excite the GFP and a band filter of 500 nm was applied. The EVs were always recorded under flow to avoid bleaching the GFP, and 10×30 seconds recordings were performed to ensure reliable recordings. For the analysis, the same settings as that used for the scattering mode were used, except for the minimum tracking distance, which was set to 5.
Sucrose gradient
DiR-labelled EVs or free DiR were loaded at the bottom of a discontinuous sucrose gradient (0.00–2.5M) and centrifuged at 200,000g for 16 hours at 4°C in a SW40 swing bucket rotor (Beckman Coulter). One millilitre fractions were then collected from the bottom and each fraction was weighed to estimate the density of that particular fraction. Each fraction was transferred to a 12-well plate that was read for fluorescence in the IVIS (in vivo imaging system) Spectrum, (PerklinElmer, Waltham, MA, USA, see below for further details) using excitation/emission wavelength of 710 nm/760 nm. Fractions 2–10 were subjected to subsequent western blotting analysis.
Western blot
Samples were treated with radioimmunoprecipitation assay (RIPA) buffer and vortexed every 5 minutes for 30 minutes to lyse the EVs; subsequently, the sample was spun at 12,000g for 12 minutes to remove any lipids and the supernatant was collected. Thirty microlitres of lysed sample was mixed with a sample buffer, containing 0.5 M dithiothreitol (DTT), 0.4 M sodium carbonate (Na2CO3), 8% Sodium dodecyl sulphate (SDS) and 10% glycerol, and heated at 65°C for 5 minutes. Samples were then loaded on a NuPAGE® Novex® 4–12% Bis-Tris Gel (Invitrogen, Life Technologies) and ran at 120 V in running buffer. The proteins on the gel were transferred to an iBlot nitrocellulose membrane (Invitrogen, Life Technologies) for 7 minutes with the iBlot system. The membranes were blocked with Odyssey blocking buffer diluted 1:1 in PBS for 60 minutes at RT with gentle shaking.
After the blocking step, the membranes were incubated with freshly prepared primary antibody solution (anti-PDC6I (Alix, ab117600) and anti-Tsg101 (ab30871), at 1:1,000 dilution from Abcam, Cambridge, UK) overnight at 4°C. Membranes were washed 4 times, 5 minutes each using washing buffer (TBS-T 0.1%) with gentle shaking before adding the secondary antibody solution (anti-mouse IgG DyLight-800 at 1:10,000 dilution for detecting Alix; anti-rabbit IgG DyLight-800 at 1:10,000 dilution for detecting Tsg101) and incubated for 1 hour at RT. After the secondary antibody incubation, membranes were washed 4 times, 5 minutes each and visualized by scanning both 700- and 800-nm channels on the LI-COR Odyssey CLx infrared imaging system.
Transmission electron microscopy
Ten microlitres of EV re-suspension in PBS was added onto formvar-carbon-coated grids (Agar Scientific, Stansted, UK) for 20 minutes. The grid was blotted with filter paper and stained with 2% uranyl acetate (UA) for 1 minute. Next, UA was removed and the grids were washed with 15 µl of distilled water for 1 minute. The grid was blotted dry and left to air-dry for 15 minutes. EVs were viewed with a JEOL 1010 transmission electron microscope (JEOL, Peabody, MA, USA).
Animal experiments
Female NMRI or C57BL/6 mice were used. Freshly purified DiR-labelled EVs were injected through the tail vein for intravenous (i.v.) injections for most experiments. Intraperitoneal (i.p.) and subcutaneous (s.c.) injections were carried out as well, where indicated. Biodistribution of DiR-labelled EVs was examined using 3 different doses: 1.5×1010 particles/gram body weight (p/g), 1.0×1010 p/g and 0.25×1010 p/g, the particle count was measured with NTA and the samples were diluted accordingly. For analysis of DiR-EVs distribution, IVIS Spectrum (Perkin Elmer) was used. IVIS spectrum is an instrument that contains a high-sensitive CCD camera, which enables both fluorescence and luminescence measurements. Here, either live (isoflurane sedated) mice were imaged or the animals were sacrificed and the organs harvested prior to analysis. For the perfusion experiment, the mice were sedated and the vascular system was flushed by transcardial perfusion for 5 minutes. The left ventricle was infused with PBS (5 ml/min) and the right atrium was perforated. The outflow liquid, liver and tail were monitored during the procedure to assure successful perfusion. After 5 minutes of perfusion, the organs were harvested and analysed. The live mice or the harvested organs were imaged for 1–2 seconds (excitation 710 nm, emission 760 nm). The data were analysed with the IVIS software (Living Image Software for IVIS®). For fluorescent micro computed tomography (µCT)-images, mice were secured in a mouse imaging shuttle, imaged in the IVIS for 3D fluorescence (FLIT) and transferred to the Quantum FX micro CT Imaging System (PerklinElmer, MA, USA) for CT-imaging. Co-registration of FLIT with the CT-scan was generated using Living Image software (Citation30, Citation31). Adobe Photoshop CS4 and Adobe Illustrator were used to crop out and align the organ images.
