Abstract
Background
Penetration of skin, migration through tissues and establishment of long-lived intravascular partners require Schistosoma parasites to successfully manipulate definitive host defences. While previous studies of larval schistosomula have postulated a function for excreted/secreted (E/S) products in initiating these host-modulatory events, the role of extracellular vesicles (EVs) has yet to be considered. Here, using preparatory ultracentrifugation as well as methodologies to globally analyse both proteins and small non-coding RNAs (sncRNAs), we conducted the first characterization of Schistosoma mansoni schistosomula EVs and their potential host-regulatory cargos.
Results
Transmission electron microscopy analysis of EVs isolated from schistosomula in vitro cultures revealed the presence of numerous, 30–100 nm sized exosome-like vesicles. Proteomic analysis of these vesicles revealed a core set of 109 proteins, including homologs to those previously found enriched in other eukaryotic EVs, as well as hypothetical proteins of high abundance and currently unknown function. Characterization of E/S sncRNAs found within and outside of schistosomula EVs additionally identified the presence of potential gene-regulatory miRNAs (35 known and 170 potentially novel miRNAs) and tRNA-derived small RNAs (tsRNAs; nineteen 5′ tsRNAs and fourteen 3′ tsRNAs).
Conclusions
The identification of S. mansoni EVs and the combinatorial protein/sncRNA characterization of their cargo signifies that an important new participant in the complex biology underpinning schistosome/host interactions has now been discovered. Further work defining the role of these schistosomula EVs and the function/stability of intra- and extra-vesicular sncRNA components presents tremendous opportunities for developing novel schistosomiasis diagnostics or interventions.
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
Schistosomes are parasitic flatworms responsible for schistosomiasis, a neglected tropical disease currently affecting approximately 230 million people found in poverty-stricken areas of the developing world (Citation1). This chronic and debilitating disorder is initiated when the aquatic cercariae stage of the parasite's lifecycle actively penetrates human skin and transforms into a tissue-migrating schistosomulum. Upon penetration, the schistosomulum releases excretory/secretory (E/S) products in a directed attempt to simultaneously manipulate the host extracellular matrix, to modulate host defensive barriers and to protect itself from oxidative stress (Citation2, Citation3). Collectively, these E/S-driven biological processes ultimately enable a proportion of infectious parasites to successfully continue their complex migration throughout the human host, develop into blood-dwelling dioecious adults and establish an infection that can last for years (Citation4). As these E/S products need to operate in the presence of harsh environmental stresses within human tissue/blood, their packaging into protective transferable units would be advantageous for sustaining bioactivity (Citation5). An evolutionarily conserved packaging system that has not been thoroughly investigated in this aspect of schistosome biology is extracellular vesicles (EVs).
Eukaryote EVs (exosomes and ectosomes) are between 30 and 1,000 nm in size (Citation6, Citation7) and are released from cells by either the fusion of multivesicular bodies with the plasma membrane (exosomes, 30–100 nm) or by directly budding from the plasma membrane (ectosomes, 100–1,000 nm) (Citation7–Citation11). During biogenesis, EVs protectively package intracellular components (e.g. proteins, RNAs, lipids and protein/RNA complexes) for extracellular release and short- or long-distance intercellular communication (Citation9, Citation12). While the molecular mechanisms underlying EV biogenesis and cellular release are generally well understood, the processes responsible for selective packaging of cargo destined for intercellular communication and the uptake of these packages by recipient cells is likely to vary among different cell types or organisms analysed (Citation13). Among EV cargos, there is growing evidence that certain miRNA, lipid and protein species are found enriched in certain vesicle types (e.g. 12). This implies that EVs (and their packages) may regulate specific cell-to-cell, organ-to-organ, individual-to-individual or pathogen-to-host signalling, potentially playing a central role in many important biological processes (Citation14). In terms of parasites, the role of protozoan-derived EVs in mediating horizontal gene transfer and immunomodulation has been hypothesized for a variety of species (Citation15). Within the metazoan endoparasites, recent investigations of Echinostoma caproni, Fasciola hepatica, Dicrocoelium dendriticum, Schistosoma japonicum, Opisthorchis viverrini, Heligmosomoides polygyrus and Trichuris suis have demonstrated that EVs are excreted/secreted from helminths and may be taken up by host cells (Citation16–Citation21). Based on the growing realization that EVs facilitate intercellular communication in eukaryotes and are excreted/secreted from related platyhelminths, we hypothesized that they are important players in the initiation and maintenance of long-lived host/parasite interactions during schistosomiasis.
Here, for the first time, we isolate EVs derived from Schistosoma mansoni schistosomula E/S products and characterize their protein and small non-coding RNA (sncRNA) components. While our findings highlight a rich repository for infection-related biomarkers, they also identify an entirely new strategy by which schistosomes likely manipulate host defensive barriers. Further exploration of these vesicles’ functions may identify novel strategies for schistosomiasis control.
Materials and methods
Ethics statement
All procedures performed on mice adhered to the United Kingdom Home Office Animals (Scientific Procedures) Act of 1986 (project license PPL 40/3700) as well as the European Union Animals Directive 2010/63/EU and were approved by Aberystwyth University's (AU) Animal Welfare and Ethical Review Body.
Parasite material
A Puerto Rican strain (NMRI) of S. mansoni was used throughout the study and passaged between Mus musculus (Tuck Ordinary, TO) and Biomphalaria glabrata (NMRI albino and pigmented hybrid) hosts. Cercariae were shed from both B. glabrata strains by exposure to light in an artificially heated room (26°C) for 2 h. Collected cercariae were mechanically transformed into schistosomula according to established methodologies (Citation22) and resuspended in culture medium containing DMEM (Dulbecco's Modified Eagle Medium, Sigma-Aldrich, Dorset, UK), 4.5 g/l glucose, 2 mM L-glutamine, 200 U/ml penicillin and 200 µg/ml streptomycin (all Sigma-Aldrich) with or without 10% v/v foetal calf serum (FCS, Sigma-Aldrich). All schistosomula (7,500 individuals/ml) were cultivated at 37°C and left to develop for 72 h in a humidified atmosphere containing 5% CO2.
EV purification
EVs were purified by differential centrifugation of 72 h schistosomula E/S products as previously described (Citation23) with slight modifications. Briefly, the schistosomula culture supernatant (containing E/S products) was collected and centrifuged at low speed (500×g for 2 min, 4°C) to remove remaining parasites and then sequentially centrifuged at a higher speed (700×g for 20 min, 4°C) to remove any residual debris. This resulting “pre-cleared” supernatant was subsequently centrifuged at 120,000×g for 80 min at 4°C on an Optima™ L-100 XP ultracentrifuge (Beckman Coulter, High Wycombe, UK) using a Type 70 Ti rotor. The EV-enriched pellet was washed 3 times in phosphate buffered saline (PBS, pH 7.4) and finally resuspended in 100 µl of PBS. The EV-depleted supernatant was concentrated to 500 µl using a 5 k MWCO filter (Amicon, Fisher Scientific, Loughborough, UK), washed 2 times by diluting with 2 ml PBS and reconcentrated to 500 µl. Both EV-enriched and EV-depleted samples were stored at −80°C until further use.
