Abstract
The Extracellular RNA (exRNA) Communication Consortium, funded as an initiative of the NIH Common Fund, represents a consortium of investigators assembled to address the critical issues in the exRNA research arena. The overarching goal is to generate a multi-component community resource for sharing fundamental scientific discoveries, protocols, and innovative tools and technologies. The key initiatives include (a) generating a reference catalogue of exRNAs present in body fluids of normal healthy individuals that would facilitate disease diagnosis and therapies, (b) defining the fundamental principles of exRNA biogenesis, distribution, uptake, and function, as well as development of molecular tools, technologies, and imaging modalities to enable these studies, (c) identifying exRNA biomarkers of disease, (d) demonstrating clinical utility of exRNAs as therapeutic agents and developing scalable technologies required for these studies, and (e) developing a community resource, the exRNA Atlas, to provide the scientific community access to exRNA data, standardized exRNA protocols, and other useful tools and technologies generated by funded investigators.
This paper is part of the Special Issue: Extracellular RNA Communication Consortium. More papers from this issue can be found at http://www.journalofextracellularvesicles.net
The concept that RNA molecules are secreted in extracellular spaces and act as endocrine signals that alter the phenotypes of target cells, both locally and at distant sites, represents a novel paradigm in intercellular signalling. Since pioneering studies by Ratajczak and colleagues (Baj-Krzyworzeka et al., Cancer Immunol. Immunother. 2006) and Lötvall and colleagues (Valadi et al., Nat Cell Biol. 2007) showed extracellular release of RNA in vesicles, extracellular RNAs (exRNAs) have been identified in all body fluids examined including blood, saliva, urine, breast milk, cerebral spinal fluid, amniotic fluid, ascites, and pleural effusions highlighting the transformative potential that secreted RNAs may have in regulating health and disease. A systematic analysis of circulating exRNAs in body fluids of healthy individuals along with a better understanding of exRNA sorting to distinct secretory pathways and carriers, regulation of exRNA secretion, mechanisms of targeting, and effector function in target cells will generate opportunities to identify novel strategies for prognosis, diagnosis, and intervention of many diseases.
The creation of an international consortium to coordinate large-scale studies to understand the role of exRNAs in regulating health and disease was encouraged by the recently established American Society of Exosomes and Microvesicles (ASEMV, www.asemv.org) and International Society of Extracellular Vesicles (ISEV, www.isev.org). Both ISEV and ASEMV promote the common goal of advancing the field of extracellular vesicle (EV) research through education, communication, and collaboration. Since EVs were initially considered the only carrier of exRNAs outside the boundaries of the cell of origin, representatives from the National Institutes of Health (NIH) Common Fund sought input from these communities. The NIH Common Fund identifies and supports emerging scientific disciplines by developing cross-cutting programmes to address problems that may seem intractable or to seize new and potentially high-impact opportunities. All Common Fund programmes are goal-driven and as such are expected to be catalytic – producing tools, technology, and/or data for the entire research community.
The goals of the Extracellular RNA Communication Consortium
The Extracellular RNA Communication Consortium (ERCC) was initiated in 2012 with the publication of 5 Requests for Applications (RFAs, www.commonfund.nih.gov/ExRNA/grants). These 5 RFAs will develop a community resource supported by a consortium of investigators to address the critical issues in this nascent field. The overarching goals are to generate fundamental scientific discoveries and innovative tools and technologies to advance the field. The 5 key initiatives are (a) defining the fundamental principles of exRNA biogenesis, distribution, uptake, and function as well as the development of the molecular tools, technologies, and imaging modalities to enable these studies, (b) generating a reference catalogue of exRNAs present in the body fluids of normal healthy individuals that would facilitate disease diagnosis and therapeutic interventions, (c) identifying exRNA biomarkers of disease, (d) demonstrating the clinical utility of exRNAs as therapeutic agents and developing the scalable technologies required for these studies, and lastly (e) developing the Data Management and Resource Repository (DMRR), a community resource to provide access to exRNA data, standardized exRNA protocols, and other useful tools and technologies generated by funded projects. Investigators from all 5 initiatives will form the ERCC with goals to discover fundamental biological principles about the mechanisms of exRNA biogenesis, to identify and develop a catalogue of exRNA in normal human body fluids; and to investigate the potential for using exRNAs as therapeutic molecules or biomarkers of disease.