Injection of CD63-EGFP EVs: CD63-EGFP EVs were generated, isolated and characterized by NanoSight as described above. Twenty four hours post-injections (2.9×1010 p/g), the organs were harvested and prepared as described below. The animal experiments were approved by the British and the Swedish local boards for laboratory animals.
Immunohistochemistry
Tissue preparation
Following euthanasia using cervical dislocation, the organs were explanted, embedded in optimal cutting temperature compounds (Histolab, Gothenburg, Sweden) and snap frozen immediately. The frozen blocks were then sectioned to 8 µm thickness and mounted on Superfrost glass slides (Gerhard Menzel, Braunschweig, Germany).
Staining and imaging
The sections were fixed for 10 minutes in 4% formaldehyde (Histolab, Sweden), washed 3 times for 5 minutes each in PBS and then blocked using PBS with 5% goat serum (Life Technologies, Sweden) and 0.3% Triton X100 (Sigma-Aldrich, Sweden) for 1 hour. The primary antibody (GFP, Ab13970, Abcam) was diluted 1:1,000 in an antibody dilution buffer consisting of PBS with 0.1% Triton X100 being added to each sample and incubated overnight at 4°C. The sections were then washed as described previously, and the secondary antibody (Alexa 594 anti-rabbit IgG, Life technologies) diluted at 1:500 in antibody dilution buffer was added and incubated for 1 hour. The sections were again washed, mounted with coverslips using Slowfade Gold Anitfade with 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies) and imaged using an inverted fluorescent microscope (Olympus X70, Olympus, Tokyo, Japan).
Statistical analysis
Statistical analyses of the data were performed using Prism 6.0 (GraphPad Software Inc.) by using the nonparametric Kruskal–Wallis test followed by the Sidak post-test for all p-values. All results are expressed as mean +SEM. All graphs were made in Prism 6.0 (GraphPad Software Inc., La Jolla, CA, USA).
Results and discussion
Assessing DiR as an EV labelling tool for biodistribution studies
In order to assess the distribution of EVs systemically delivered in mice, the near-infrared (NIR) dye DiR used in earlier studies (Citation24, Citation25) was used for general EV labelling. DiR is a lipophilic dye that only fluoresces intensely when inserted into a lipid-membrane. NIR dyes are ideal for in vivo applications owing to their high tissue penetrance and low autofluorescence (Citation32–Citation36). HEK293T EVs labelled with DiR were analysed by western blot (WB) following differential UC, and the common EV markers (Citation37) Alix and Tsg101 were both present (A and Supplementary Fig. 1A). To further confirm that the purified material indeed consisted of EVs, NTA and EM analyses were performed. The NTA data showed a typical EV-like shaped curve with a mode size of around 100 nm in diameter (B) and the EM analysis revealed vesicles with a diameter of approximately 100 nm (C). Furthermore, the EM shows predominantly intact vesicles with classical EV morphology; hence, the DiR labelling seems to have no effect on the vesicle morphology.
Fig. 1. DiR labelling does not affect EV morphology and co-localizes with EVs. (A) Western Blot (WB) stained for Alix and Tsg101 on HEK293T purified, DiR-labelled EVs and cell lysate (CL) (whole WB-membrane shown in Supplementary Fig. 1A). (B) NTA profile of DiR-labelled HEK293T EVs showing an EV-like distribution with a mode size around 100 nm. The curve has been normalized to dilution factors. (C) Electron microscopy picture of DiR-labelled HEK-293T EVs. Scale bar represents 100 nm. (D) Image depicting DiR-EVs and free DiR sucrose gradients in UC tubes and subsequent 12-well plates containing the different fractions. (E) Fluorescent values (normalized to background) of the DiR-EVs and free DiR gradients (assayed from the 12-well plates depicted in 1D). (F) WB stained for Alix for DiR-EVs from fractions 2–10 (whole WB-membrane shown in Supplementary Fig. 1B).
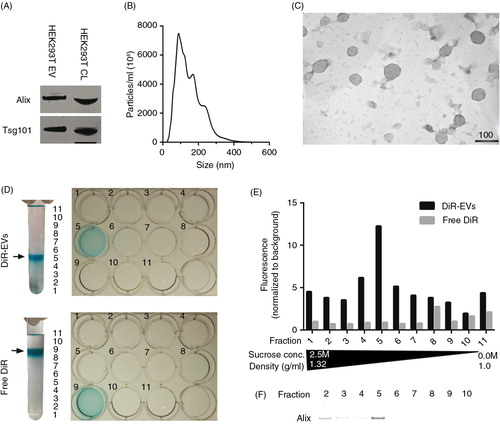
Similar to any dye-based system, there is a risk that the signal originates from free dye rather than the labelled construct itself, and since free dye, when injected, might bind serum proteins, circulating cells and blood vessels, several control experiments were performed to exclude that. Sucrose gradient fractionation of DiR-labelled EVs reveals that the dye, when bound to EVs, is visible (D) and fluoresces (E) at the expected density of exosomes (1.11–1.21 g/ml) (Citation5), which also correlates with the presence of Alix (F and Supplementary Fig. 1B). On the contrary, and as expected, free DiR without EVs had a much lower buoyant density (D), and since the free DiR sample is absent for any form of lipid membranes, the fluorescent values are, as expected, much lower compared to the DiR-EV sample (E). This indicates that the DiR-EV samples after UC purification contains DiR bound to EVs and minuscule amounts of free DiR, since DiR is only found in the same fraction as the EVs and in no other fraction.