Scanning electron microscopy
Intact cercariae were prepared as previously described (Citation24). In brief, parasites were fixed in 4% glutaraldehyde/4% paraformaldehyde for 24 h at 4°C, washed in PIPES buffer (pH 7.2) for 4 h at 4°C, incubated in 1% osmium tetroxide (1 h at 4°C), rewashed in PIPES buffer and deposited on poly-L-lysine coated coverslips prior to dehydration and critical point drying. After critical point drying, parasite samples were mounted onto scanning electron microscope (SEM) stubs, sputter coated with gold/palladium and visualized with a Philips XL 30 FEG SEM.
Transmission electron microscopy
Schistosomula EV preparations were qualitatively assessed for size and shape by transmission electron microscopy (TEM). Briefly, a 25 µl aliquot of each EV-enriched sample was fixed with an equal volume of 4% glutaraldehyde, adsorbed onto Formvar/carbon-coated copper grids and stained with 2% uranyl acetate (pH 4). Processed EV samples were subsequently visualized on a Jeol 1010 transmission electron microscope operated at 60 kV and 30,000× magnification. Images were recorded with a Kodak MegaPlus camera Model 1.4i, visualized by analySIS 3.1 software and processed on ImageJ.
Helminth fluorescent bioassay and fluorescent microscopy
Schistosomula viability in each culture condition (incomplete medium, complete medium or heat inactivated at 65°C for 10 min) was assessed at 72 h as previously described (Citation25). Briefly, after washing the parasites with DMEM, propidium iodide (PI; 544 nm excitation/620 nm emission; 2.0 µg/ml) and fluorescein diacetate (FDA; 485 nm excitation/520 nm emission; 0.5 µg/ml) were simultaneous added to each schistosomula sample. Fluorescent data were collected from a BMG Labtech Polarstar Omega plate reader (Durham, NC, USA) and converted into percent viability values. Fluorescent images of FDA/PI co-stained parasites were obtained from an ImageXpress micro widefield high content screening system (Molecular Devices, Wokingham, UK) with bright field and FITC-TRITC filter set. FDA and PI images were merged and analysed using ImageJ. Micron-sized vesicles released from in vitro cultured schistosomula labelled with FDA were visualized at 40× magnification using a Leica Axioplan (Milton Keynes, UK) microscope equipped with an FITC (494 nm excitation) filter and a mercury vapour light source. A Hamamatsu CA74295 camera (Welwyn Garden City, UK) with Wasabi Version 1.4 software was used to capture photographic images of stained schistosomula.
Proteomics
EV-enriched fractions (n=4) were centrifuged for 1 h at 100,000×g at 4°C (TL-100 ultracentrifuge, Beckman Coulter, Inc., Fullerton, CA, USA). Pellets were dissolved in 50 mM TEAB buffer (triethylammonium bicarbonate, pH 8.5) containing 1% RapiGest (Waters, Milford, MA, USA). Reduction and alkylation of cysteine was performed using 2 mM TCEP (tris(2-carboxyethyl)phosphine, 1 h at 60°C) and 4 mM MMTS (methyl methanethiosulfonate, 15 min at room temperature with constant shaking), respectively. Then, 5 µg of trypsin (Sequencing Grade Modified Trypsin, Promega, Madison, WI, USA) was added and proteins were digested overnight at 37°C. Samples were subsequently acidified to pH<2 using 10% TFA, incubated at 37°C for 30 min and centrifuged at 16,000×g for 10 min. The supernatant was concentrated to approximately 20 µl in a vacuum concentrator and stored at −20°C prior to mass spectrometry analysis.
Peptides were injected onto a trap column (Acclaim PepMap100, 100 µm×2 cm; nanoViper C18, 5 µm, 100 Å; Thermo Fisher Scientific, Waltham, MA, USA) equilibrated with 0.1% formic acid and separated on an analytical reverse phase column (Acclaim PepMap RSLC 75 µm×50 cm; nanoViper C18, 2 µm, 100 Å; Thermo Fisher Scientific) coupled to an ion trap mass spectrometer (amaZon Speed, Bruker Daltonics, Bremen, Germany) at a flow rate of 300 nl/min. The liquid chromatography was performed with a piece-linear gradient (0–10 min at 2% B, 10–25 min to 5% B, 25–165 min to 25% B, 165–175 min to 30% B, 175–190 min to 35% B, 190–195 min to 100% B, 195–210 min at 100% B, where B is 95% acetonitrile/0.1% formic acid). For mass spectrometric analysis, ions were generated using CaptiveSpray (Bruker Daltonics) at a spray voltage of 1.3 kV. The temperature of the heated capillary was set to 150°C. Tandem mass spectrometry was performed in a data-dependent manner, selecting the 10 highest peaks in an MS spectrum. Masses were excluded for 1 min after MS/MS was performed.
Peak lists from the 4 EV samples were generated using Data analysis 4.0 (Bruker Daltonics) with default settings and were exported as Mascot Generic Files. Peptides were identified in the S. mansoni database (v4.0; 13,193 sequences; 5,894,147 residues) using the Mascot algorithm (Mascot 2.5, Matrix Science, London, UK). An MS tolerance of 0.5 Da (with the number of 13C=1) and an MS/MS tolerance of 0.5 Da were used. Trypsin was designated as the enzyme and up to 1 missed cleavage site was allowed. Beta-methylthiolation of cysteine was selected as a fixed modification and oxidation of methionine as a variable modification. Data were exported as XML files at a target false discovery rate for peptide identification of 1% (using a target/decoy approach). The XML files were then analysed using an in-house script, selecting only peptides corresponding to the highest peptide-spectrum match (Rank 1 peptides) with a Mascot score above 25. For the generation of the overall list of proteins identified (Supplementary file 1), only proteins with at least 2 unique peptides were selected. From this list, only proteins identified in 3 out of the 4 biological replicates were considered as schistosomula extracellular vesicle proteins.
DNA microarray transcription profiles
Data from the 37,632 element S. mansoni long-oligonucleotide DNA microarray studies of Fitzpatrick et al. (Citation26) were interrogated to find the transcription pattern corresponding to the 72 h schistosomula EV proteome across 14 life-cycle stages. Briefly, log2 normalized gene expression data was obtained for 102 of the 109 EV proteome components (unique 50-mer oligonucleotides for 7 Smps were not present on the DNA microarray; Supplementary file 2) and subjected to hierarchical agglomerative clustering (Euclidean distance and complete linkage) using the R statistical programming language (Citation27) and the Bioconductor (Citation28) package gplots. A full set of raw and normalized data is available via ArrayExpress under the experimental accession number E-MEXP-2094.