The Extracellular RNA Communication Consortium
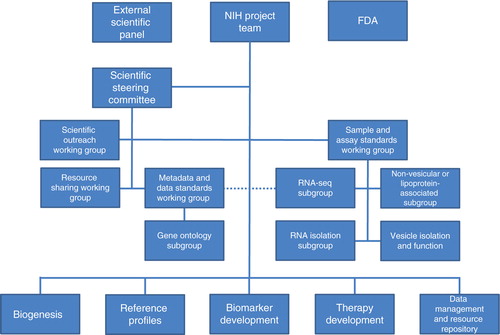
As depicted in , the ERCC structure enables not only governance and oversight but also cross-Consortium working groups to generate best practices, standard operating procedures, and cross-validation. The 5 research initiatives of the ERCC () are a) Biogenesis, b) Reference Profiles, c) Biomarker Development, d) Therapy Development and e) the Data Management and Resource Repository. A brief introduction to these collaborative scientific components is described below.
More detailed scientific discussions are found in accompanying articles in the Journal of Extracellular Vesicles. In “Biogenesis, Delivery, and Function of Extracellular RNA,” Patton et al. describe current knowledge and the projects seeking to understand the biogenesis, delivery, and functional impact of exRNA. In “Extracellular RNAs: Development as Biomarkers of Human Disease,” Quinn and Saugstad et al. discuss the 10 exRNA biomarker discovery projects. The therapeutic potential of exRNAs is detailed by Quesenberry et al. in “Consideration of Extracellular Vesicle Functional Potential.” Finally, as the central data coordinating centre, the activities of the DMRR are described by Milosavljevic et al. in “Integration of extracellular RNA profiling data using metadata, biomedical ontologies, and Linked Data technologies.”
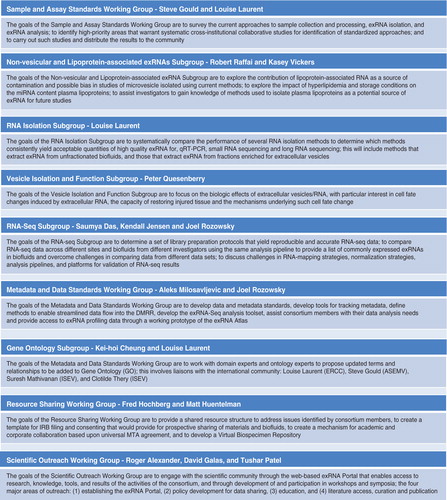
The ERCC Steering Committee is the main governing board for the Consortium. Each of the 30 awards from the 5 research initiatives has one representative on the Steering Committee. The Steering Committee is led by two Consortium investigators. The Steering Committee considers the collective goals for the Consortium, discusses the progress made towards these goals, and coordinates the improvement of exRNA research activities. Currently, there are four Consortium Working Groups: a) Sample and Assay Standards, b) Metadata and Data Standards, c) Resource Sharing and d) Scientific Outreach. These Consortium Working Groups are designed to address both the collective goals of the Consortium and the needs of individual projects through cross-collaborations. Current Working Groups are phased out when goals are met and new ones formed as needs arise. Accordingly, additional subgroups have formed to address issues with Gene Ontology, Non-vesicular or Lipoprotein-associated exRNA, RNA-Seq, RNA Isolation, and Vesicle Isolation and Function. More details of the current Working Groups and Subgroups are described in .
The External Scientific Panel (ESP) is responsible for reviewing and evaluating the progress of the ERCC towards meeting the individual and collective goals of the programme. ESP members are appointed by the NIH and provide recommendations to the NIH about continued support of the components of the Consortium. The Panel is composed of senior scientists with relevant expertise, but who are not awardees within the ERCC. Members of the ESP provide their individual advice concerning the overall direction and progress of the ERCC towards its goals, and make recommendations to the NIH about changes or improvements. In this manner, the NIH can incorporate community feedback and ensure that the programme is meeting the goals of the community it serves.
The United States Food and Drug Administration (FDA) provides guidance on the development of exRNA-based biomarkers and therapeutics. Specific expertise and guidance has been and will continue to be given by scientists at the Center for Drug Evaluation and Research and Center for Biologics Evaluation and Research. The NIH and FDA have a memorandum of understanding such that the FDA will provide guidance as projects mature towards pre-clinical development.