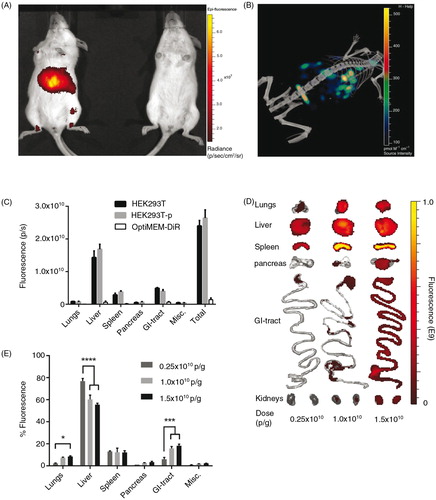
DiR-labelled HEK293T EVs were subsequently injected via the tail vein of NMRI mice. To assess the biodistribution, live mice were imaged 24 hours after injection using an in vivo imaging system (IVIS) (A) and subsequent µCT (B). From these representative images, the levels of detail from fluorescent whole mouse imaging did not allow for enough accuracy to conclude from which tissue the signal originates. Thus, to elucidate from which organ the fluorescent signal (i.e. the EVs) originated and to minimize signal interference, the organs were harvested and imaged ex vivo for the succeeding experiments. As mentioned above, a possible downside with the in vivo tracking of EVs is the risk of signal originating from the blood in the organs and from free dye per se. To affirm that we were in fact analysing DiR-labelled EVs in the organs, mice were treated with HEK293T-EVs with and without perfusion prior to organ harvest. In addition, to rule out the possibility of monitoring free dye, EV-free medium (OptiMEM) was incubated with DiR and subjected to UC in the same fashion as when labelling EVs. As shown in C, perfusion did not affect the distribution and injection of OptiMEM-DiR resulted in negligible signal in all organs (less than 6% of the total tissue fluorescence compared to DiR-HEK293T-EVs). Importantly, these results suggest that we are indeed tracking labelled EVs and not merely free dye in the tissues, corroborating the sucrose gradient fractionation results seen in D–F. Using this methodology, it is possible to determine tissue accumulation but it is not possible to discriminate what is truly intracellular. However, it provides an indication as to whether a certain EV formulation is likely to be therapeutically active.
Fig. 2. Systemically delivered EVs display a dose-dependent increase in tissue uptake in mice. (A) Representative IVIS image 24 hours post-injection of a live mouse injected i.v. with 1.0×1010 p/g DiR-EVs (left) and PBS-treated control (right). (B) Representative co-registration of FLIT with CT-scan 24 hours post-injection of a mouse injected i.v. with 1.0×1010 p/g DiR-EVs. (C) Fluorescent signal in each organ 24 hours post-iv injection of HEK293T-EVs (1.0×1010 p/g) with (HEK293T-p) and without (HEK293T) perfusion and free OptiMEM-DiR. Misc.=brain, heart, kidneys and quadriceps muscle. N=4. The results represent mean+SEM. (D) Representative IVIS images of organs (24 hours post-injection) from mice injected i.v. with 1.5×1010 p/g, 1.0×1010 p/g and 0.25×1010 p/g of HEK293T EVs. (E) The percentage of fluorescent signal in each organ for the same experiment as in D (Absolute fluorescent signals are plotted in Supplementary Fig. 3). N=4, in E *represents p=0.05, **p=0.001 and ****p=0.0001. The results represent mean+SEM.
To further confirm that we indeed tracked EVs and not only the dye, CD63-EGFP-labelled EVs were injected in mice. HEK293T cells were transfected with a CD63-EGFP plasmid (transfection efficiency of approximately 90%, data not shown) and the subsequent EVs were harvested. According to NTA analysis, approximately 20% of the EVs were EGFP-positive (Supplementary Fig. 2A). Twenty four hours post-i.v. injection of EGFP-positive EVs, the organs were harvested, snap frozen, sectioned and stained with an anti-EGFP antibody. EGFP-positive EVs could be detected in the parenchyma of the liver and spleen (Supplementary Fig. 2B). However, no or only negligible levels of EGFP could be detected in lungs and kidneys (data not shown). This is probably because the levels of the EVs in other organs are lower and thus below the detection threshold of the technique. Overall, these results corroborate with our results obtained by the DiR-labelled EVs, with the highest accumulation of EVs detected in the liver and spleen.