RNA extraction, sncRNA library preparation and sequencing
Total RNA was extracted from 100 µl EV-enriched (n=3) and 100 µl EV-depleted (n=3) schistosomula E/S samples using the miRNeasy kit (Qiagen, Dorking, UK) according to the manufacturer's instructions. The profile of the small RNA samples was assessed with a Bioanalyzer small RNA chip (Agilent Technologies, Santa Clara, CA, USA). Small RNA concentrations ranged from 60 pg/ml (EV-enriched fraction) to 750 pg/ml (EV-depleted fraction) as determined by the Bioanalyzer.
sncRNA libraries were prepared using the NEBNext Multiplex Small RNA Library Prep Set and NEBNext index primer for Illumina (New England Biolabs, Ipswich, MA, USA), according to the manufacturer's protocol. Size selection of the polymerase chain reaction (PCR) products was performed using 6% non-denaturing polyacrylamide gel electrophoresis, and bands between 130 and 200 bp were recovered following gel extraction. HiSeq 100 bp single end Illumina sequencing was performed at the AU Translational Genomics facility using the HiScanSQ platform. Raw reads were deposited at the NCBI sequence read archives under the accession number SRP057687.
microRNA detection and annotation
The miRDeep2 package (Citation29) was used to identify known and putative novel miRNA present in the EV-enriched and EV-depleted compartments of schistosomula E/S. After cleaning the reads with Trimmomatic (Citation30), trimmed sequences of at least 16 nucleotides long were pre-processed (i.e. aligned) against the S. mansoni genome v5.2 (Citation31) using the mapper module of miRDeep2 (utilizing bowtie) with default options that included discarding reads shorter than 18 bp, collapsing reads and mapping reads without any mismatches. These stringent criteria resulted in low numbers of mapped reads. Weakening the criteria to allow mismatches greatly increases the proportion of mapped reads identified (e.g. 60% or more depending upon the criteria selected). Unmapped reads (at any criteria) included a high proportion of low complexity regions. Identification of miRNAs was performed with the miRDeep2.pl script, including quantification and expression profiling performed by the Quantifier module. Known precursor S. mansoni miRNAs as well as mature miRNA sequences derived from 5 platyhelminth species (Schmidtea mediterranea, Echinococcus granulosus and Echinococcus multilocularis, Schistosoma japonicum and S. mansoni) were provided to the miRDeep2.pl script from miRBase (release 21). Outputs from both the Quantifier and miRDeep2 were merged and 2 filtering steps were applied to generate a final list of identified miRNAs: (a) only those miRNAs displaying a log-odd score above 0 and (b) only those miRNAs displaying at least 10 reads in 2 out of the 3 biological replicates (EV-enriched or EV-depleted fractions) were kept (Supplementary file 3). Library size scaling factors were calculated for each sample and miRNA data were normalized following the protocol of Anders et al. (Citation32) using the R statistical programming language (Citation27) and the Bioconductor (Citation28) package DESeq2 (Citation33). The Quantifier module maps reads to predefined miRNA precursors derived from miRBase; however, some of these miRNAs could not be mapped to the current S. mansoni genome assembly (v5.2).
tRNA-derived small RNA detection and annotation
Trimmed reads (passing QC metrics) ranging from 1 to 95 nucleotides (size range of inserts in our sncRNA libraries) were mapped to the S. mansoni tRNA database (depleted of mitochondrial and pseudo-tRNA sequences) obtained from GeneDB (Citation34; accessed Nov-2014) using BLAST. As a first filter, only reads displaying a top BLAST hit with 100% sequence identity and an alignment covering 90% of their sequence length were retained (dashed line, ). Reads mapping to tRNA sequences encoding for the same tRNA type, but containing different anticodon sequences, were combined. This processing step was performed to reduce the complexity of mapping reads to 1,099 genomic loci encoding tRNAs (current number of tRNA genes in the S. mansoni genome, v5.2). It allowed us to identify the predominant tRNA types (from the 20 ubiquitous tRNAs, selenocysteine tRNA and suppressor tRNA) present in our E/S fractions. As a second filter, these reads were processed using an in-house script to identify both 5′ and 3′ tRNA-derived small RNAs (tsRNA) composed of both tRNA halves (tiRNAs) and tRNA fragments (tRFs) (Citation35, Citation36). Here, reads of at least 11 bp were classified as follows: (a) as a 5′ tsRNA if the alignment started at Position 1 in the mature tRNA and ended at or before Position 35; (b) as a 3′ tsRNA if the alignment started at the last position in the tRNA and extended 11–30 bp or 40–50 bp towards the 5′ end (solid line, ). Mature 3′ tsRNA reads (containing a terminal CCA-3′ anchor) were subsequently quantified using similar processing, except that the tRNAs from GeneDB were modified by adding a CCA anchor to their 3′ end before performing the BLAST alignments. Only tsRNA fragments containing at least 10 reads in 2 out of the 3 biological replicates (EV-enriched or EV-depleted fractions) were kept. Data normalization on these tsRNAs was performed following the procedures described for the miRNA analysis above.
Results and discussion
In vitro cultivated S. mansoni schistosomula excrete/secrete EVs
To investigate whether S. mansoni produces exosome-like EVs, we specifically examined a lifecycle stage that actively excretes/secretes copious amounts of parasite products, the tissue-migrating schistosomula (Citation37). This choice maximized our chances of finding excreted/secreted nanosized vesicles in culture supernatants derived from in vitro cultivated schistosomes (). Epifluorescent microscopy of FDA (an esterase substrate) labelled schistosomula as well as TEM of cercaria clearly demonstrated that these lifecycle stages produce micron-sized vesicles from their anterior ends (Fig. 1a), a phenomenon that has been previously documented (Citation38). It is these secretory products, originating from schistosomula unicellular acetabular or head glands (Citation39) and accumulating throughout the cultivation period, that represented a rich source of starting material (in addition to tegumental contributions) for the first isolation and characterization of nanosized exosome-like EVs in S. mansoni.
Fig. 1. Schistosoma mansoni excretes/secretes exosome-like extracellular vesicles (EVs) during in vitro cultivation. (a) Microscopic analysis of infective stage cercariae (scanning electron microscopy – left panel) and tissue-migrating schistosomulum (epifluorescent microscopy of fluorescein diacetate labelled parasites – right panel) demonstrates the excretion/secretion of micron-sized vesicles (highlighted by white rectangle boxes). To determine whether EVs were present in these (or other) excreted/secreted (E/S) products, we specifically examined the supernatant of 72 h in vitro cultivated schistosomula. (b) Parasite viability was assessed at 72 h using the helminth fluorescent bioassay (HFB) (Citation25) (n=11) in cultures containing or lacking foetal calf serum (FCS, a source of contaminating bovine EVs (Citation40)) compared to heat-killed controls. Error bars represent the standard deviation of the mean. Student's t-test was used to assess differences in viability between the schistosomula cultures. High content imaging (FITC and TRITC filters, 10×magnification) of fluorescently co-labelled schistosomula (fluorescein diacetate, green and propidium iodide, red) supports the HFB measurements. (c) EVs were isolated from E/S products by ultracentrifugation of the 72 h schistosomula culture supernatant (-FCS) and analysed by transmission electron microscopy at 30,000×magnification.
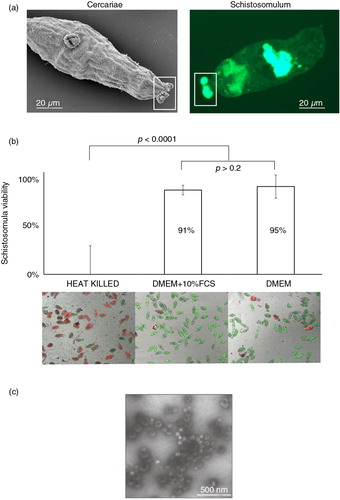
As FCS is a potential source of contaminating bovine EVs (Citation40) and a downstream confounding variable for the identification of parasite-derived EV-associated proteins and sncRNAs, we first determined the viability of schistosomula cultivated in media lacking FCS (Fig. 1b). Here, parasites cultured in the absence of FCS for 72 h were phenotypically normal (motile, non-granular) and demonstrated similar viability values (95%) when compared to the control (91%) schistosomes cultured in the presence of FCS (Student's t-test, one-tailed; p>0.2). Therefore, the lack of FCS in the cell culture media did not negatively affect the viability of 72 h cultivated schistosomula. Preparatory ultracentrifugation of E/S products derived from these 72 h schistosomula cultures (-FCS) and TEM of the pelleted vesicles demonstrated morphological features (sizes ranging between 30 and 100 nm in diameter) consistent with exosomes (Fig. 1c). These vesicles were not observed (according to the limits and resolution of TEM) in the EV-depleted, schistosomula E/S fractions. Because these schistosomula nanovesicles potentially represent novel modulators of host/parasite interrelationships, their proteomic and sncRNA content was explored by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and sncRNA-seq analyses.