Research initiatives of the Extracellular RNA Communication Consortium
Initiative 1: exRNA biogenesis, biodistribution, uptake and effector function
A major goal of the ERCC is to determine the regulatory principles that guide the selection of specific RNA and/or classes of RNA molecules for extracellular transport and the role of exRNA (including exRNAs from food and the environment) in human health and disease. It is expected that enabling tools, technologies, bioreagents, and innovative analytical approaches will be developed in the process of achieving these research goals. This aspect of the ERCC will address four overarching scientific areas of interest.
exRNA biogenesis
Both EVs and RNA-binding carriers, such as lipoproteins and ribonucleoproteins, are reported to transport molecules in the extracellular space. Investigators will determine which RNA classes are targeted for export, describe the subcellular processing pathways by which exRNAs are directed to specific secretory pathways, and identify molecular mechanisms that regulate the secretion of exRNAs.
exRNA biodistribution
Mechanisms that regulate exRNA stability and distribution in the extracellular environment need to be identified. For example, it is currently unknown if exRNA from food or breast milk can survive the gastrointestinal tract of the host. The range, targets, possible co-factors, or any other influences on the biodistribution of exRNA need to be elucidated. Here, novel tools and technologies to track exRNA distribution in vivo will be generated.
exRNA uptake
How exRNAs, whether encapsulated in EVs or complexed with RNA-binding carriers, gain access to target tissues or cells, either locally or systemically, is a critical unknown that is being addressed. Mechanisms by which cell surface-targeting motifs mediate contact between exRNA carriers and target cells, as well as regulate the uptake of exRNAs, will be determined.
exRNA effector function
exRNAs are thought to regulate target cell phenotypes and contribute to the onset, regulation, and resolution of various diseases. There is a critical need to identify and validate molecular targets and outcomes for exRNA, as well as determine the influences of any possible co-factors on the function of the target cell. Lastly, the mechanisms by which exRNAs access specific subcellular compartments, such as the nucleus, need to be determined.
Initiative 2: defining a comprehensive reference profile of circulating human exRNA
To understand the functions of exRNAs, and then use that knowledge to leverage them as biomarkers and diagnostic and therapeutic tools, it is of fundamental importance to establish normative distribution profiles from healthy populations. Hence, this initiative focuses on generating a comprehensive reference profile of exRNAs, including any environmentally derived exRNA (e.g. from diet or microbiome) from a diversity of healthy human body fluids including blood, saliva, urine, breast milk, semen, amniotic fluid, cerebrospinal fluid, ascites, and pleural effusions. The body fluids are obtained from healthy human subjects to serve as reference data for future analyses of the role of exRNAs in normal biological processes and in disease conditions. The profiles obtained through this discovery phase will include exRNAs from vesicles and those bound to carrier proteins such as lipoproteins and RNA-binding proteins, and environmentally derived exRNA species such as from the diet. Furthermore, the initiative encourages the development of reagents, tools, and approaches for profiling both short and long non-coding exRNA species.
Initiative 3: exRNA biomarkers
The ability to study and treat disease, and to develop effective interventions is hampered by a lack of unique, reliable, quantifiable, and easily measured biomarkers that correlate well with disease progression. Biomarkers are physical, functional, or biochemical indicators of a normal physiological or disease process that has diagnostic and/or prognostic utility and can replace symptomatic or phenotypic clinical end points as a measure of the effect of new therapies.
These awards were made using the UH2/UH3 mechanism. During the UH2 phase, investigators will identify novel exRNA biomarkers obtained from human biofluids such as serum, plasma, saliva, urine, and cerebrospinal fluid. Investigators will identify biomarkers with characteristics unique to human disease conditions that may lead to potential clinical utility. ExRNA biomarkers developed may include biomarkers of risk, diagnosis, and toxicity; they will also include biomarkers that predict drug response, prognosis and/or pharmacodynamics, and biomarkers as surrogate endpoints of disease. Ideally, biomarkers will be measured in a minimally invasive way. They will also be reproducible over time, indicative of disease prognosis, and well correlated with progression and responsiveness to therapy. The successful completion of confirmatory studies with specific goals and milestones will allow the investigators to transition into the second phase of award, the UH3 phase. Upon evaluation of milestone completion, it is anticipated that not all UH2 awards will progress to the second phase.
The UH3 phase will support validation and qualification studies of biomarkers from a successful UH2 phase. Validation has been described as the process of linking a biomarker to clinical or behavioural endpoints. The process of validation is complex, but includes assay characterization and establishing the clinical utility and superiority of a new biomarker relative to standard tests. It also involves an understanding of the employed technology and clinical knowledge of the disease of interest. The expertise needed to validate clinical biomarkers may include assay development, quality control, epidemiology, and biostatistics.
Initiative 4: exRNA therapy
The goal of these UH2/UH3 awards is to demonstrate the therapeutic potential of exRNAs and to develop methods and tools that will enable targeting to specific cells. These studies will capitalize on the early gains in knowledge of exRNA biology by incorporating cutting-edge molecular, genomic, and bioengineering principles to develop exRNAs, EVs, and RNA-binding complexes for the treatment of human diseases.