Dose titration of DiR-labelled EVs
To investigate whether the dose of EVs injected may affect the biodistribution, a dose-titration study was carried out. All injection doses were determined by using a fixed number of EVs (measured by NTA) per gram body weight of the mice, hence particles/gram body weight (p/g). A positive dose-dependent response of the total tissue fluorescence was observed following i.v. injection of 3 different doses: 0.25×1010, 1.0×1010 and 1.5×1010 p/g of DiR-labelled HEK293T EVs. EVs accumulated mainly in liver, spleen, gastrointestinal tract (GI)-tract and lungs. However, even though an increase in dose resulted in an increased fluorescent signal in the analysed organs (D and E and Supplementary Fig. 3), the relative distribution among the organs shifted. Relative liver accumulation decreased from 77%±2.4 to 60%±3.9 (p<0.0001) to 56%±1.3 (p<0.0001) when treated with 0.25×1010 p/g, 1.0×1010 p/g and 1.5×1010 p/g, respectively. This might be due to a low signal (close to the detection limit) observed in all organs except liver and spleen analysed for the lowest dose. The relative dose-dependent decrease in liver signal may also result from a saturation of the mononuclear phagocyte system (also known as the reticuloendothelial system, RES) resulting in more effective bypass of the liver at higher doses. The intermediate dose (1×1010 p/g) was determined to be sufficient for detection in all organs analysed with minimal saturation and thus this dose was used in subsequent experiments.
In line with earlier findings on the biodistribution of synthetic liposomes, which are similar to EVs in size and also contain a lipophilic outer layer (Citation38–Citation41), we found that EVs generally distribute to organs of the mononuclear phagocyte system with highest accumulation in the liver, followed by spleen, GI-tract and lungs. Many compounds, when injected i.v., will be taken up by patrolling macrophages in the RES organs, for example, by Küpffer cells in liver or splenic macrophages (Citation42–Citation44). However, several nanoparticle studies have indicated that the size of the particles affects the distribution and clearance. For instance, small nanoparticles (≤100 nm) are less prone to opsonisation and penetrate the fenestrations in the hepatic sinusoidal endothelium, thus increasing the hepatocyte uptake of the total hepatic accumulation. Similarly, in spleen, small particles avoid the filtration process at the interendothelial slits and extravasation occurs due to the discontinuous endothelium, allowing entry into the red pulp (Citation42–Citation44).
EV distribution kinetics
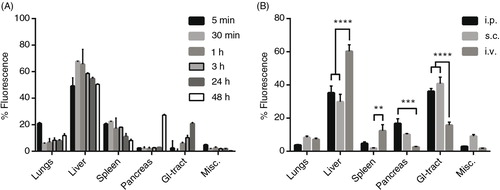
A potential pitfall when using lipophilic dyes to track EVs is that the dye itself has a much longer half-life compared to the EVs. Hence, there is a possibility that, at longer time points, the distribution of remaining dye is being analysed and not the EVs per se. Therefore, EV distribution at different time points was examined following i.v. injection of DiR-labelled HEK293T EVs. The overall EV biodistribution profile remained largely unchanged during the first 24 hours, whereas greater changes were detected at the 48-hour time point (A and Supplementary Fig. 4). The distribution pattern amongst the analysed organs is in line with previous reports (Citation19, Citation20) (Citation21, Citation22) (Citation24, Citation25) . However, in these articles, the authors assess the biodistribution at earlier time points as well as at 24 hours. Nonetheless, our results indicate a similar distribution pattern at 24 hours compared to the earlier time points. At 48 hours, the fluorescent signal started to decline in heart, GI-tract and kidneys (heart and kidneys included in the Misc. group in A and Supplementary Fig. 4) and the signal increased dramatically in pancreas. These findings may be explained by possible re-distribution of the EVs, a late phase uptake of remaining EVs or artefacts due to the long half-life of the dye. Due to the change in distribution pattern at 48 hours, the potential risk of unspecific staining and possible tissue accumulation of the dye, the organs were harvested at 24 hours post-i.v. injection in the succeeding experiments.
Fig. 3. Distribution kinetics and effect of different injection routes of DiR-labelled EVs on tissue distribution. (A) Percentage fluorescent signal of each organ ex vivo at different time points after injection of 1.0×1010 p/g DiR-labelled HEK293T EVs (fluorescent signal in absolute numbers in Supplementary Fig. 4). (B) Percentage fluorescent signal in each organ following i.p., s.c. and i.v. injection of 1.0×1010 p/g DiR-labelled HEK293T EVs (Absolute fluorescent signals are plotted in Supplementary Fig. 5b). Misc.=brain, heart, kidneys and quadriceps muscle. N=3–4, **represents p<0.01, ***p<0.001 and ****p<0.0001. The results represent mean+SEM.
Different delivery routes alter EV distribution pattern
To assess whether the route of injection influences the distribution pattern, mice were given the same amount of HEK293T-DiR-EVs (1.0×1010 p/g) using 3 different systemic delivery routes, i.v., i.p. and s.c. (B). The different injection routes rendered different distribution patterns. In contrast to i.v. injections, i.p. and s.c. injections resulted in significantly (p<0.0001) lower EV accumulation in liver (i.v.: 60%±3.9; i.p.: 35%±4.2; s.c.: 30%±4.5) and spleen (i.v.: 12%±3.6; i.p.: 5%±1.0; s.c.: 2%±0.3; p<0.01 for i.v. vs. s.c.) whereas increased accumulation was observed in pancreas (i.v.: 2.6%±0.3; i.p.: 17%±2.9; s.c.: 10%±0.5; p<0.001 for i.v. vs. i.p) and GI (i.v.: 16%±1.8; i.p.: 36%±1.7; s.c.: 41%±3.9; p<0.0001). Furthermore, i.p. injections displayed slightly higher total tissue fluorescence compared to i.v. injections, whereas s.c. injections resulted in much lower signals (i.v.: 2.0×1010 p/s; i.p.: 3.0×1010 p/s; s.c.: 0.5×1010 p/s; Supplementary Fig. 5A and B). These results indicate that the route of injection influences tissue distribution of infused EVs and the site of injection alternations may thus be used to increase the EV distribution to a potential tissue target.