Schistosomula EV proteomes contain a collection of conserved vesicle biomarkers and unique parasite components
Proteomic analyses of schistosomula EV-enriched fractions identified 109 proteins that were reproducibly detected (see Materials and methods) in 4 independently collected parasite samples ( and Supplementary file 1). When we compared them to a collated set of previously annotated eukaryote EV proteomes (Citation12), we found entries in common with our schistosomula dataset including (not exclusive) tetraspanins (Smp_181530, Smp_173150), heat shock proteins (Smp_106930), annexins (Smp_077720, Smp_045500, Smp_074140, Smp_045560 and Smp_045550), Rab11 proteins (Smp_005670, Smp_104310 and Smp_173990), 14-3-3 isoforms (Smp_009760, Smp_002410 and Smp_034840.1), cytoskeletal proteins (e.g. Smp_066760, Smp_046600 and Smp_202970.1) and metabolic enzymes (e.g. Smp_054160, Smp_056970.1 and Smp_102070). These proteomic data strongly support our TEM interpretations (Fig. 1) and indicate that the nanovesicles isolated from in vitro schistosomula cultures are indeed exosome-like EVs (in comparison to apoptotic bodies or ectosomes). Measuring these data against previous proteomic investigations of schistosomula E/S products (Citation37, Citation41), we find an overlapping subset (~20%; 22/109) of atypical E/S proteins (e.g. absence of a signal peptide, as predicted by SignalP 4.1 (Citation42); performed May 2015) also present in our EV dataset (e.g. Smp_046600, actin; Smp_106930, hsp70; Smp_054160, GST28; Smp_008070, thioredoxin; Smp_024110, enolase; Smp_095360.1, fatty acid binding protein; Smp_040130, cyclophilin; Smp_056970.1, GAPDH; Smp_214190, calpain; Smp_042160.2, fructose bisphosphate aldolase; Smp_086480, SmTAL2; Smp_086530, SmTAL3; Smp_003990, triosephosphate isomerase 1a). While holocrine secretion, schistosomula damage or cultivation conditions could be responsible for the presence of these “cytosolic” proteins in E/S proteomes, our results now provide an alternative explanation and indicate that schistosome EVs are a vehicle for non-classical protein excretion/secretion.
Table I. The top 35 protein hits in schistosomula EVsa
Further sequence comparisons to EV proteomes derived from related F. hepatica (hermaphroditic liver fluke), E. caproni (hermaphroditic intestinal fluke) and D. dendriticum (hermaphroditic liver fluke) trematodes (Citation16, Citation17) revealed a common subset of proteins involved in metabolic processes (e.g. enolase, glyceraldehyde 3-phosphate dehydrogenase, fructose bisphosphate aldolase), calcium binding (e.g. calcium binding proteins) and structure (e.g. actin and annexin). When differences were found (e.g. F. hepatica EVs contain large numbers of proteases whereas E. caproni, D. dendriticum and S. mansoni schistosomula do not), divergent life histories (hermaphroditic vs. dioecious lifecycles; liver vs. intestine vs. blood residency) or compared developmental stages (F. hepatica, D. dendriticum and E. caproni adults vs. S. mansoni larvae) were likely responsible. Indeed, it is well-established that EVs derived from different cellular sources contain distinct subsets of proteins linked to specific cell-associated activities (Citation6, Citation43). Therefore, functional characterization of the 15 hypothetical (not conserved) proteins identified in the schistosomula EV proteome (Supplementary file 1) may provide important insight into how these particular vesicle components specifically influence schistosome infection, migration and development.
Extending our schistosomula EV proteome interrogation further, we utilized both gene ontology (GO) (Citation44) and existing DNA microarray (Citation26) datasets to quantify functional category enrichment as well as to predict whether these particular proteins could be found in EVs derived from other schistosome lifecycle stages (). GO analyses of these 109 schistosomula EV proteins using g:Profiler (Citation45) indicated that 11 categories (5 within biological processes, 6 within molecular function but none within cellular component) were significantly over-represented when compared to a background dataset comprised of all electronically GO-annotated S. mansoni proteins (ENA assembly version ASM23792v2) (a). Interestingly, 2 of these 5 “biological process” categories (small GTPase-mediated signal transduction and protein transport) were also found over-represented in a recent GO meta-analysis of proteins commonly found in human EVs (Citation12). While this analysis provides additional evidence supporting the classification of these schistosomula vesicles as EVs, characterization of EV proteomes derived from other schistosome lifecycle stages is necessary to fully describe their functional diversity.
Fig. 2. Schistosomula EVs contain vesicular-enriched functional components and include proteins likely to be found in EVs derived from other schistosome lifecycle stages. EVs were isolated from 72 h schistosomula cultures using preparative ultracentrifugation (Citation23). Pelleted EVs (n=4) were digested with trypsin and subsequently prepared for liquid chromatography–tandem mass spectrometry. Only proteins represented by 1 or more unique peptides in 3 out of the 4 replicates (with 1 of these replicates having to contain at least 2 unique peptides) were considered as schistosomula EV proteins. (a) Bar chart representation of significantly enriched gene ontology (GO) categories found in EV proteomes analysed by g:Profiler (Citation45) using Benjamini–Hochberg false discovery rate detection and best per-parent GO identifier (moderate). (b) Expression profiling of the genes (derived from 102/109 Smps) encoding EV proteome components throughout the schistosome life cycle (Supplementary file 2). DNA microarray data (log2 normalized gene expression values) obtained from Fitzpatrick et al. (Citation26) was subjected to agglomerative hierarchical clustering (Euclidean distance and complete linkage) and illustrated as a heat map. The dashed rectangular box represents the 72 h schistosomula lifecycle stage.
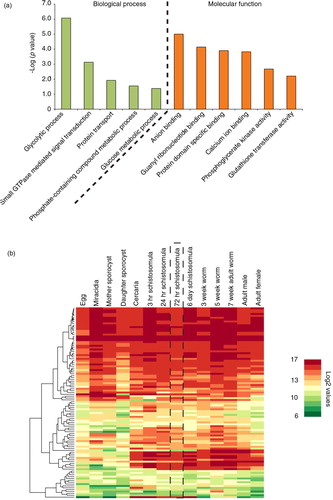
To determine if the 72 h schistosomula EV proteome (Supplementary file 1) simply reflected the most abundantly expressed genes found within this lifecycle stage, we quantified transcription data for these 109 gene products from a recently published schistosome DNA microarray study (Citation26). This analysis (b, dashed vertical box, and Supplementary file 2) revealed that 72 h schistosomula EVs contained proteins derived from both highly and lowly expressed genes. While caution must be taken in extrapolating the abundance of transcripts derived from whole schistosomula transcriptomes to schistosomula EV proteomes, our finding is in line with current thinking about the biogenesis of EVs, which indicates that protein packaging is governed by controlled endovesicular sorting mechanisms, independent of abundance (Citation46). These sorting mechanisms involve the co-operation of hsp70/hsp90 chaperones, 14-3-3 epsilons, tetraspanins and protein kinases (Citation47). The presence of these proteins in EVs derived from 72 h cultivated schistosomula as well as their ubiquitous expression throughout the parasite's lifecycle (Supplementary file 2) indicates that this conserved packaging mechanism may also be operational within schistosomes. How many additional proteins identified in our current study are also found in EVs derived from other schistosome lifecycle stages awaits further investigation. Our gene expression analyses predict that there may be overlap (b).