During the UH2 phase of award, support will be provided for proof of principle studies to develop and advance the therapeutic utility of exRNAs, RNA-containing EVs or exosomes, and/or exRNA bound to other carrier molecules, such as lipoproteins and RNA-binding complexes. UH2 studies will examine the biological rationale for the proposed therapeutic use, identify the dose or exposure of the therapeutic agent in the proposed patient group, develop appropriate methods for in vivo tracking, and inform patient selection. The second UH3 phase will support latter stage pre-clinical studies to further findings from the UH2 phase and may involve safety, tolerability, pharmacokinetic and pharmacodynamic studies in response to the therapeutic agent.
Initiative 5: data management and resource repository
The Data Management Resource and Repository (DMRR) will integrate the efforts of all of the funded components of the ERCC and serve as a community-wide resource for ERCC data. Critical to these efforts, the DMRR will work with consortium members and other scientific stakeholders to ensure that all ERCC-generated data have standardized associated metadata that use standardized ontologies and quality metrics to ensure the data are interoperable, transportable, and maximally useful to the public. ERCC data sets will be deposited in appropriate public or controlled-access databases to provide user-friendly access. In addition, computational analysis tools will be developed to enable investigators to query, integrate, analyse, and model the data. The functions of the DMRR require three distinct stand-alone components in the areas of Scientific Outreach, Data Coordination, and Data Integration and Analysis.
The DMRR Scientific Outreach Component (DMRRSOC) has established the exRNA Research Portal (exRNA.org), which will eventually include an exRNA Atlas. The exRNA Atlas will provide the scientific community with user-friendly access to exRNA data (exRNAs from normal human body fluids), standardized exRNA protocols (e.g. vesicle isolation, RNA-seq, small RNA-seq), information about exRNA biogenesis, biomarkers, therapeutics, software, and other useful tools and technologies generated by the ERCC. The SOC will develop an outreach strategy to advertise available ERCC protocols and resources to the scientific community.
The DMRR Data Coordination Component (DMRR DCC) will work closely with consortium investigators to obtain protocols, metadata, data, etc. to provide the scientific community with user-friendly and publicly accessible exRNA information. The DCC will help develop data pipelines and data quality metrics, standardized ontologies, metadata, and data formats, they will incorporate data sets from beyond the ERCC and establish an export pipeline to permit timely transfer of ERCC data from consortium data freezes or publications to appropriate public repositories and community databases.
The DMRR Data Integration and Analysis Component (DMRR DIAC) will facilitate analysis of the data obtained by consortium investigators. The DIAC will coordinate multiple teams for analysing consortium data in different ways. The DIAC will facilitate data analysis efforts of smaller projects and also provide substantial support for consortium-wide integrative analysis efforts. The majority of data sets that will be analysed will likely be RNA-seq, small RNA-seq, or related data types; however, other data types (e.g. proteomic, metabolomic, and individual variation) may also require analysis. The DIAC will generate comprehensive catalogues of exRNAs for different body fluids and/or define biological roles for these exRNAs, coordinate and facilitate integrative analyses of exRNA consortium data, and provide resources and coordination activities to enable the ERCC to share useful information about exRNA species and profiles to the larger biomedical research community.
Detailed information on the projects of all 5 of these research initiatives may be found on the NIH RePORTER and the NIH Common Fund's exRNA Communication webpage (www.commonfund.nih.gov/exrna/fundedresearch). Information on RFAs may be found under archived funding opportunities (www.commonfund.nih.gov/Exrna/grants and www.grants.nih.gov/grants/guide/rfa-files).
In summary, the objective of this paper is to describe the overall scope and vision of the NIH ERCC, to inform the research community about the goals of the Consortium, and to describe outreach strategies to share the fundamental scientific discoveries, protocols, data standards, quality control, and innovative tools and technologies. Ultimately, these deliverables and resources will be found on the exRNA Research Portal (exRNA.org).
Additional information for the reviewers
As requested, here are the non-American laboratories (names and location) involved in the consortium: Giovanni Camussi (University of Torino), Kyoung Mee Kim (Samsung Medical Center), Suresh Mathivanan (La Trobe University), Olivier Voinnet (Eidgenossische Technische Hochschule Zurich), Andreas von Deimling (Heidelberg University Hospital), Paul Wilmem (University of Luxembourg), and Matthew Wood (University of Oxford).
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
Notes
This paper is part of the Special Issue: Extracellular RNA Communication Consortium. More papers from this issue can be found at http://www.journalofextracellularvesicles.net