Cell source influences the biodistribution of EVs
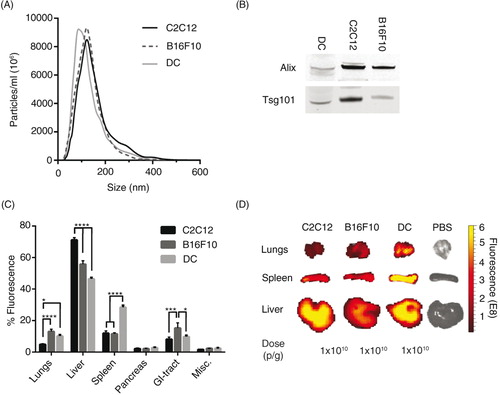
Cells of different origin are known to home in on varying locations in vivo, for example immunological cells preferentially target sites with immunological activity, such as the spleen, to a greater extent than control cells (Citation45). EVs from different cell sources could potentially also show different innate homing capabilities in vivo. To compare intrinsic tropisms, a comparison of the biodistribution of EVs derived from 3 different mouse cell sources – a muscle cell line; C2C12, a melanoma cell line; B16F10 and primary immature bone marrow-derived DCs – was undertaken. Furthermore, xenotransplantation of EVs from rat cells (oligodendrocytes; OLN-93) and from cells of human origin (HEK293T and primary human MSCs) were included for a cross-species comparison. The NTA profile and mode size of the EVs was similar when comparing the EVs from the different cell sources, with a mode size of approximately 100 nm (A and Supplementary Fig. 6A), except for the MSC-EVs that displayed a very homogenous size distribution and a mode size of around 60 nm (Supplementary Fig. 6A). To further characterize the EVs, WB was performed. EVs from the different cell sources stained positive for the common EV markers Alix and Tsg101 (B and Supplementary Fig. 7), indicating that the purified material indeed contained EVs. The in vivo distribution of DiR-labelled EVs from the different mouse cell sources was similar with liver, spleen, GI-tract and lungs being the highest sites of accumulation (C and D). Yet, importantly, distinct significant differences were also detected. C2C12-derived EVs displayed greater liver accumulation (C2C12-EVs, 71%±1.5), compared to B16F10-EVs (56%±2.2) and DC-derived EVs, which had the lowest liver distribution (DC-EVs, 46%±0.9). Inversely, the lung accumulation was found to be lower for C2C12-EVs (5%±0.5) compared to B16F10-EVs (13%±1.5) and DC-EVs (10%±0.9). B16F10-EVs were more readily found in the GI-tract (15%±3.3) compared to C2C12-EVs (8%±1.3) and DC-EVs (10%±0.8). Interestingly, DC-EVs showed increased accumulation in spleen (28%±1.3) compared to EVs derived from the other sources, with C2C12-EVs and B16F10-EVs displaying similar splenic distribution (12%±1.5 and 12%±0.6, respectively) (C and D). The xenotransplanted EVs displayed the same common distribution trend as the allotransplanted (mouse derived) EVs (Supplementary Fig. 6B and C). MSC-EVs had the highest liver accumulation (71%±4.0) compared to the other 2 sources (OLN93-EVs 65%±2.5 and HEK293T-EVs 60%±3.9). Inversely, the GI-tract accumulation was significantly lower for the MSC-EVs (3%±1.5) compared to OLN93-EVs (11%±0.4) and HEK293T-EVs (16%±1.8). Graphs displaying the biodistribution of EVs in absolute values from all 6 cell sources are shown in Supplementary Fig. 8A–F.
Fig. 4. The biodistribution of EVs differs between cell sources. (A) Particle distribution of DiR-labelled EVs from mouse C2C12-, B16F10- and DC-cells measured by NTA. (B) Western Blot stained against Alix and Tsg101 of EVs from C2C12-, B16F10- and DC-cells (whole WB-membranes in Supplementary Fig. 7). (C) The percentage of fluorescent signal in each organ after injection of 1.0×1010 p/g DiR-labelled EVs from the 3 different mouse cell types (Absolute fluorescent signals are plotted in Supplementary Fig. 8). Misc.=brain, heart, kidneys and quadriceps muscle. (D) Representative spleen, liver and lung IVIS images from mice injected with EVs from the 3 different mouse cell sources and PBS-treated control. N=4, in C *represents p=0.05, ***p=0.001 and ****p=0.0001. The results represent mean+SEM.