Schistosomula E/S products contain miRNAs found within and outside EVs
Recent studies have demonstrated that helminth parasites excrete/secrete sncRNAs (Citation17, Citation18), with the most detailed analysis to date indicating that 78% of the sncRNA E/S fraction of H. polygyrus (<30 nucleotides) is composed of miRNAs (Citation18). Because of these findings, we hypothesized that schistosomula E/S products would also contain detectable miRNA and, potentially, other sncRNA classes that could be present within or outside the pelleted EVs produced during our in vitro culture conditions (). Therefore, we performed a sncRNA RNA-seq analysis of schistosomula EV-enriched (c) and EV-depleted E/S fractions.
Applying standard quality control measures and stringent S. mansoni genome (version 5.2) mapping (see Materials and methods) to these 6 RNA-seq libraries (EV-enriched fraction, n=3; EV-depleted fraction, n=3) led to similar average numbers of reads/fraction available for miRNA analyses (). Within the complete (EV-enriched and EV-depleted fractions) schistosomula E/S miRNA-ome, we identified 35 previously characterized platyhelminth miRNAs (known) and 170 potentially novel S. mansoni miRNAs (sma-miRNAs, having no seed sequence similarity to any previously characterized platyhelminth miRNA as determined by miRDeep2) in total ( and Supplementary file 3). These 205 sma-miRNAs could be segregated into those that were found in comparable abundance (114 miRNAs) between EV-enriched and EV-depleted fractions (Cluster 1), those more abundant (51 miRNAs) in EV-enriched fractions (Cluster 2) and those more abundant (40 miRNAs) in EV-depleted fractions (Cluster 3) (a). The genomic origin of these 205 E/S-derived schistosomula sma-miRNAs (vertical black lines) was distributed across both autosomes and sex-defining chromosomes (allosomes) in a proportion no different than the distribution of all known sma-miRNAs deposited in miRBase (vertical grey lines) (b) (p>0.05,Fisher's exact test). This suggests that, while excretion/secretion of these gene-regulatory RNAs may be a directed and carefully controlled process (Citation48), E/S inclusion of sma-miRNAs is independent of autosome/allosome source.
Fig. 3. Schistosomula excrete/secrete extracellular miRNAs. Supernatant from 72 h in vitro cultured schistosomula was separated into EV-enriched and EV-depleted fractions (n=3) by preparatory ultracentrifugation. Total RNA from each fraction was extracted using the miRNeasy kit (Qiagen) and prepared for RNA-seq using the NEBNext Small RNA (for Illumina HiSeq sequencing) kit. To identify sma-miRNAs in our samples, the miRDeep2 package was utilized (29). Only miRNAs with at least 10 reads in 2 out of the 3 biological replicates (in at least one of the fractions, EV-enriched OR EV-depleted) were considered in the study. A total of 205 putative sma-miRNAs passed this criterion. sma-miRNA read count data were normalized using the DESeq2 package (33) as described in the Materials and methods. (a) Heatmap depiction of sma-miRNA abundance (represented by Z-scores) found within EV-enriched and EV-depleted supernatant fractions after agglomerative hierarchical clustering and standardization. Each row represents a miRNA and its relative Z-score value in EV-enriched and EV-depleted fractions is displayed in the 2 columns. All sma-miRNA specifics (name, sequence, raw/normalized read counts and cluster location) are included in Supplementary file 3. (b) sma-miRNA localization found throughout the S. mansoni karyotype (v5.2). Vertical grey bars represent the position of known sma-miRNAs available in miRBase (v.21). Vertical black bars represent the localization of all extracellular sma-miRNAs (within EV-depleted and EV-enriched fractions) newly identified in our study. Black stars above the vertical lines represent 13 sma-miRNAs found in our study that are also present in miRBase. Eighteen miRNAs localized on unmapped scaffolds (16 not mapped at all; 1 unplaced on Ch 7, 4 and 3; 5 unplaced on Ch 1) as well as 15 miRNAs not yet mapped to the current S. mansoni genome assembly were not included in this analysis. All available sma-miRNA localization coordinates are available in Supplementary file 3.
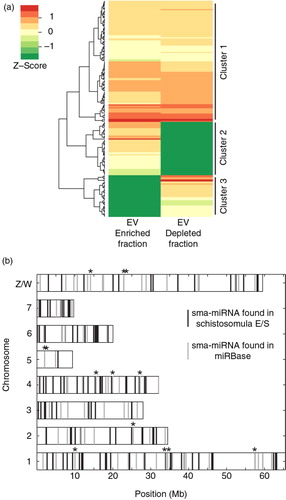
Table II. Small non-coding (snc) RNA-seq library statisticsa
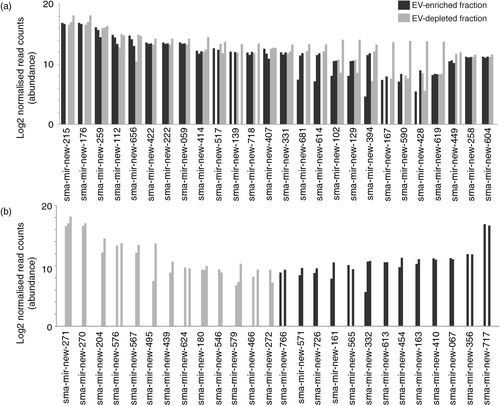
Among the 35 known sma-miRNAs identified in the schistosomula E/S products (), sma-bantam, sma-miR-10, sma-miR-3479 and sma-miR-n1 were all previously detected in sera obtained from individuals (mice, humans or rabbits) chronically infected with S. mansoni (Citation49, Citation50). While none of the newly identified 170 sma-miRNAs have yet been detected in infected sera, their presence should be investigated due to their high abundance in schistosomula E/S products ( and Supplementary file 3). Nevertheless, our findings now indicate a potential mechanism, larval excretion/secretion of known and novel miRNAs, by which these sma-miRNAs are released from schistosomes and accumulate in host blood. The discovery that sma-bantam, sma-miR-10, sma-miR-3479 and sma-miR-n1 (among numerous novel sma-miRNAs) are found in similar abundance between EV-enriched and EV-depleted E/S fractions (a and 5a) would suggest that, in addition to EV encapsulation, serum-detectable sma-miRNAs could be stabilized by protein interactions. Alternatively, the comparable abundance of these (and other, Supplementary file 3) miRNAs found within EV-enriched and EV-depleted E/S fractions could also be indicative of vesicle lysing during the 72 h in vitro cultures. Until a time in which these 2 scenarios are resolved (using differential proteinase K and RNase A processing of EVs), quantitative reverse-transcription PCR detection of the most abundantly excreted/secreted 72 h schistosomula sma-miRNAs (sma-mir-new-215, sma-mir-new-271 and sma-mir-new-717; bold miRNAs highlighted in a and b) in infected mouse sera is currently being planned to confirm their presence in EV-enriched and EV-depleted host biofluids.
Fig. 4. Most of the 35 known miRNAs released by schistosomula are found in comparable abundance between EV-enriched and EV-depleted fractions. Outputs from the miRDeep2.pl script within the miRDeep2 package were used to identify known platyhelminth miRNAs in schistosomula E/S samples. Manual curation against the recent literature (49,51,53) was also performed to identify previously described miRNAs that have not yet been uploaded to miRBase. A total of 35 known miRNAs were subsequently identified. (a) Known sma-miRNAs (26) found at similar abundance between EV-enriched (dark bars) and EV-depleted (light bars) fractions. (b) Known sma-miRNAs (9) found at a higher relative proportion in the EV-enriched (black bars) or the EV-depleted (grey bars) fraction (i.e. containing reads only in the EV-enriched or EV-depleted fractions after normalization). The asterisk (*) represents sma-miRNAs (sma-mir-76 and sma-mir-2149) identified by seed sequence similarity to Schmidtea mediterranea sme-miRNAs. Bar charts represent the miRNA log2 normalized read counts for each sample where the indicated sma-miRNA was found (see Materials and methods). Raw and processed sma-miRNA read data are found in Supplementary file 3.
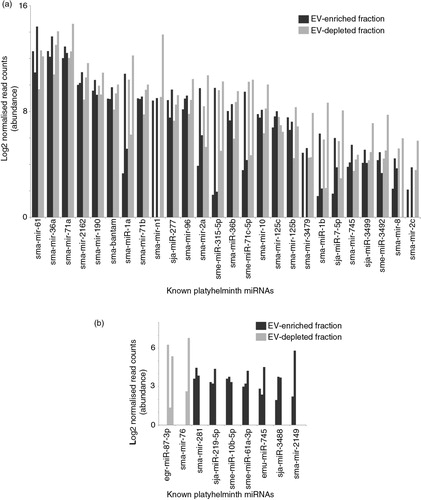
Fig. 5. Schistosomula E/S products contain large numbers of novel miRNAs found within and outside EVs. The miRDeep2 module was used to identify 170 potential novel sma-miRNAs found within schistosomula E/S products (see Materials and methods for details). (a) The 26 most abundant (out of 88 in total) novel sma-miRNAs found in both EV-enriched (black bars) and EV-depleted (grey bars) fractions. (b) The 26 most abundant miRNAs more likely to be found either in the EV-enriched (44 in total) or the EV-depleted (38 in total) E/S fractions (i.e. containing reads only in the EV-enriched or EV-depleted fractions after normalization). Bar charts represent the miRNA log2 normalized read counts for each sample where the indicated sma-miRNA was found (see Materials and methods). Raw and processed sma-miRNA read data are found in Supplementary file 3. Bold sma-miRNAs in (a) and (b) represent the most abundant miRNAs found in this study.
Polycistronic transcription of both sma-mir-71a/sma-mir-2a and sma-mir-71b/sma-mir-2c paralogous gene clusters could account for the presence of these known miRNAs in schistosomula E/S products (Citation51). This possibility prompted us to investigate whether any other schistosomula miRNAs present in the E/S products were found arranged in gene clusters (same orientation in the genome, less than 10 kb apart and on the same chromosome or scaffold). Only one other miRNA cluster was identified in our study, sma-mir-new-207/sma-mir-new-208 (both miRNAs predominantly found in the EV-depleted E/S fraction, Supplementary file 3). These data, along with those previously reported (Citation51), would indicate that most S. mansoni miRNAs are not transcribed as polycistronic clusters. This finding is consistent with polycistronic analyses of other animal miRNAs (Citation52).
A previous study has recently reported that sma-bantam, sma-mir-61 and the sma-mir-71b/sma-mir-2c cluster all display gender-biased expression in adult schistosomes (Citation53). Our identification of these gene products in schistosomula E/S products suggests that either gender-associated miRNA expression is also found in larval schistosomes (Citation54) or that gender-biased expression of these (and other) specific sma-miRNAs develops in time as also occurs for protein coding RNAs (Citation26). Until schistosome or host mRNA targets are experimentally identified, the functional significance of gender-biased sma-miRNA expression in either larval or adult schistosomes is currently unknown.
tsRNAs are found within and outside schistosomula EVs
It is apparent that other classes of sncRNAs, in addition to miRNAs, can play gene silencing roles and also participate in a variety of regulatory functions in eukaryotic transcription and translation (Citation35, Citation55, Citation56). One particular class of sncRNA gaining recognition in this regard is the tsRNAs, which is a term used to define both tRNA halves (tiRNAs, 30–35 nucleotide 5′ tRNA halves or 40–50 nucleotide 3′ tRNA halves generated by cleavage in the anticodon loop) and other tRFs (12–30 nucleotide 5′ or 3′ fragments) (Citation35). These tsRNAs have previously been found in exosomal cargo derived from mammalian neuronal and dendritic cells (Citation57, Citation58) and, more recently, in Leishmania exosomes (Citation36). We therefore postulated that schistosomula E/S products derived from 72 h culture supernatants would contain tsRNAs and that these would be found in EV-enriched and, perhaps, EV-depleted fractions ().
Fig. 6. Schistosomula release tRNA-derived small RNAs (tsRNA). Reads were mapped against the S. mansoni tRNA database depleted of mitochondrial and pseudo-tRNAs available from geneDB (34) and, using an in-house script, were characterized as 5′ or 3′ tsRNAs. (a) Secondary structure of smtRNA-SeC depicting characteristic cloverleaf shape composed of D-, anticodon, variable and T-loops. Below, the coverage of RNA-seq reads mapping to smtRNA-SeC (85 nucleotides) are quantified to give an example of how 5′ and 3′ tsRNAs were identified for all tsRNAs (using our processing criteria, no full-length tRNAs were identified in our study). Dashed and solid lines represent the reads mapping to smtRNA-SeC and illustrate the sequential steps (Filter 1 and Filter 2) employed by our in-house script as outlined in the Materials and methods. The arrow represents Position 35 (U in bold), which is the terminal nucleotide (initiating from the 5′ end) used to define 5′ tsRNAs (which happens to be in the anticodon loop for smtRNA-SeC). To define 3′ tsRNAs, a maximal length of 50 nt (also represented by the black arrow) initiating from the 3′ tRNA end was used. Clear read peaks can be visualized mapping to both 5′ and 3′ ends of tRNA-SeC. (b) Bar chart represents the tsRNAs (log2 normalized read counts) found in EV-enriched (black bars) or EV-depleted (grey bars) supernatant fractions. G1–G6 represent tsRNA groups displaying specific associations (see text for details). (c) Stacked histogram representation of 3′ tsRNAs derived from precursor tRNAs (lacking the 3′ CCA trinucleotide motif; white or light grey bars, EV-enriched and EV-depleted samples, respectively) or mature tRNAs (containing the 3′ CCA trinucleotide motif; black or dark grey bars, EV-enriched and EV-depleted samples, respectively).