As aforementioned, the most prominent shift was displayed by the DC-EVs, which showed increased distribution to the spleen. It is thus intriguing to speculate that EVs may adopt the homing pattern of the parental cell of origin, which suggests that EVs acquire the same repertoire of surface receptors and extracellular matrix-binding proteins as their parent cell. This natural tropism emphasizes the importance of choosing the appropriate cell source for derivation of EVs when initiating an in vivo therapeutic project aiming at delivery to a given organ. Others have shown that the tetraspanin-repertoire of EVs can influence the in vivo biodistribution and uptake in different cells (Citation46, Citation47). Surprisingly, the species origin (human, rat or mouse) did not seem to affect the biodistribution. HEK293T-, OLN-93- and C2C12-EVs had similar distribution patterns despite the different species origin of the cells.
Untargeted EVs accumulate in tumour tissue
Using engineered EVs to treat cancer is an attractive idea and several laboratories worldwide are currently pursuing EVs as potential cancer treatments. To efficiently target cancer directly with EVs, and not only by immune-activation using, for example, DC-derived exosomes, the EVs need to localize to the tumour tissue in vivo. To investigate whether systemically injected EVs can home into tumour tissue, we designed the following proof-of-principle experiment. Briefly, B16-F10 cells, a mouse melanoma cell line, were injected subcutaneously in C57BL/6 mice. Fourteen days post-implantation, 1×1010 p/g of HEK293T EVs labelled with DiR were injected i.v. Organs were harvested 24 hours post-injection and scanned using the IVIS. Generally, the fluorescence signals detected in the liver and spleen were the highest amongst all the organs; this was consistent in both tumour- and non-tumour-bearing mice (A and B and Supplementary Fig. 9). Interestingly, 3% of the total tissue fluorescence was detected in the tumour (A and B).
Fig. 5. EVs distribute to tumour tissue and can be targeted to the brain. (A) The percentage of fluorescent signal in each organ of tumour-bearing mice after injection of 1.0×1010 p/g HEK293T DiR-labelled EVs (Absolute fluorescent signals are plotted in Supplementary Fig. 9). (B) Fluorescent signal from tumour, kidneys and brain shown from the same mice described in A. (C) NTA of RVG-positive (+RVG) and RVG-negative (-RVG) DC-EVs. (D) The percentage of fluorescent signal in each organ after injection of 1.0×1010 p/g of +RVG and –RVG DC-EVs (Absolute fluorescent signals are plotted in Supplementary Fig. 10). (E) Representative image of mouse brains, harvested 24 hour post-injection of 1.0×1010 p/g of +RVG and –RVG DC-EVs. (F) The percentage of fluorescence signal in the brain, heart and muscle from the same mice as shown in D (Absolute fluorescent signals are plotted in Supplementary Fig. 11). Misc.=brain, heart, kidneys and quadriceps muscle. N=4, **represents p<0.01 and ***represents p<0.001, the results represent mean+SEM.
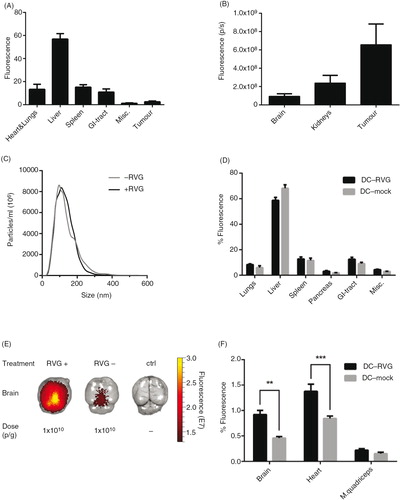
In line with the hepatic and splenic uptake observed here and by others, tumour vasculature is known to be leaky, the permeation and retention effect, allowing entry for nanometre-sized particles. Similar to other nanoparticles, such as liposomes, the size of EVs potentially makes them ideal for anti-tumour treatments (Citation48). The tumour accumulation can probably be optimised with different cell sources and target moieties; however, these data showing tumour accumulation suggests that the EVs are a viable option as a delivery modality to tumours, supporting earlier findings by others that it is feasible to harness the delivery capacity of EVs for tumour treatment (Citation25).
RVG-targeted EVs accumulate in acetylcholine receptor (AChR)-rich tissues
Another intriguing possibility is to use EVs as a delivery vector for therapeutics. To optimise their efficacy, targeted therapies are being developed. So far, there have been a few successful targeting strategies; however, the biodistribution of the targeted EVs has not been fully investigated. In 2011, Alvarez-Erviti et al. showed knockdown of an Alzheimer's gene in the brain by siRNA-loaded RVG-EVs (Citation17). Furthermore, the RVG ligand has been used in several studies demonstrating that RVG-coupled species more readily localize to brain, due to the presence of nicotinic AChR (nAChR), the receptor for the full length RVG, which the RVG-peptide ligand is derived from (Citation17),Citation49–(Citation51). Therefore, the influence of RVG ligand on biodistribution was assessed by comparing RVG- and non-RVG-targeted DC-EVs. Before EV harvesting, half of the DCs were transfected with the fusion protein LAMP2b-RVG, which targets the RVG peptide to the EV membrane, and the other half was mock transfected. The NTA profiles of RVG- and non-RVG DC-EVs were very similar with a mode size of around 100 nm (C). The targeted and untargeted EVs were subsequently labelled with DiR and injected i.v. (1.0×1010 p/g) in C57BL/6 (syngeneic) mice. The organs were harvested 24 hours post-injection and imaged using the IVIS. The distribution of both RVG and non-RVG-EVs followed the distribution of un-modified EVs with the strongest signal detected in liver, spleen, GI-tract and lungs (D and Supplementary Fig. 10). Interestingly, the RVG-EVs displayed a significantly increased signal in brain (p<0.01), heart (p<0.001) and (higher but not significant) muscle, compared to the non-RVG EV group (E and F and Supplementary Fig. 11). In line with previous reports of RVG-labelled liposomes and siRNA (Citation17),Citation49–(Citation51), a 2-fold greater accumulation in brain was observed with RVG-EVs as compared to non-targeted EVs. Besides the brain, the muscle and heart also express high levels of the nAChR and muscarinergic AChR, respectively (Citation52–Citation55); hence, further supporting our findings of the higher signal intensities when applying RVG-EVs in these 2 organs. The small but significant change in the distribution of RVG-EVs shows the sensitivity of the labelling technique and furthermore corroborates the fact that we are indeed tracking EVs as opposed to free dye.
Conclusion
EVs and particularly exosomes are being increasingly studied, as they appear to play an important role in intercellular communication, complementing the endocrine and paracrine systems. This has led to a number of reported attempts to use EVs as therapeutic agents or disease biomarkers in vivo. However, their biodistribution has been less studied despite knowledge of this being essential for understanding EV biology and for potential therapeutic applications. To our knowledge, this is the first time a comparative study is performed evaluating how the biodistribution of EVs in vivo is affected by different EV doses, routes of injection and cellular origin of EVs. We also assessed how targeted EVs as well as tumour burden influenced biodistribution. These results highlight the important considerations for the design of in vivo therapeutic studies using EVs.
Authors' contributions
JN, OW, IM, AO, YG, HS, GC, PV, YL, NH and SEA designed, performed the experiments and wrote the manuscript. PM and PJ supervised the immunohistochemistry experiments, KLB provided primary MSCs, and LAE, MJW and CIE provided inputs on the manuscript draft. All authors contributed with correction of the manuscript.
Conflict of interest and funding
The authors declare no competing financial interests. This project was funded by the Swedish Research Council (VR-Med and EuroNanoMedII), Swedish Cancer Society and SEA holds a research fellowship form the Swedish Society of Medical Research (SSMF). JN and OW are both recipients of Karolinska Institutet MD/PhD grants. AO is supported by the Wellcome Trust and YL is supported by the Agency for Science, Technology and Research (A*STAR), Singapore. IM is supported by a Postdoctoral MOBILITAS Fellowship of the Estonian Science Foundation and by the EU IMI (Innovative Medicines Initiative) project COMPACT (Collaboration on the optimisation of macromolecular pharmaceutical access to cellular targets). HS is supported by the national scholarship program Kristjan Jaak, which is funded and managed by Archimedes Foundation in collaboration with the Estonian Ministry of Education and Research. NH and KLB are supported by Swedish Research Council (VR K2014-64X-20742-07-5) and Vinnova (2010-00501).
Supplementary Figures
Download PDF (15.7 MB)Notes
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
References
- EL Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013; 12: 347–57. [PubMed Abstract].
- Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006; 20: 847–56.
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012; 18: 883–91.
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008; 10: 619–24.
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007; 9: 654–9.
- Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011; 9: 86.
- Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012; 10: 5.
- Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, Bossios A et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011; 9: 9.
- Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004; 101: 13368–73.
- Yamada T, Inoshima Y, Matsuda T, Ishiguro N. Comparison of methods for isolating exosomes from bovine milk. J Vet Med Sci. 2012; 74: 1523–5.
- Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases?. Front Physiol. 2012; 3: 124.
- Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH et al. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010; 30: 6838–51.
- Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med. 2011; 6: 481–92.
- Timmers L, Lim SK, Hoefer IE, Arslan F, Lai RC, Van Oorschot AA et al. Human mesenchymal stem cell-conditioned medium improves cardiac function following myocardial infarction. Stem Cell Res. 2011; 6: 206–14.
- Gatti S, Bruno S, Deregibus MC, Sordi A, Cantaluppi V, Tetta C et al. Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia-reperfusion-induced acute and chronic kidney injury. Nephrol Dial Transplant. 2011; 26: 1474–83.
- Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998; 4: 594–600.
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011; 29: 341–5.
- Kordelas L, Rebmann V, Ludwig AK, Radtke S, Ruesing J, Doeppner TR et al. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014; 28: 970–3. [PubMed Abstract].
- Grange C, Tapparo M, Bruno S, Chatterjee D, Quesenberry PJ, Tetta C et al. Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a model of acute kidney injury monitored by optical imaging. Int J Mol Med. 2014; 33: 1055–63. [PubMed Abstract] [PubMed CentralFull Text].
- Jang SC, Kim SR, Yoon YJ, Park KS, Kim JH, Lee J et al. In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small. 2014; 11: 456–61.
- Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire CA, Chen JW et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014; 8: 483–94.
- Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol. 2013; 165: 77–84.
- Ignowski JM, Schaffer DV. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol Bioeng. 2004; 86: 827–34.
- Smyth T, Kullberg M, Malik N, Smith-Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release. 2014; 199C: 145–55.
- Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013; 21: 185–91.
- Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014; 35: 2383–90.
- Le Blanc K, Frassoni F, Ball L, Locatelli F, Roelofs H, Lewis I et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 2008; 371: 1579–86.
- Le Blanc K, Samuelsson H, Gustafsson B, Remberger M, Sundberg B, Arvidson J et al. Transplantation of mesenchymal stem cells to enhance engraftment of hematopoietic stem cells. Leukemia. 2007; 21: 1733–8.
- Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P et al. Sizing and phenotyping of cellular vesicles using Nanoparticle tracking analysis. Nanomedicine. 2011; 7: 780–8.
- Niska JA, Meganck JA, Pribaz JR, Shahbazian JH, Lim E, Zhang N et al. Monitoring bacterial burden, inflammation and bone damage longitudinally using optical and muCT imaging in an orthopaedic implant infection in mice. PLoS One. 2012; 7: e47397.
- Patel AR, Lim E, Francis KP, Singh M. Opening up the optical imaging window using nano-luciferin. Pharm Res. 2014; 31: 3073–84.
- Herschman HR. Molecular imaging: looking at problems, seeing solutions. Science. 2003; 302: 605–8.
- Hilderbrand SA, Weissleder R. Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol. 2010; 14: 71–9.
- Luo S, Zhang E, Su Y, Cheng T, Shi C. A review of NIR dyes in cancer targeting and imaging. Biomaterials. 2011; 32: 7127–38.
- Ntziachristos V, Bremer C, Weissleder R. Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur Radiol. 2003; 13: 195–208. [PubMed Abstract].
- Kalchenko V, Shivtiel S, Malina V, Lapid K, Haramati S, Lapidot T et al. Use of lipophilic near-infrared dye in whole-body optical imaging of hematopoietic cell homing. J Biomed Optics. 2006; 11: 050507.
- Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009; 9: 4997–5000.
- Lopez-Berestein G, Kasi L, Rosenblum MG, Haynie T, Jahns M, Glenn H et al. Clinical pharmacology of 99mTc-labeled liposomes in patients with cancer. Cancer Res. 1984; 44: 375–8. [PubMed Abstract].
- Rank A, Nieuwland R, Crispin A, Grutzner S, Iberer M, Toth B et al. Clearance of platelet microparticles in vivo. Platelets. 2011; 22: 111–16.
- Ostro MJ, Cullis PR. Use of liposomes as injectable-drug delivery systems. Am J Hosp Pharm. 1989; 46: 1576–87. [PubMed Abstract].
- Liu D, Mori A, Huang L. Role of liposome size and RES blockade in controlling biodistribution and tumor uptake of GM1-containing liposomes. Biochim Biophys Acta. 1992; 1104: 95–101.
- Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol Rev. 2001; 53: 283–318. [PubMed Abstract].
- Sarin H. Physiologic upper limits of pore size of different blood capillary types and another perspective on the dual pore theory of microvascular permeability. J Angiogenesis Res. 2010; 2: 14.
- Garnett MC, Kallinteri P. Nanomedicines and nanotoxicology: some physiological principles. Occup Med. 2006; 56: 307–11.
- Kupiec-Weglinski JW, Austyn JM, Morris PJ. Migration patterns of dendritic cells in the mouse. Traffic from the blood, and T cell-dependent and -independent entry to lymphoid tissues. J Exp Med. 1988; 167: 632–45.
- Rana S, Yue S, Stadel D, Zoller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol. 2012; 44: 1574–84.
- Rana S, Zoller M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem Soc Trans. 2011; 39: 559–62.
- Bae YH, Park K. Targeted drug delivery to tumors: myths, reality and possibility. J Control Release. 2011; 153: 198–205.
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007; 448: 39–43.
- Mazarakis ND, Azzouz M, Rohll JB, Ellard FM, Wilkes FJ, Olsen AL et al. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum Mol Genet. 2001; 10: 2109–21.
- Pulford B, Reim N, Bell A, Veatch J, Forster G, Bender H et al. Liposome-siRNA-peptide complexes cross the blood–brain barrier and significantly decrease PrP on neuronal cells and PrP in infected cell cultures. PLoS One. 2010; 5: e11085.
- Boksa P, Quirion R. [3H]N-methyl-carbamylcholine, a new radioligand specific for nicotinic acetylcholine receptors in brain. Eur J Pharmacol. 1987; 139: 323–33.
- Lentz TL, Burrage TG, Smith AL, Crick J, Tignor GH. Is the acetylcholine receptor a rabies virus receptor?. Science. 1982; 215: 182–4.
- Lentz TL, Burrage TG, Smith AL, Tignor GH. The acetylcholine receptor as a cellular receptor for rabies virus. Yale J Biol Med. 1983; 56: 315–22. [PubMed Abstract] [PubMed CentralFull Text].
- Peralta EG, Ashkenazi A, Winslow JW, Smith DH, Ramachandran J, Capon DJ. Distinct primary structures, ligand-binding properties and tissue-specific expression of four human muscarinic acetylcholine receptors. EMBO J. 1987; 6: 3923–9. [PubMed Abstract] [PubMed CentralFull Text].