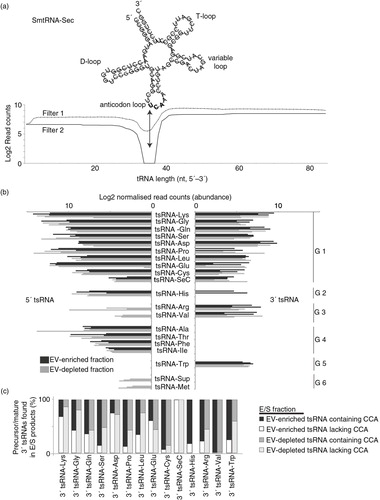
Putative tsRNA reads from 6 (EV-enriched, n=3; EV-depleted, n=3) schistosomula RNA-seq libraries were aligned to full-length tRNA sequences and, as expected, large numbers mapped to either the 5′ or 3′ ends (representing 5′ tsRNAs and 3′ tsRNAs, respectively) as displayed for tRNA-SeC (a). Due to the small insert sizes contained within our sncRNA libraries (range: 1–95 nucleotides; average: 24 nucleotides with less than 0.5% of the reads greater than 70 nucleotides), there were no reads that mapped with 100% coverage to full-length S. mansoni tRNAs (the longest read found was 67 nucleotides mapping to the 72 nucleotide SmtRNA-LysCTT, representing 93% coverage). Using our strategy, we identified tsRNAs derived from 20 tRNAs (18 out of the 20 ubiquitous tRNA classes, tRNA-AA; selenocysteine tRNA, tRNA-SeC; suppressor tRNA, tRNA-Sup) (b). Notably, tsRNA reads mapping to tRNA-Tyr and tRNA-Asn were not abundant (as defined in the Materials and methods) and, therefore, not included in our analyses.
Both 5′ and 3′ tsRNAs derived from 10 tRNA classes were found in relative equal abundance between EV-enriched and EV-depleted fractions (b, G1). The remaining 10 tRNA classes displayed contrasting patterns. Within EV-enriched fractions, 5′ tsRNAs derived from tRNA-Arg and tRNA-Val were missing (b, G3), as were 5′ tsRNAs for tRNA-Sup and tRNA-Met (b, G6). Within the EV-depleted fraction, 3′ tsRNAs derived from tRNA-His (b, G2), tRNA-Sup (b, G6) and tRNA-Met (b, G6) were missing. Noticeably, 3′ tsRNAs derived from tRNA-Ala, tRNA-Thr, tRNA-Phe and tRNA-Ile as well as 5′ tsRNA derived from tRNA-Trp were neither found in EV-enriched nor EV-depleted E/S products (b, G4 and b, G5 respectively). Among the 8 most abundant tsRNAs (tsRNA-Lys, tsRNA-Gly, tsRNA-Gln, tsRNA-Ser, tsRNA-Asp, tsRNA-Pro, tsRNA-Leu and tsRNA-Glu) found in schistosomula EVs (b), 5 of these (tsRNA-Gly, tsRNA-Gln, tsRNA-Glu, tsRNA-Asp and tsRNA-Leu) are also the most abundant tsRNAs found in Leishmania exosomes (Citation36). The single most abundant schistosomula E/S tsRNA (including both 5′ and 3′ tsRNAs) derived from tRNA-Lys is also the most abundant tsRNA found in HeLa cell sncRNA pools (Citation59) and dendritic cell extracellular RNAs (Citation58). The mechanism by which these abundant tsRNAs are processed among these different cells/organisms is currently unknown. The current S. mansoni genome assembly (v5.2) does not contain obvious homologs of the tRNA-processing enzymes angiogenin or RNASE T2 (except the egg-specific omega-1 (Citation60)). Therefore, other molecular pathways to generate tsRNAs may be operational within schistosomula (as reviewed in Ref. 61).
We further quantified schistosomula 3′ tsRNAs for the presence/absence of a terminal 3′ CCA tri-nucleotide motif in order to identify those derived from mature vs. precursor tRNAs (c). While differences exist in the exact percentages of 3′ tsRNAs containing a terminal 3′ CCA across the 20 identified tRNA classes, our results suggest that 54% of E/S schistosomula 3′ tsRNA reads contain this distinguishing feature. Therefore, at least half of all E/S 3′ tsRNAs are likely derived from the cleavage of a mature tRNA. Further studies are necessary to understand the significance and ubiquity (occurrence in schistosomula sncRNA intracellular pools, presence in other schistosome lifecycle stages, etc.) of our observations.
It is currently unclear why large numbers of tsRNAs are found in schistosomula E/S products or, indeed, how they are generated. While this could represent a consequence of in vitro culture (stress can sometimes, but not always, induce the cleavage of eukaryotic tRNAs (Citation35, Citation61)), we contend that release of tsRNAs (and indeed miRNAs) represents a bona fide adaptation of schistosome parasites to subvert host defensive barriers. Determining whether schistosomula E/S tsRNAs (containing both tiRNAs and tRFs) can be taken up by host cells (within or outside EVs) and affect translational inhibition and/or transcriptional repression (reviewed in Ref. 56) will help shed light on this fascinating new area of host/parasite biology.
Conclusions
S. mansoni schistosomula excrete/secrete EVs during in vitro development and these 30–100 nm sized vesicles contain potential host-modulating proteins (109 in total), miRNAs (205 in total) and tsRNAs (derived from 20 tRNA classes). The description of these vesicles and their biological components should now promote studies aiming to functionally characterize these new players in host interactions.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
Authors’ contributions
FCN performed the schistosomula cultures, viability assays, EV isolations, TEM, sncRNA library preparations/sequence analyses, proteome classifications and drafting of the manuscript. OIK, PJH and CHH all participated in the EV proteome analyses and drafting of the manuscript. MTS, UN and AI all contributed to the development of bespoke sncRNA informatics pipelines, and MTS also participated in the interpretation of the results and drafting of the manuscript. JFQ assisted in sncRNA library construction. AHB assisted in the design of the study and interpretation of the results. KFH conceived the study and participated in the design of the experiments, interpretation of the results and drafting of the manuscript. All authors read and approved the final manuscript.
Supplementary file 1
Download MS Excel (76.5 KB)Supplementary file 2
Download MS Excel (41.1 KB)Supplementary file 3
Download MS Excel (159 KB)Acknowledgements
This work was supported by an IBERS PhD studentship awarded to FCN. We acknowledge all members of the Hoffmann laboratory for assisting in the maintenance of the schistosome lifecycle, Dr Kathy Geyer and Ms Jennifer Edwards for helminth fluorescent bioassay set-up and Dr Steven Wade for helping with TEM visualization of schistosomula EVs. We also thank Drs Jennifer Fitzpatrick and Emily Peak for contributing the TEM and epifluorescent images of cercaria and schistosomula samples included in this study.
Notes
To access the supplementary material to this article, please see Supplementary files under ‘Article Tools’.
References
- Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014; 383: 2253–64.
- Hansell E, Braschi S, Medzihradszky KF, Sajid M, Debnath M, Ingram J et al. Proteomic analysis of skin invasion by blood fluke larvae. PLoS Negl Trop Dis. 2008; 2: e262.
- Ramaswamy K, Kumar P, He YX. A role for parasite-induced PGE2 in IL-10-mediated host immunoregulation by skin stage schistosomula of Schistosoma mansoni. J Immunol. 2000; 165: 4567–74.
- Fulford AJ, Butterworth AE, Ouma JH, Sturrock RF. A statistical approach to schistosome population dynamics and estimation of the life-span of Schistosoma mansoni in man. Parasitology. 1995; 110: 307–16.
- Siles-Lucas M, Morchon R, Simon F, Manzano-Roman R. Exosome-transported microRNAs of helminth origin: new tools for allergic and autoimmune diseases therapy?. Parasite Immunol. 2015; 37: 208–14.
- Simons M, Raposo G. Exosomes – vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009; 21: 575–81.
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009; 9: 581–93.
- Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012; 23: 91–7.
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010; 73: 1907–20.
- Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002; 3: 321–30.
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006; 140: 13–21.
- Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013; 13: 1554–71.
- Shifrin DA Jr., Demory Beckler M, Coffey RJ, Tyska MJ. Extracellular vesicles: communication, coercion, and conditioning. Mol Biol Cell. 2013; 24: 1253–9.
- EL Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013; 12: 347–57.
- Barteneva NS, Maltsev N, Vorobjev IA. Microvesicles and intercellular communication in the context of parasitism. Front Cell Infect Microbiol. 2013; 3: 49.
- Marcilla A, Trelis M, Cortes A, Sotillo J, Cantalapiedra F, Minguez MT et al. Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS One. 2012; 7: e45974.
- Bernal D, Trelis M, Montaner S, Cantalapiedra F, Galiano A, Hackenberg M et al. Surface analysis of Dicrocoelium dendriticum. The molecular characterization of exosomes reveals the presence of miRNAs. J Proteomics. 2014; 105: 232–41.
- Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014; 5: 5488.
- Wang L, Li Z, Shen J, Liu Z, Liang J, Wu X et al. Exosome-like vesicles derived by Schistosoma japonicum adult worms mediates M1 type immune- activity of macrophage. Parasitol Res. 2015; 114: 1865–73.
- Chaiyadet S, Sotillo J, Smout M, Cantacessi C, Jones MK, Johnson MS et al. Carcinogenic liver fluke secretes extracellular vesicles that promote cholangiocytes to adopt a tumorigenic phenotype. J Infect Dis. 2015. http://dx.doi.org/10.1093/infdis/jiv291.
- Hansen EP, Kringel H, Williams AR, Nejsum P. Secretion of RNA-containing extracellular vesicles by the porcine whipworm, Trichuris suis. J Parasitol. 2015; 101: 336–40.
- Colley DG, Wikel SK. Schistosoma mansoni: simplified method for the production of schistosomules. Exp Parasitol. 1974; 35: 44–51.
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006. Chapter 3:Unit 3 22.
- Fitzpatrick JM, Hirai Y, Hirai H, Hoffmann KF. Schistosome egg production is dependent upon the activities of two developmentally regulated tyrosinases. Faseb J. 2007; 21: 823–35.
- Peak E, Chalmers IW, Hoffmann KF. Development and validation of a quantitative, high-throughput, fluorescent-based bioassay to detect Schistosoma viability. PLoS Negl Trop Dis. 2010; 4: e759.
- Fitzpatrick JM, Peak E, Perally S, Chalmers IW, Barrett J, Yoshino TP et al. Anti-schistosomal intervention targets identified by lifecycle transcriptomic analyses. PLoS Negl Trop Dis. 2009; 3: e543.
- The R project for statistical computing. Available from: http://www.r-project.org/ [cited 1 November 2014]..
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5: R80.
- Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012; 40: 37–52.
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30: 2114–20.
- Protasio AV, Tsai IJ, Babbage A, Nichol S, Hunt M, Aslett MA et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis. 2012; 6: e1455.
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013; 8: 1765–86.
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15: 550.
- Logan-Klumpler FJ, De Silva N, Boehme U, Rogers MB, Velarde G, McQuillan JA et al. GeneDB – an annotation database for pathogens. Nucleic Acids Res. 2012; 40: D98–108.
- Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014; 588: 4297–304.
- Lambertz U, Oviedo Ovando ME, Vasconcelos E, Unrau PJ, Myler PJ, Reiner NE. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genomics. 2015; 16: 151.
- Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol Cell Proteomics. 2005; 4: 1862–75.
- Paveley RA, Aynsley SA, Cook PC, Turner JD, Mountford AP. Fluorescent imaging of antigen released by a skin-invading helminth reveals differential uptake and activation profiles by antigen presenting cells. PLoS Negl Trop Dis. 2009; 3: e528.
- Dorsey CH, Cousin CE, Lewis FA, Stirewalt MA. Ultrastructure of the Schistosoma mansoni cercaria. Micron. 2002; 33: 279–323.
- Shelke GV, Lasser C, Gho YS, Lotvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2014; 3: 24783. doi: http://dx.doi.org/10.3402/jev.v3.24783.
- Curwen RS, Ashton PD, Sundaralingam S, Wilson RA. Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Mol Cell Proteomics. 2006; 5: 835–44.
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011; 8: 785–6.
- Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008; 8: 4083–99.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000; 25: 25–9.
- Reimand J, Arak T, Vilo J. g:Profiler – a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 2011; 39: W307–15.
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013; 200: 373–83.
- Buschow SI, van Balkom BW, Aalberts M, Heck AJ, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol Cell Biol. 2010; 88: 851–6.
- Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012; 13: 328–35.
- Hoy AM, Lundie RJ, Ivens A, Quintana JF, Nausch N, Forster T et al. Parasite-derived microRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl Trop Dis. 2014; 8: e2701.
- Cheng G, Luo R, Hu C, Cao J, Jin Y. Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology. 2013; 140: 1751–61.
- de Souza Gomes M, Muniyappa MK, Carvalho SG, Guerra-Sa R, Spillane C. Genome-wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni. Genomics. 2011; 98: 96–111.
- Marco A, Ninova M, Ronshaugen M, Griffiths-Jones S. Clusters of microRNAs emerge by new hairpins in existing transcripts. Nucleic Acids Res. 2013; 41: 7745–52.
- Marco A, Kozomara A, Hui JH, Emery AM, Rollinson D, Griffiths-Jones S et al. Sex-biased expression of microRNAs in Schistosoma mansoni. PLoS Negl Trop Dis. 2013; 7: e2402.
- Fitzpatrick JM, Protasio AV, McArdle AJ, Williams GA, Johnston DA, Hoffmann KF. Use of genomic DNA as an indirect reference for identifying gender-associated transcripts in morphologically identical, but chromosomally distinct, Schistosoma mansoni Cercariae. PLoS Negl Trop Dis. 2008; 2: e323.
- Falaleeva M, Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. BioEssays. 2013; 35: 46–54.
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013; 10: 1798–806.
- Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012; 40: 10937–49.
- Nolte-'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012; 40: 9272–85.
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009; 15: 2147–60.
- Fitzsimmons CM, Schramm G, Jones FM, Chalmers IW, Hoffmann KF, Grevelding CG et al. Molecular characterization of omega-1: a hepatotoxic ribonuclease from Schistosoma mansoni eggs. Mol Biochem Parasitol. 2005; 144: 123–7.
- Megel C, Morelle G, Lalande S, Duchene AM, Small I, Marechal-Drouard L. Surveillance and cleavage of eukaryotic tRNAs. Int J Mol Sci. 2015; 16: 1873–93.
